Asymmetric Bioreduction of Ethyl 4-Chloroacetoacetate into Ethyl 4-Chloro-3-hydroxybutyrate by Recombinant Escherichia coli CgCR in Ethyl Acetate-Betaine:Lactic Acid-Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of DES Betaine/Lactic Acid
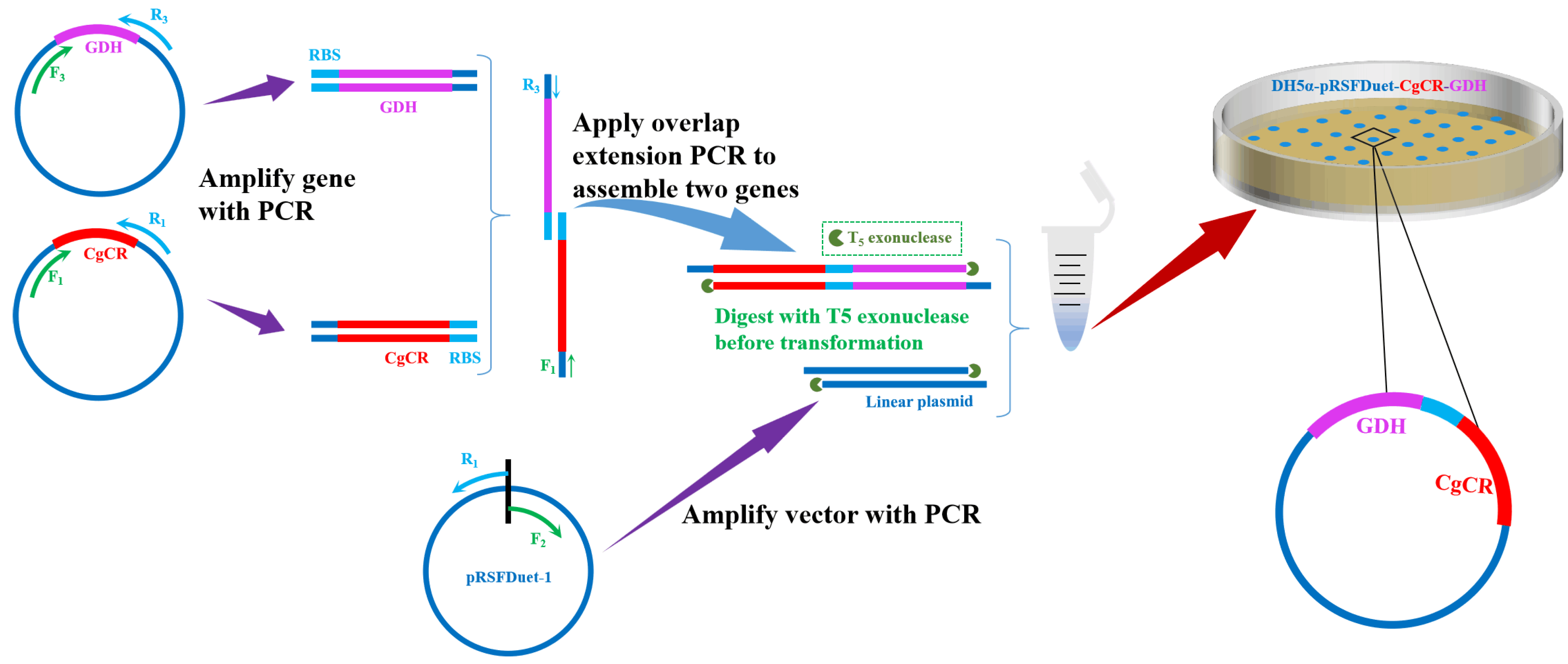
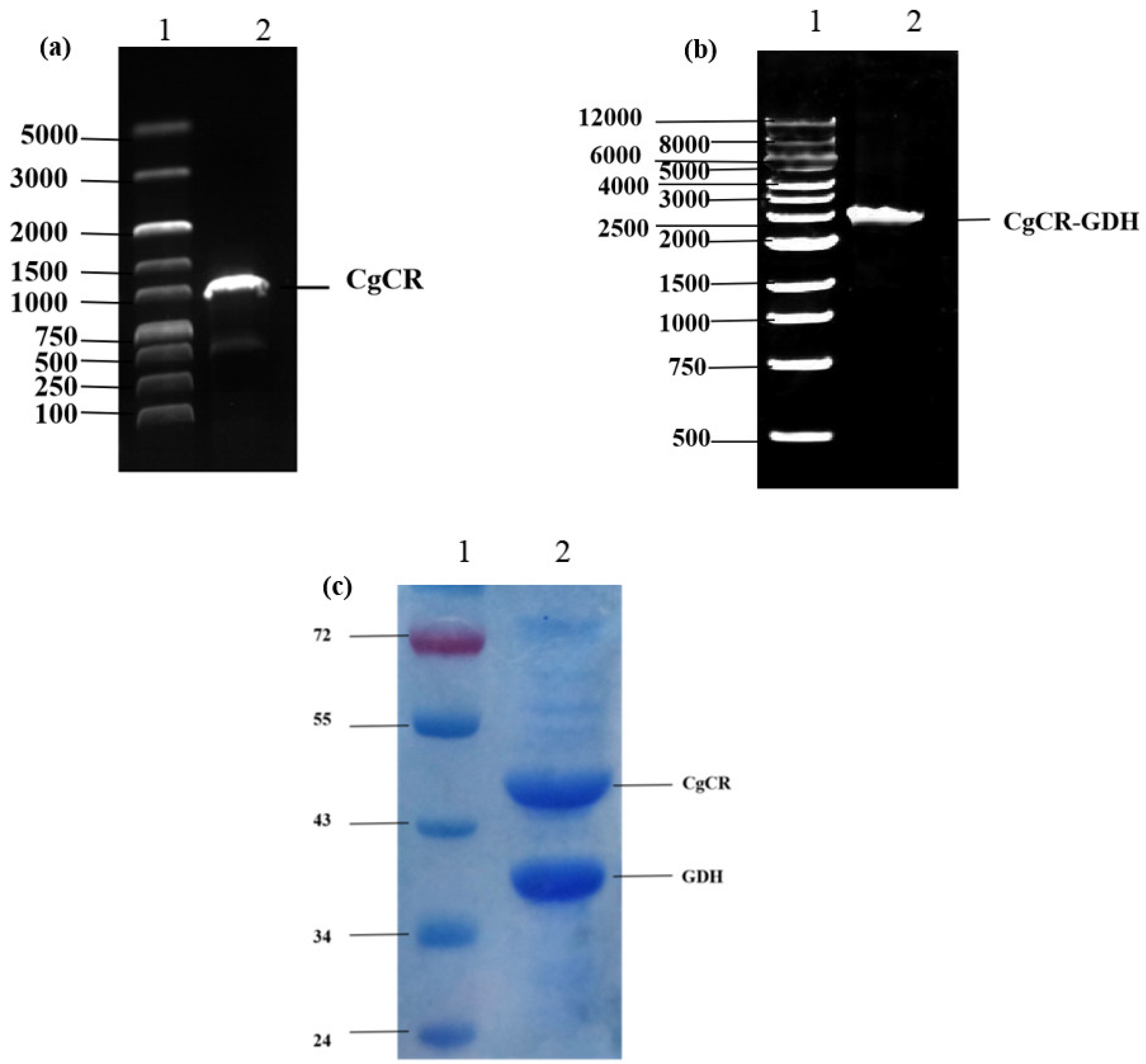
2.3. Construction and Expression of Reductase
2.4. Bioreduction of COBE with CgCR Cell in Water System
2.5. Bioreduction of COBE with CgCR Cells in Organic Solvent-Betaine/Lactic Acid-Water System
2.6. Effect of Cell Permeability on the Bioreduction of COBE
2.7. Analytical Method
3. Result and Discussion
3.1. Construction of Recombinant E. coli Expressing CgCR and GDH
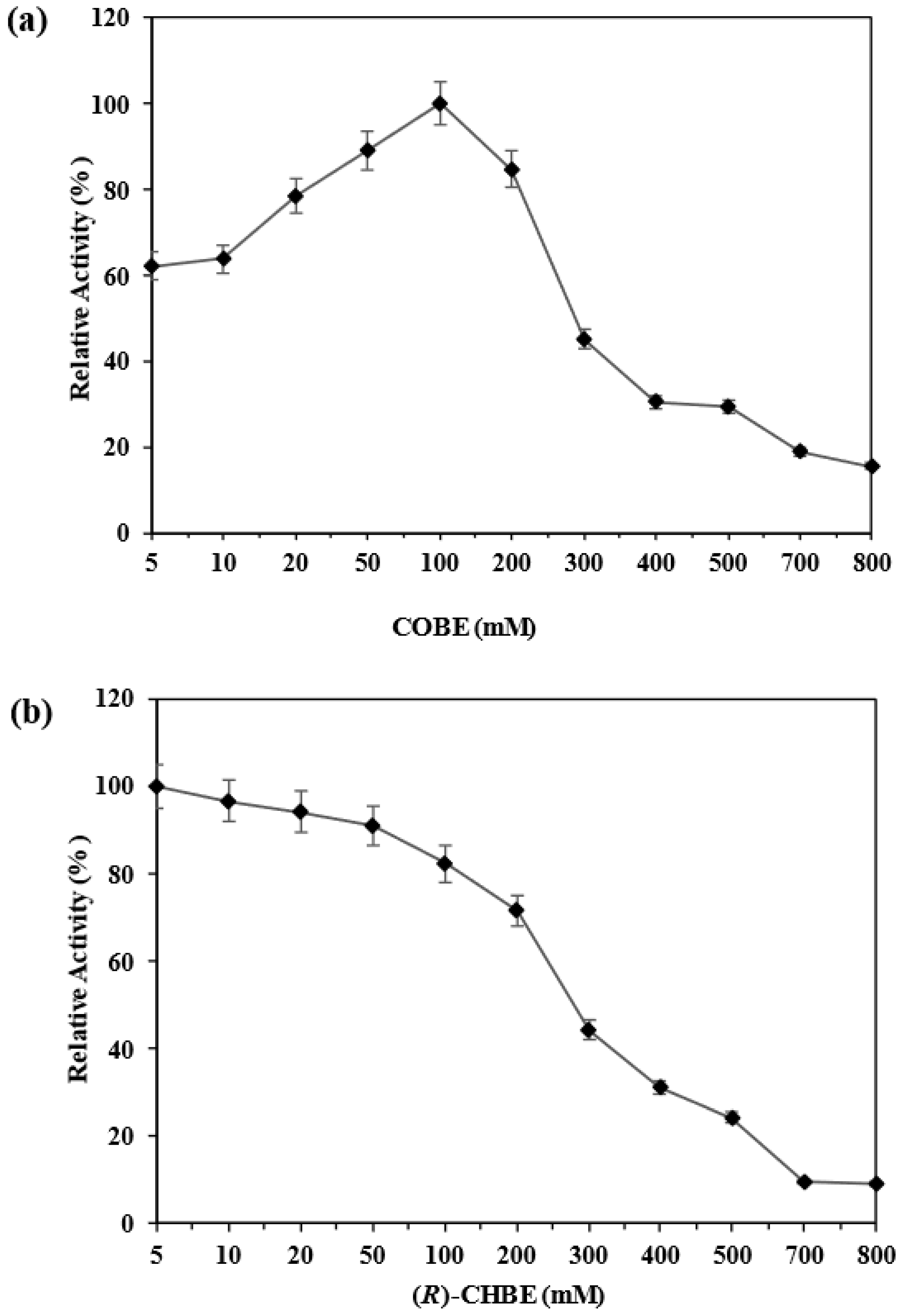
3.2. Biotransformation of COBE in the Aqueous System
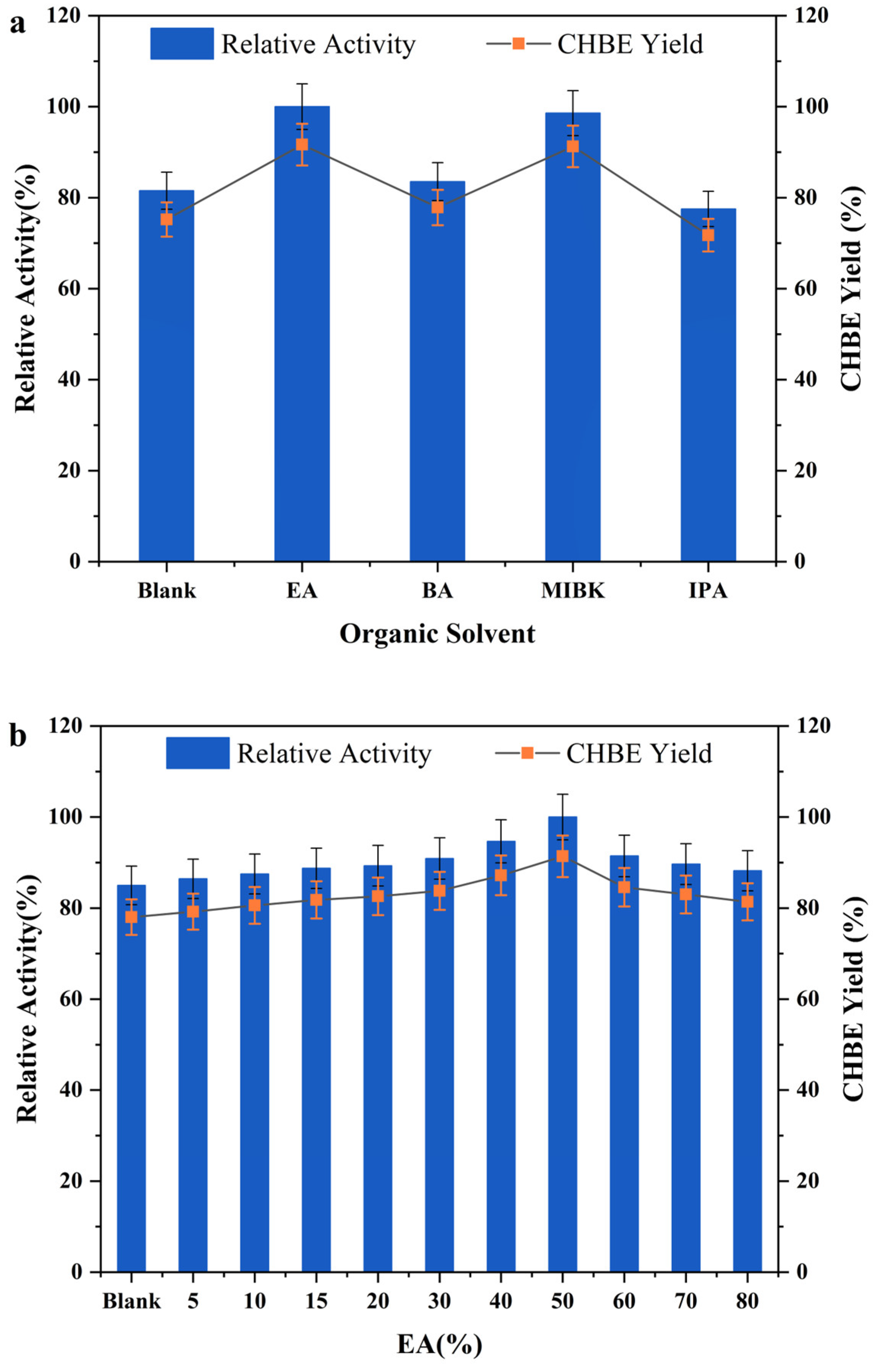
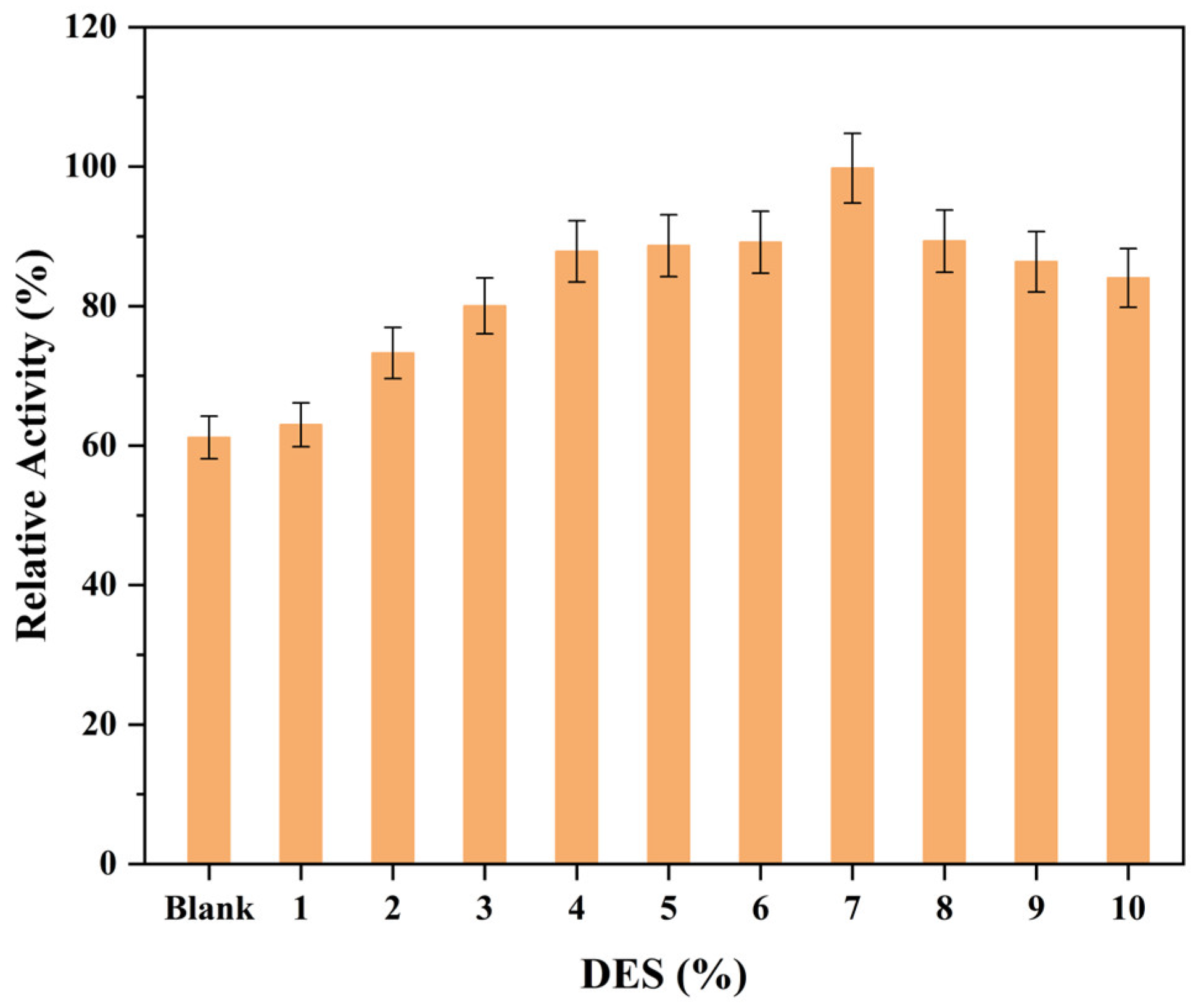
3.3. Enhancement in COBE Bioreduction through Supply of Organic Solvent and Betaine/Lactic Acid
3.4. The Effect of Cell Permeability and Damage Degree on Biocatalysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinto, M.M.; Fernandes, C.; Tiritan, M.E. Chiral Separations in Preparative Scale: A Medicinal Chemistry Point of View. Molecules 2020, 25, 1931. [Google Scholar] [CrossRef] [PubMed]
- Hancu, G.; Modroiu, A. Chiral Switch, between therapeutical benefit and marketing strategy. Pharmaceuticals 2022, 15, 240. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Almalki, W.H.; Afzal, O.; Altamimi, A.S.A.; Ullah, S.N.M.N.; Barkat, M.A.; Beg, S. Chiral-engineered supraparticles, emerging tools for drug delivery. Drug Discov. Today 2022, 28, 103420. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulou, I.; Varriale, S.; Topakas, E.; Rova, U.; Christakopoulos, P.; Faraco, V. Enzymatic synthesis of bioactive compounds with high potential for cosmeceutical application. Appl. Microbiol. Biotechnol. 2016, 100, 6519–6543. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liang, S.; Xiao, Z.; Ge, C. Research progress on extraction technology and biological activity of polysaccharides from Edible Fungi: A review. Food Rev. Int. 2022, 39, 4909–4940. [Google Scholar] [CrossRef]
- Fernandes, C.; Ribeiro, R.; Pinto, M.; Kijjoa, A. Absolute Stereochemistry Determination of Bioactive Marine-Derived Cyclopeptides by Liquid Chromatography Methods: An Update Review (2018–2022). Molecules 2023, 28, 615. [Google Scholar] [CrossRef] [PubMed]
- Frontana-Uribe, B.A.; Little, R.D.; Ibanez, J.G.; Palma, A.; Vasquez-Medrano, R. Organic electrosynthesis, a promising green methodology in organic chemistry. Green Chem. 2010, 12, 2099–2119. [Google Scholar] [CrossRef]
