Comparative Genomic Analysis of Extracellular Electron Transfer in Bacteria
Abstract
1. Introduction
2. Materials and Methods
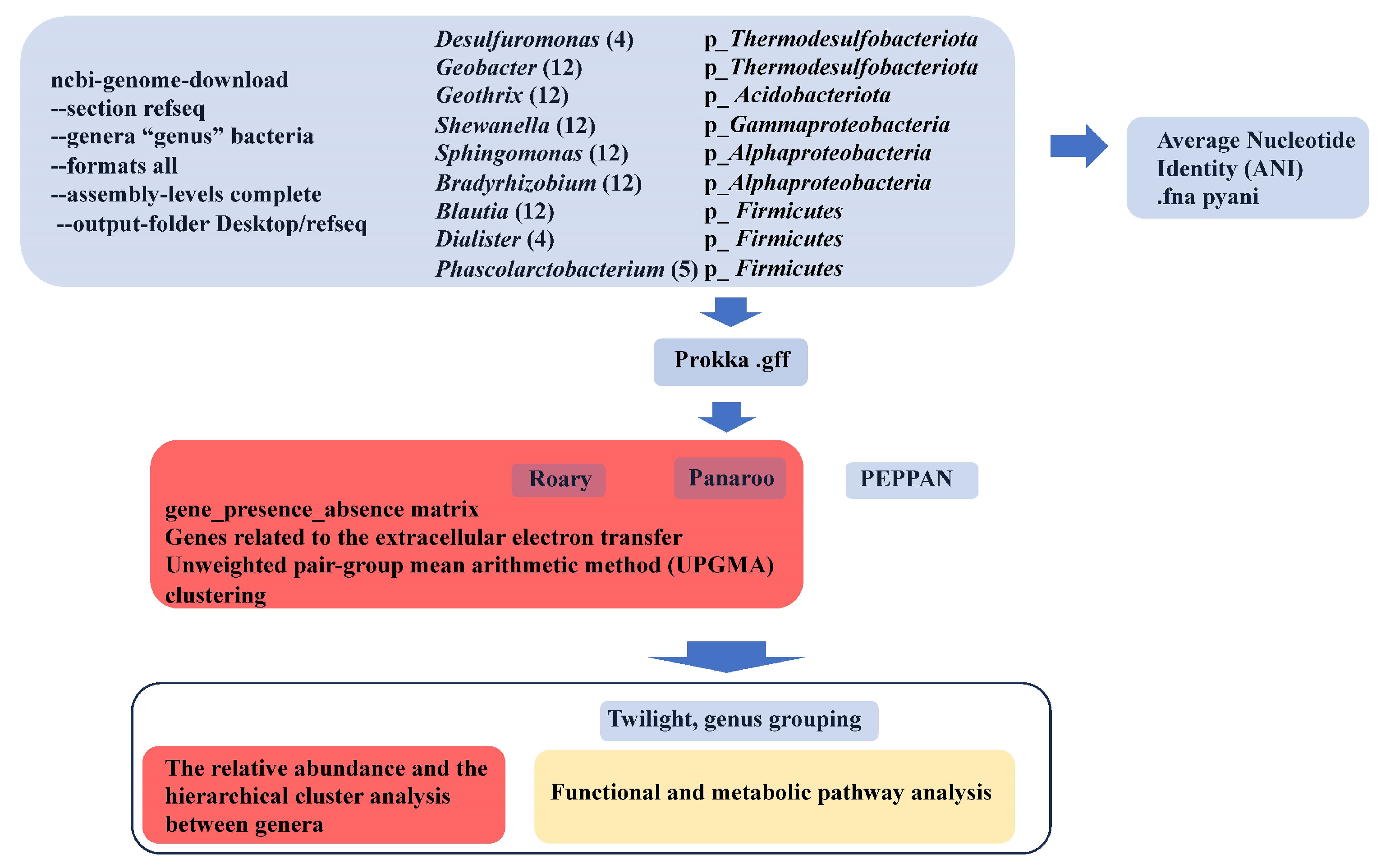
2.1. Strains in This Study
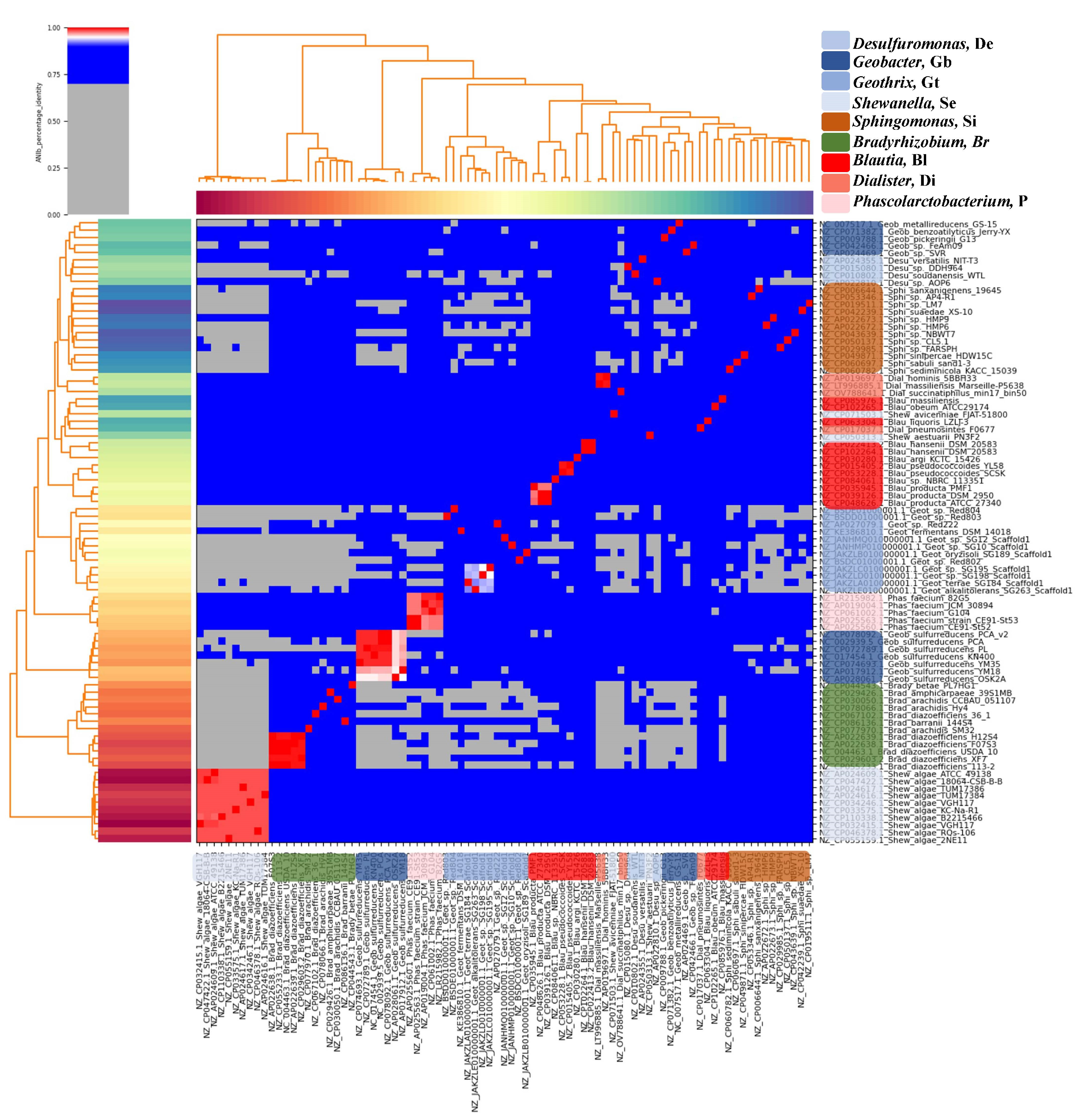
2.2. Average Nucleotide Identity (ANI) Analysis
2.3. Genome Annotation and Pangenome Analyses
2.4. Retrieval of Extracellular Electron Transfer Genes
2.5. Functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses
3. Results
3.1. Strategy
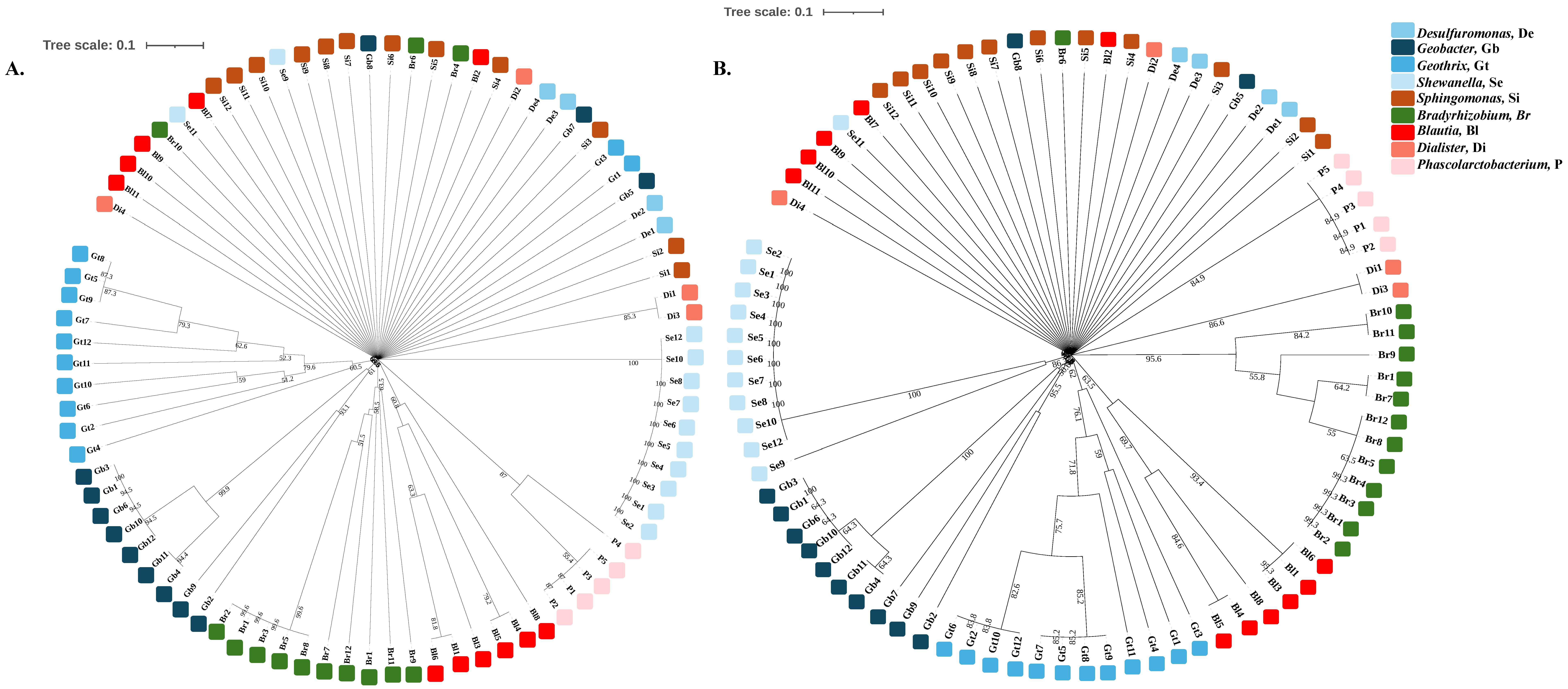
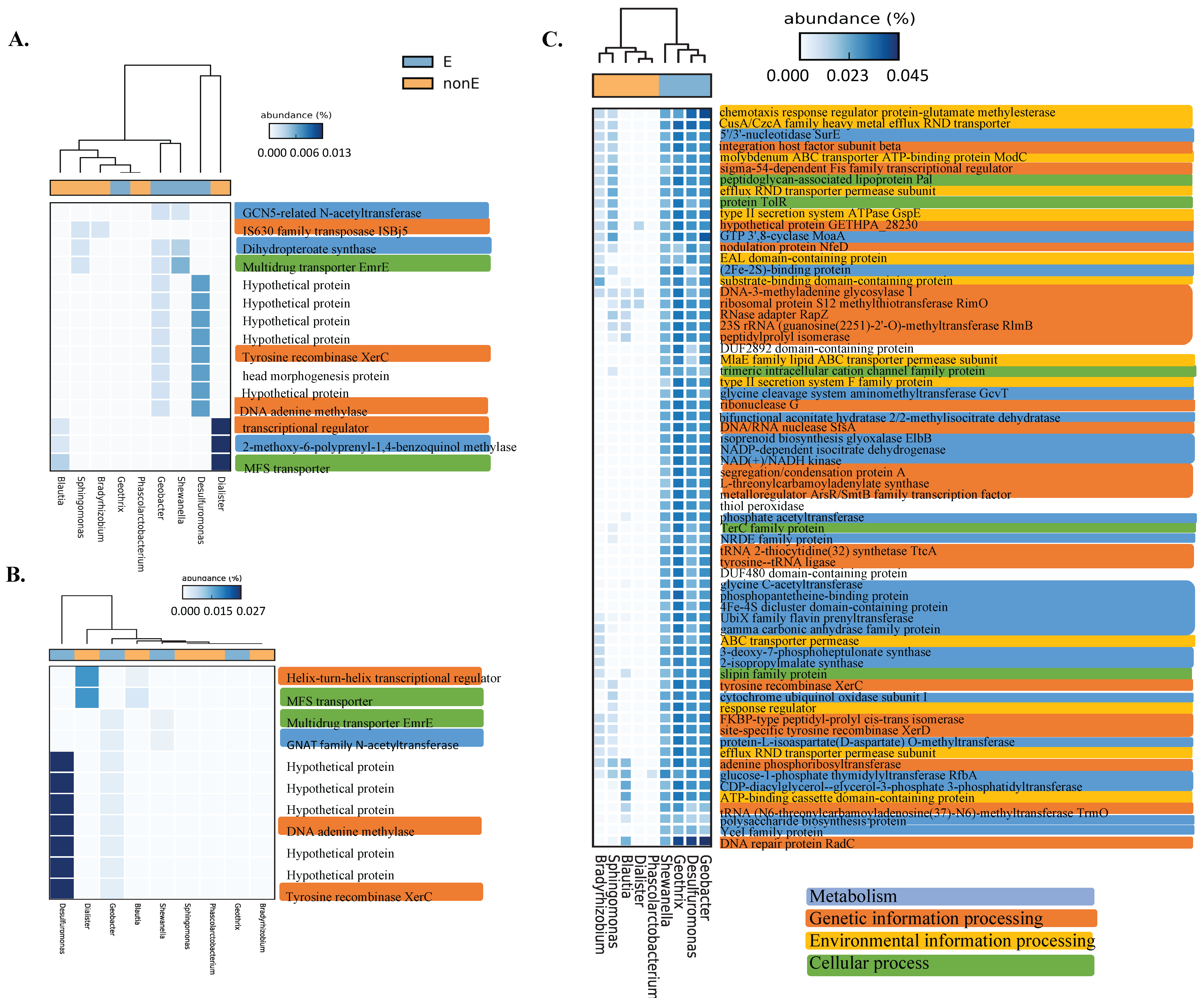
3.2. The Genes Related to Extracellular Electron Transfer May Cluster Well at the Genus Level
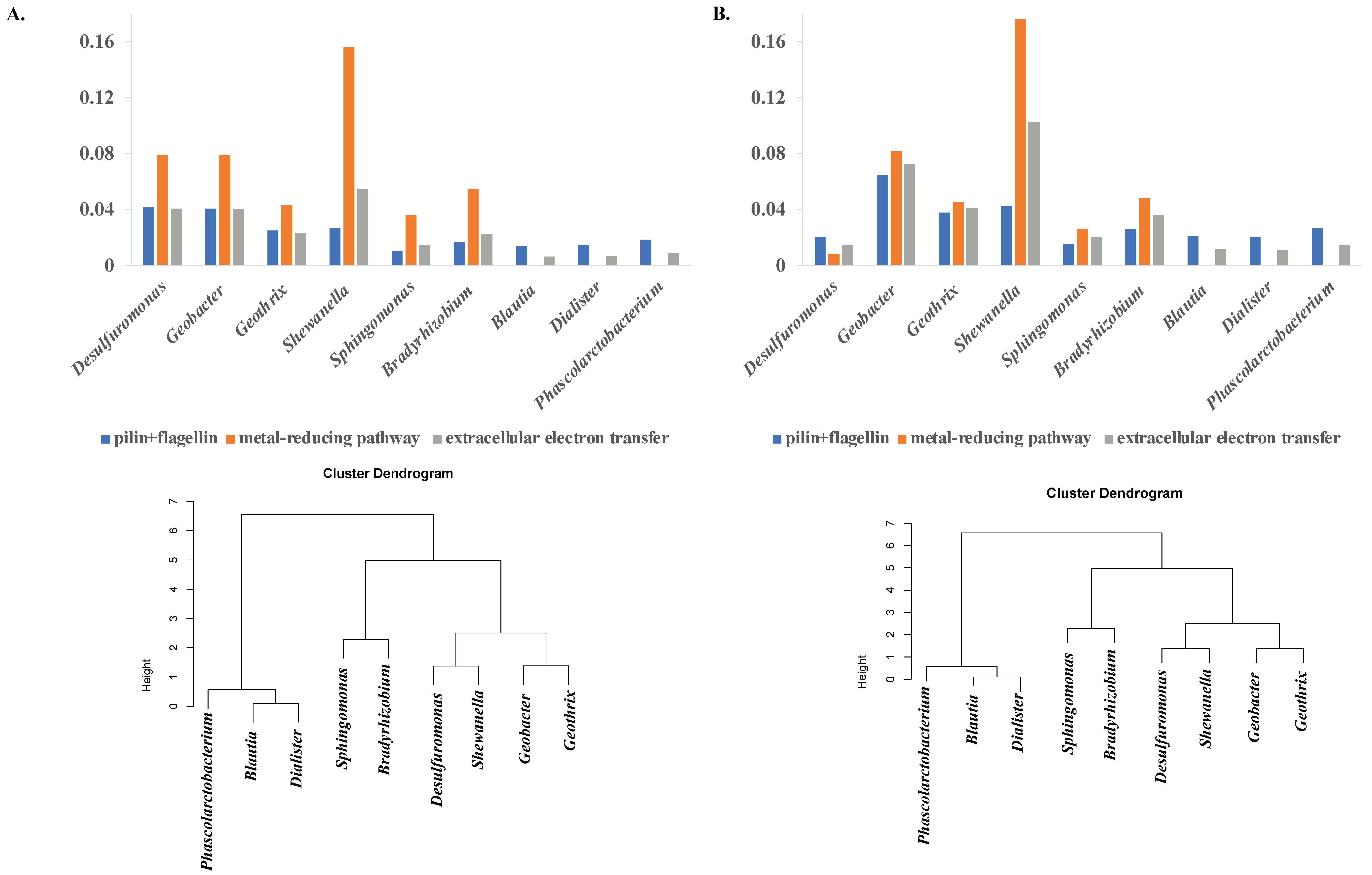
3.3. Bacteria with Extracellular Electron Transfer Ability Have a Greater Abundance of Related Genes and Cluster Together
3.4. Functional and Metabolic Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Logan, B.E. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 2009, 7, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Mu, H.; Liu, W.; Zhang, R.; Guo, J.; Xian, M.; Liu, H. Electricigens in the anode of microbial