- He, T.; Wang, D.; Wu, Z.; Huang, C.; Xu, X.; Xu, X.; Liu, C.; Hashimoto, K.; Yang, C. A bibliometric analysis of research on (R)-ketamine from 2002 to 2021. Neuropharmacology 2022, 218, 109207. [Google Scholar] [CrossRef]
- Wan, X.; Eguchi, A.; Fujita, Y.; Fujita, L.; Ma, X.; Wang, Y.; Yang, Y.; Qu, L.; Chang, J.; Zhang, C.; et al. Effects of (R)-ketamine on reduced bone mineral density in ovariectomized mice, A role of gut microbiota. Neuropharmacology 2022, 213, 109139. [Google Scholar] [CrossRef]
- Pomara, C.; Cassano, T.; D’Errico, S.; Bello, S.; Romano, A.D.; Riezzo, I.; Serviddio, G. Data available on the extent of cocaine use and dependence, biochemistry, pharmacologic effects and global burden of disease of cocaine abusers. Curr. Med. Chem. 2012, 19, 5647–5657. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, J.; Bischoff, B.; Becker, K.; Baronian, K.; Bode, R.; Schauer, F.; Vorbrodt, H.M.; Kunze, G. Synthesis of 1-(S)-phenylethanol and ethyl (R)-4-chloro-3-hydroxybutanoate using recombinant Rhodococcus erythropolis alcohol dehydrogenase produced by two yeast species. Biochem. Eng. J. 2016, 106, 107–117. [Google Scholar] [CrossRef]
- Hoyos, P.; Pace, V.; Alcántara, A.R. Biocatalyzed synthesis of statins, A sustainable strategy for the preparation of valuable drugs. Catalysts 2019, 9, 260. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Ye, W.-J.; Xie, Y.-Y.; Liu, Q.-H.; Chen, R.; Wang, H.-L.; Wei, D.-Z. Efficient asymmetric synthesis of ethyl (S)-4-chloro-3-hydroxybutyrate using alcohol dehydrogenase SmADH31 with high tolerance of substrate and product in a monophasic aqueous system. Org. Process Res. Dev. 2020, 24, 1068–1076. [Google Scholar] [CrossRef]
- Kluson, P.; Stavarek, P.; Penkavova, V.; Vychodilova, H.; Hejda, S.; Jaklova, N.; Curinova, P. Stereoselective synthesis of optical isomers of ethyl 4-chloro-3-hydroxybutyrate in a microfluidic chip reactor. J. Flow Chem. 2019, 9, 221–230. [Google Scholar] [CrossRef]
- Otani, Y.; Taguchi, A.; Hamada, K.; Hayashi, Y.; Yamaguchi, Y.; Baba, H. Influence of novel readthrough agents on myelin protein zero translation in the peripheral nervous system. Neuropharmacology 2022, 211, 109059. [Google Scholar] [CrossRef] [PubMed]
- Purwadini, A.K.N.; Lusiani, C.E. Literature studt of L-carnitine preparation methods 1000 tons of annual production. DISTILAT J. Teknol. Separasi 2022, 8, 394–401. [Google Scholar] [CrossRef]
- Tadiparthi, K.; Anand, P. A Journey toward the Syntheses of γ-Amino-β-hydroxybutyric Acid (GABOB) and Carnitine. Org. Process Res. Dev. 2021, 25, 2008–2019. [Google Scholar] [CrossRef]
- Cano, A.; Turowski, P.; Ettcheto, M.; Duskey, J.T.; Tosi, G.; Sanchez-Lopez, E.; Garcia, M.L.; Camins, A.; Souto, E.B.; Ruiz, A. Nanomedicine-based technologies and novel biomarkers for the diagnosis and treatment of Alzheimer’s disease, from current to future challenges. J. Nanobiotechnol. 2021, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- González De Aguilar, J.L. Lipid biomarkers for amyotrophic lateral sclerosis. Front. Neurol. 2019, 10, 284. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, L.; Liu, Y.; Li, J. Recent advances on formation mechanism and functionality of chitosan-based conjugates and their application in o/w emulsion systems: A review. Food Chem. 2022, 380, 131838. [Google Scholar] [CrossRef]
- Hamada, K.; Omura, N.; Taguchi, A.; Baradaran-Heravi, A.; Kotake, M.; Arai, M.; Takayama, K.; Taniguchi, A.; Roberge, M.; Hayashi, Y. New Negamycin-Based Potent Readthrough Derivative Effective against TGA-Type Nonsense Mutations. ACS Med. Chem. Lett. 2019, 10, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-K.; Tseng, P.-Y. Total synthesis of (–)-negamycin from a chiral advanced epoxide. Synth. Commun. 2023, 53, 119–126. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, S.; Ahmad, I.; Patel, H. In silico validation of anti-viral drugs obtained from marine sources as a potential target against SARS-CoV-2 Mpro. J. Indian Chem. Soc. 2021, 98, 100272. [Google Scholar] [CrossRef]
- List, B.; Grossmann, O. Biocatalytic Reduction of Keto Esters and Heterocyclic Ketones in Continuous Flow. Synfacts 2020, 16, 1131. [Google Scholar] [CrossRef]
- Kwon, J.H.; Lee, J.; Kim, J.; Kirchner, V.A.; Jo, Y.H.; Miura, T.; Kim, N.; Song, G.-W.; Hwang, S.; Lee, S.-G.; et al. Upregulation of Carbonyl Reductase 1 by Nrf2 as a Potential Therapeutic Intervention for Ischemia/Reperfusion Injury during Liver Transplantation. Mol. Cells 2019, 42, 672–685. [Google Scholar] [CrossRef]
- Simić, S.; Zukić, E.; Schmermund, L.; Faber, K.; Winkler, C.K.; Kroutil, W. Shortening synthetic routes to small molecule active pharmaceutical ingredients employing biocatalytic methods. Chem. Rev. 2021, 122, 1052–1126. [Google Scholar] [CrossRef]
- Kong, L.; Fan, B.; He, Y.-C. Efficient whole-cell biosynthesis of (S)-2-chloro-1-(3,4-difluorophenyl)-ethanol from 2-chloro-1-(3,4-difluorophenyl) ethanone in a sustainable reaction system. Mol. Catal. 2023, 550, 113570. [Google Scholar] [CrossRef]