fuel cells: Pure cultures versus mixed communities. Microb. Cell Fact. 2019, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.A.; Hagerhall, C.; Gorton, L. Electron transfer mechanisms between microorganisms and electrodes in bioelectrochemical systems. Bioanal. Rev. 2012, 4, 159–192. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, M.; Guo, J.; Sun, G. Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochem. 2012, 47, 8. [Google Scholar] [CrossRef]
- Tremblay, P.L.; Aklujkar, M.; Leang, C.; Nevin, K.P.; Lovley, D. A genetic system for Geobacter metallireducens: Role of the flagellin and pilin in the reduction of Fe(III) oxide. Environ. Microbiol. Rep. 2012, 4, 82–88. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Liu, J.; Levar, C.; Edwards, M.J.; Babauta, J.T.; Kennedy, D.W.; Shi, Z.; Beyenal, H.; Bond, D.R.; et al. A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ. Microbiol. Rep. 2014, 6, 776–785. [Google Scholar] [CrossRef]
- Logan, B.E.; Rossi, R.; Ragab, A.; Saikaly, P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Microbiol. 2019, 17, 307–319. [Google Scholar] [CrossRef]
- Kuo, J.; Liu, D.; Wang, S.H.; Lin, C.H. Dynamic Changes in Soil Microbial Communities with Glucose Enrichment in Sediment Microbial Fuel Cells. Indian. J. Microbiol. 2021, 61, 497–505. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020, 21, 180. [Google Scholar] [CrossRef]
- Zhou, Z.; Charlesworth, J.; Achtman, M. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 2020, 30, 1667–1679. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 1 August 2024).
- Horesh, G.; Taylor-Brown, A.; McGimpsey, S.; Lassalle, F.; Corander, J.; Heinz, E.; Thomson, N.R. Different evolutionary trends form the twilight zone of the bacterial pan-genome. Microb. Genom. 2021, 7, 000670. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Guo, Y.; Aoyagi, T.; Inaba, T.; Sato, Y.; Habe, H.; Hori, T. Complete Genome Sequence of Desulfuromonas sp. Strain AOP6, an Iron(III) Reducer Isolated from Subseafloor Sediment. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Guo, Y.; Aoyagi, T.; Hori, T. Comparative insights into genome signatures of ferric iron oxide- and anode-stimulated Desulfuromonas spp. strains. BMC Genom. 2021, 22, 475. [Google Scholar] [CrossRef]