- Pennacchio, A.; Giordano, A.; Pucci, B.; Rossi, M.; Raia, C.A. Biochemical characterization of a recombinant short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius. Extremophiles 2010, 14, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Bai, Y.; Fan, T.-P.; Zheng, X.; Cai, Y. Characterization of acid-resistant aldo–keto reductases capable of asymmetric synthesis of (R)-CHBE from Lactobacillus plantarum DSM20174. Syst. Microbiol. Biomanuf. 2022, 3, 634–646. [Google Scholar] [CrossRef]
- Zhuang, W.; Liu, H.; Zhang, Y.; He, J.; Wang, P. Effective asymmetric preparation of (R)-1-[3-(trifluoromethyl) phenyl] ethanol with recombinant E. coli whole cells in an aqueous Tween-20/natural deep eutectic solvent solution. AMB Express 2021, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, Q.; Tao, Y.; Ma, C.; Chai, H.; Ai, Y.; He, Y.-C. An efficient chemoenzymatic strategy for valorisation of corncob to furfuryl alcohol in CA:Betaine-water. Ind. Crops Prod. 2022, 186, 115203. [Google Scholar] [CrossRef]
- Qin, L.; Di, J.; He, Y. Efficient Synthesis of Furfuryl Alcohol from Corncob in a Deep Eutectic Solvent System. Processes 2022, 10, 1873. [Google Scholar] [CrossRef]
- Jiang, W.; Pei, R.; Wu, W.; Zhao, P.; Tian, L.; Zhou, S.F. Catalytic site analysis and characterization of a solvent-tolerant aldo-keto reductase. BioPharm Int. 2019, 32, 34–40. [Google Scholar]
- Dong, F.; Chen, H.; Malapit, C.A.; Prater, M.; Li, M.; Yuan, M.; Lim, K.; Minteer, S. Biphasic bioelectrocatalytic synthesis of chiral β-hydroxy nitriles. J. Am. Chem. Soc. 2020, 142, 8374–8382. [Google Scholar] [CrossRef]
- Xie, P.; Zhou, X.; Zheng, L. Stereoselective synthesis of a key chiral intermediate of (S)-Rivastigmine by AKR-GDH recombinant whole cells. J. Biotechnol. 2019, 289, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Panić, M.; Bubalo, M.C.; Redovniković, I.R. Designing a biocatalytic process involving deep eutectic solvents. J. Chem. Technol. Biotechnol. 2021, 96, 14–30. [Google Scholar] [CrossRef]
- Zhu, L.; Di, J.; Li, Q.; He, Y.-C.; Ma, C. Enhanced conversion of corncob into furfurylamine via chemoenzymatic cascade catalysis in a toluene–water medium. J. Mol. Liq. 2023, 380, 121741. [Google Scholar] [CrossRef]
- Liang, S.; Wu, X.-L.; Xiong, J.; Zong, M.-H.; Lou, W.-Y. Metal–organic frameworks as novel matrices for efficient enzyme immobilization: An update review. Coord. Chem. Rev. 2020, 406, 213149. [Google Scholar] [CrossRef]
- Wang, X.; Lan, P.C.; Ma, S. Metal-organic frameworks for enzyme immobilization, beyond host matrix materials. ACS Cent. Sci. 2020, 6, 1497–1506. [Google Scholar] [CrossRef]
- Thangaraj, B.; Solomon, P.R. Immobilization of lipases–a review. Part I, enzyme immobilization. ChemBioEng Rev. 2019, 6, 157–166. [Google Scholar] [CrossRef]
- Abbott, A.P.; Boothby, D.; Capper, G.; Davies, D.L.; Rasheed, R.K. Deep Eutectic Solvents Formed between Choline Chloride and Carboxylic Acids: Versatile Alternatives to Ionic Liquids. J. Am. Chem. Soc. 2004, 126, 9142–9147. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef]
- Cao, J.; Wang, C.; Shi, L.; Cheng, Y.; Hu, H.; Zeng, B.; Zhao, F. Water based-deep eutectic solvent for ultrasound-assisted liquid–liquid microextraction of parabens in edible oil. Food Chem. 2022, 383, 132586. [Google Scholar] [CrossRef]
- Li, Q.; Ma, C.-L.; He, Y.-C. Effective one-pot chemoenzymatic cascade catalysis of biobased feedstock for synthesizing 2,5-diformylfuran in a sustainable reaction system. Bioresour. Technol. 2023, 378, 128965. [Google Scholar] [CrossRef] [PubMed]
- Cotroneo-Figueroa, V.P.; Gajardo-Parra, N.F.; López-Porfiri, P.; López-Porfiri, P.; Leiva, Á.; Gonzalez-Miquel, M.; Garrido, J.M.; Canales, R.I. Hydrogen bond donor and alcohol chain length effect on the physicochemical properties of choline chloride based deep eutectic solvents mixed with alcohols. J. Mol. Liq. 2022, 345, 116986. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, X.-F.; Wang, Z.; Zheng, T.; Yao, J. Deep eutectic solvent with bifunctional Brønsted-Lewis acids for highly efficient lignocellulose fractionation. Bioresour. Technol. 2022, 347, 126723. [Google Scholar] [CrossRef] [PubMed]
- Nian, B.; Cao, C.; Liu, Y. Synergistic Catalytic Synthesis of Gemini Lipoamino Acids Based on Multiple Hydrogen-Bonding Interactions in Natural Deep Eutectic Solvents-Enzyme System. J. Agric. Food Chem. 2020, 68, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Pätzold, M.; Siebenhaller, S.; Kara, S.; Liese, A.; Syldatk, C.; Holtmann, D. Deep eutectic solvents as efficient solvents in biocatalysis. Trends Biotechnol. 2019, 37, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhao, N.; Tang, S.; Zhu, X.; Ma, C.; Fan, V.B.; Liang, J.; Yu, B.; Yang, L.; He, Y.C. A hybrid strategy for efficient valorization of bulrush into furoic acid in water-ChCl-based deep eutectic solvent. Industrial Crops Prod. 2022, 177, 114434. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zheng, G.-W.; Zong, M.-H.; Li, N.; Lou, W.-Y. Recent progress on deep eutectic solvents in biocatalysis. Bioresour. Bioprocess. 2017, 4, 34. [Google Scholar] [CrossRef]