- Xie, L.; Yoshida, N.; Ishii, S.; Meng, L. Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture. Microorganisms 2021, 9, 1953. [Google Scholar] [CrossRef] [PubMed]
- Mehta-Kolte, M.G.; Bond, D.R. Geothrix fermentans secretes two different redox-active compounds to utilize electron acceptors across a wide range of redox potentials. Appl. Environ. Microbiol. 2012, 78, 6987–6995. [Google Scholar] [CrossRef] [PubMed]
- Weidenhaupt, M.; Rossi, P.; Beck, C.; Fischer, H.M.; Hennecke, H. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch. Microbiol. 1996, 165, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Martins da Costa, E.; Azarias Guimaraes, A.; Pereira Vicentin, R.; de Almeida Ribeiro, P.R.; Ribas Leao, A.C.; Balsanelli, E.; Lebbe, L.; Aerts, M.; Willems, A.; de Souza Moreira, F.M. Bradyrhizobium brasilense sp. nov., a symbiotic nitrogen-fixing bacterium isolated from Brazilian tropical soils. Arch. Microbiol. 2017, 199, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Mania, D.; Mousavi, S.A.; Lycus, P.; Arntzen, M.O.; Woliy, K.; Lindstrom, K.; Shapleigh, J.P.; Bakken, L.R.; Frostegard, A. Competition for electrons favours N(2) O reduction in denitrifying Bradyrhizobium isolates. Environ. Microbiol. 2021, 23, 2244–2259. [Google Scholar] [CrossRef]
- Wasai-Hara, S.; Itakura, M.; Fernandes Siqueira, A.; Takemoto, D.; Sugawara, M.; Mitsui, H.; Sato, S.; Inagaki, N.; Yamazaki, T.; Imaizumi-Anraku, H.; et al. Bradyrhizobium ottawaense efficiently reduces nitrous oxide through high nosZ gene expression. Sci. Rep. 2023, 13, 18862. [Google Scholar] [CrossRef]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef]
- Afouda, P.; Dubourg, G.; Tomei, E.; Raoult, D.; Fournier, P.E. Dialister massiliensis sp. nov., a new bacterium isolated from the human gut. New Microbes New Infect. 2020, 34, 100657. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tang, T.; Deng, D.; Wang, Y.; Pei, D. Isolation and Electrochemical Analysis of a Facultative Anaerobic Electrogenic Strain Klebsiella sp. SQ-1. Pol. J. Microbiol. 2024, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Philippon, T.; Ait-Itto, F.Z.; Monfort, A.; Barriere, F.; Behan, J.A. Fe(III) oxide microparticles modulate extracellular electron transfer in anodic biofilms dominated by bacteria of the Pelobacter genus. Bioelectrochemistry 2023, 151, 108394. [Google Scholar] [CrossRef] [PubMed]
- Ait-Itto, F.Z.; Behan, J.A.; Martinez, M.; Barriere, F. Development of bioanodes rich in exoelectrogenic bacteria using iron-rich palaeomarine sediment inoculum. Bioelectrochemistry 2024, 156, 108618. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Ma, Z.; Li, X.; Qiu, Y.; Liu, D.; Liu, S. N-Doped Carbon Nanowire-Modified Macroporous Carbon Foam Microbial Fuel Cell Anode: Enrichment of Exoelectrogens and Enhancement of Extracellular Electron Transfer. Materials 2023, 17, 69. [Google Scholar] [CrossRef]
- Hemdan, B.A.; El-Taweel, G.E.; Naha, S.; Goswami, P. Bacterial community structure of electrogenic biofilm developed on modified graphite anode in microbial fuel cell. Sci. Rep. 2023, 13, 1255. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Liu, Y.; Wang, S.; Lee, C.K.; Huang, Y.; Duan, X. High power density redox-mediated Shewanella microbial flow fuel cells. Nat. Commun. 2024, 15, 8302. [Google Scholar] [CrossRef]