- Yadav, N.; Venkatesu, P. Current understanding and insights towards protein stabilization and activation in deep eutectic solvents as sustainable solvent media. Phys. Chem. Chem. Phys. 2022, 24, 13474–13509. [Google Scholar] [CrossRef]
- He, Y.-C.; Tao, Z.-C.; Zhang, X.; Yang, Z.-X.; Xu, J.-H. Highly efficient synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate and its derivatives by a robust NADH-dependent reductase from E. coli CCZU-K14. Bioresour. Technol. 2014, 161, 461–464. [Google Scholar] [CrossRef]
- Kolayli, S.; Keskin, M. Natural bee products and their apitherapeutic applications. Stud. Nat. Prod. Chem. 2020, 66, 175–196. [Google Scholar]
- Li, Y.-Y.; Li, Q.; Zhang, P.-Q.; Ma, C.-L.; Xu, J.-H.; He, Y.-C. Catalytic conversion of corncob to furfuryl alcohol in tandem reaction with tin-loaded sulfonated zeolite and NADPH-dependent reductase biocatalyst. Bioresour. Technol. 2021, 320, 124267. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhong, W.; Liu, X. Recent developments in electrochemical investigations into iron carbonyl complexes relevant to the iron centres of hydrogenases. Dalton Trans. 2022, 51, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Fu, H.; Ye, W.; Xie, Y.; Liu, Q.; Wang, H.; Wei, D. Efficient asymmetric synthesis of chiral alcohols using high 2-propanol tolerance alcohol dehydrogenase SmADH2 via an environmentally friendly TBCR system. Catal. Sci. Technol. 2020, 10, 70–78. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Z.Q.; Lin, C.P.; Zheng, Y.G. Efficient biosynthesis of ethyl (R)-4-chloro3-hydroxybutyrate using a stereoselective carbonyl reductase from Burkholderia gladioli. BMC Biotechnol. 2016, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, Z.; Rother, D. Reductive amination of ketones catalyzed by whole cell biocatalysts containing imine reductases (IREDs). J. Biotechnol. 2017, 258, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xie, B.; Zhou, L.; Maria-Solano, M.A.; Liu, W.D.; Li, J.; Feng, J.H.; Liu, X.T.; Osuna, S.; Guo, R.T.; et al. A tailor-made self-sufficient whole-cell biocatalyst enables scalable enantioselective synthesis of (R)-3-quinuclidinol in a high space-time yield. Org. Process Res. Dev. 2019, 23, 1813–1821. [Google Scholar] [CrossRef]
- Gu, T.; Wang, B.; Zhang, Z.; Wang, Z.; Chong, G.; Ma, C.; Tang, Y.-J.; He, Y. Sequential pretreatment of bamboo shoot shell and biosynthesis of ethyl (R)-4-chloro-3-hydroxybutanoate in aqueous-butyl acetate media. Process Biochem. 2019, 80, 112–118. [Google Scholar] [CrossRef]
- Lu, Y.; Dai, H.; Shi, H.; Tang, L.; Sun, X.; Ou, Z. Synthesis of ethyl (R)-4-chloro-3-hydroxybutyrate by immobilized cells using amino acid-modified magnetic nanoparticles. Process Biochem. 2022, 99, 9–20. [Google Scholar] [CrossRef]
- Perveen, S.; Zhang, S.; Wang, L.; Ong, P.; Ouyang, Y.; Jiao, J.; Li, P. Synthesis of axially chiral biaryls via enantioselective ullmann coupling of ortho-chlorinated aryl aldehydes enabled by a chiral 2,2′-bipyridine ligand. Angew. Chem. Int. Ed. 2022, 134, e202212108. [Google Scholar] [CrossRef]
- Liang, X.; Deng, H.; Xiong, T.; Bai, Y.; Fan, T.P.; Zheng, X.; Cai, Y. Overexpression and biochemical characterization of a carboxyspermidine dehydrogenase from Agrobacterium fabrum str. C58 and its application to carboxyspermidine production. J. Sci. Food Agric. 2022, 102, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Liu, Y.; Meng, J.; Cheng, W.; Chen, W.; Liu, S.; Liu, Y.; Li, J.; Yu, H. Multiple hydrogen bond coordination in three-constituent deep eutectic solvents enhances lignin fractionation from biomass. Green Chem. 2018, 20, 2711–2721. [Google Scholar] [CrossRef]
- Luo, W.; Du, H.-J.; Bonku, E.M.; Hou, Y.-L.; Li, L.-L.; Wang, X.-Q.; Yang, Z.-H. An Alkali-tolerant Carbonyl Reductase from Bacillus subtilis by Gene Mining: Identification and Application. Catal. Lett. 2019, 149, 2973–2983. [Google Scholar] [CrossRef]
- Yuan, L.; Qin, Y.-L.; Zou, Z.-C.; Appiah, B.; Huang, H.; Yang, Z.-H.; Qun, C. Enhancing intracellular NADPH bioavailability through improving pentose phosphate pathway flux and its application in biocatalysis asymmetric reduction reaction. J. Biosci. Bioeng. 2022, 134, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Plž, M.; Petrovičová, T.; Rebroš, M. Semi-continuous flow biocatalysis with affinity co-immobilized ketoreductase and glucose dehydrogenase. Molecules 2020, 25, 4278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duan, Y.; Zhong, L.; Li, Z.; Cui, Z.; Huang, Y. Characterization of a novel aldo-keto reductase with anti-Prelog stereospecificity from Corallococcus sp. EGB. Int. J. Biol. Macromol. 2020, 146, 36–44. [Google Scholar] [CrossRef]
- Xu, S.; Lin, Q.; Chen, W.; Lin, R.; Shen, Y.; Tang, P.; Yu, S.; Du, W.; Li, J. Efficient Biosynthesis of (S)-1-chloro-2-heptanol Catalyzed by a Newly Isolated Fungi Curvularia hominis B-36. Catalysts 2022, 13, 52. [Google Scholar] [CrossRef]