- Klein, E.M.; Heintz, H.; Wurst, R.; Schuldt, S.; Hahl, H.; Jacobs, K.; Gescher, J. Comparative analysis of the influence of BpfA and BpfG on biofilm development and current density in Shewanella oneidensis under oxic, fumarate- and anode-respiring conditions. Sci. Rep. 2024, 14, 23174. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Kuo, J.; Lin, C.-H. Comparative Genomic Analysis of Extracellular Electron Transfer in Bacteria. Processes 2024, 12, 2636. https://doi.org/10.3390/pr12122636
Liu D, Kuo J, Lin C-H. Comparative Genomic Analysis of Extracellular Electron Transfer in Bacteria. Processes. 2024; 12(12):2636. https://doi.org/10.3390/pr12122636
Chicago/Turabian StyleLiu, Daniel, Jimmy Kuo, and Chorng-Horng Lin. 2024. "Comparative Genomic Analysis of Extracellular Electron Transfer in Bacteria" Processes 12, no. 12: 2636. https://doi.org/10.3390/pr12122636
APA StyleLiu, D., Kuo, J., & Lin, C.-H. (2024). Comparative Genomic Analysis of Extracellular Electron Transfer in Bacteria. Processes, 12(12), 2636. https://doi.org/10.3390/pr12122636