- Bekler, F.M.; Güven, K.; Gül Güven, R. Purification and characterization of novel α-amylase from Anoxybacillus ayderensis FMB1. Biocatal. Biotransform. 2021, 39, 322–332. [Google Scholar] [CrossRef]
- Bu, C.; Qiao, H.; Zou, L.; Tao, Y.; Fu, J.; Liu, C.; Zheng, Z.; Ouyang, J. Hydrogenation of biomass pyrolysis oil using enhanced reduction catalytic capacity cellular by expressing excavated carbonyl reductase. Fuel 2023, 332, 126108. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Alcántara, A.R.; de María, P.D.; Sánchez-Montero, J.M. Biocatalysis for the asymmetric synthesis of Active Pharmaceutical Ingredients (APIs): This time is for real. Expert Opin. Drug Discov. 2022, 17, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Sunder, A.V.; Singh, P.; Wangikar, P.P. Characterization and Application of a Robust Glucose Dehydrogenase from Paenibacillus pini for Cofactor Regeneration in Biocatalysis. Indian J. Microbiol. 2020, 60, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Di, J.; Liao, X.; Li, Q.; He, Y.-C.; Ma, C. Significantly enhanced bioconversion of high titer biomass-derived furfural to furfuryl alcohol by robust endogenous aldehyde reductase in a sustainable way. Green Chem. 2023, in press. [Google Scholar] [CrossRef]
- Ye, Q.; Ouyang, P.K.; Ying, H.J. A review—biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate ester: Recent advances and future perspectives. Appl. Microbiol. Biotechnol. 2011, 89, 513–522. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Xu, D.; Jiang, L.; He, Y. Asymmetric Bioreduction of Ethyl 4-Chloroacetoacetate into Ethyl 4-Chloro-3-hydroxybutyrate by Recombinant Escherichia coli CgCR in Ethyl Acetate-Betaine:Lactic Acid-Water. Processes 2023, 11, 3144. https://doi.org/10.3390/pr11113144
Yang L, Xu D, Jiang L, He Y. Asymmetric Bioreduction of Ethyl 4-Chloroacetoacetate into Ethyl 4-Chloro-3-hydroxybutyrate by Recombinant Escherichia coli CgCR in Ethyl Acetate-Betaine:Lactic Acid-Water. Processes. 2023; 11(11):3144. https://doi.org/10.3390/pr11113144
Chicago/Turabian StyleYang, Linsong, Daozhu Xu, Luyao Jiang, and Yucai He. 2023. "Asymmetric Bioreduction of Ethyl 4-Chloroacetoacetate into Ethyl 4-Chloro-3-hydroxybutyrate by Recombinant Escherichia coli CgCR in Ethyl Acetate-Betaine:Lactic Acid-Water" Processes 11, no. 11: 3144. https://doi.org/10.3390/pr11113144
APA StyleYang, L., Xu, D., Jiang, L., & He, Y. (2023). Asymmetric Bioreduction of Ethyl 4-Chloroacetoacetate into Ethyl 4-Chloro-3-hydroxybutyrate by Recombinant Escherichia coli CgCR in Ethyl Acetate-Betaine:Lactic Acid-Water. Processes, 11(11), 3144. https://doi.org/10.3390/pr11113144






