High Molecular Weight α-Galactosidase from the Novel Strain Aspergillus sp. D-23 and Its Hydrolysis Performance
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Medium Formula
2.3. UV-DES Mutagenesis Procedure
2.4. α-Galactosidase Assay and Protein Concentration Assay
2.5. Purification of α-Galactosidase
2.6. Electrophoretic Analysis
2.6.1. SDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis)
2.6.2. Native-PAGE and Activity Staining
2.7. α-Galactosidase Characterization
2.8. Kinetic Parameters and Substrate Specificity
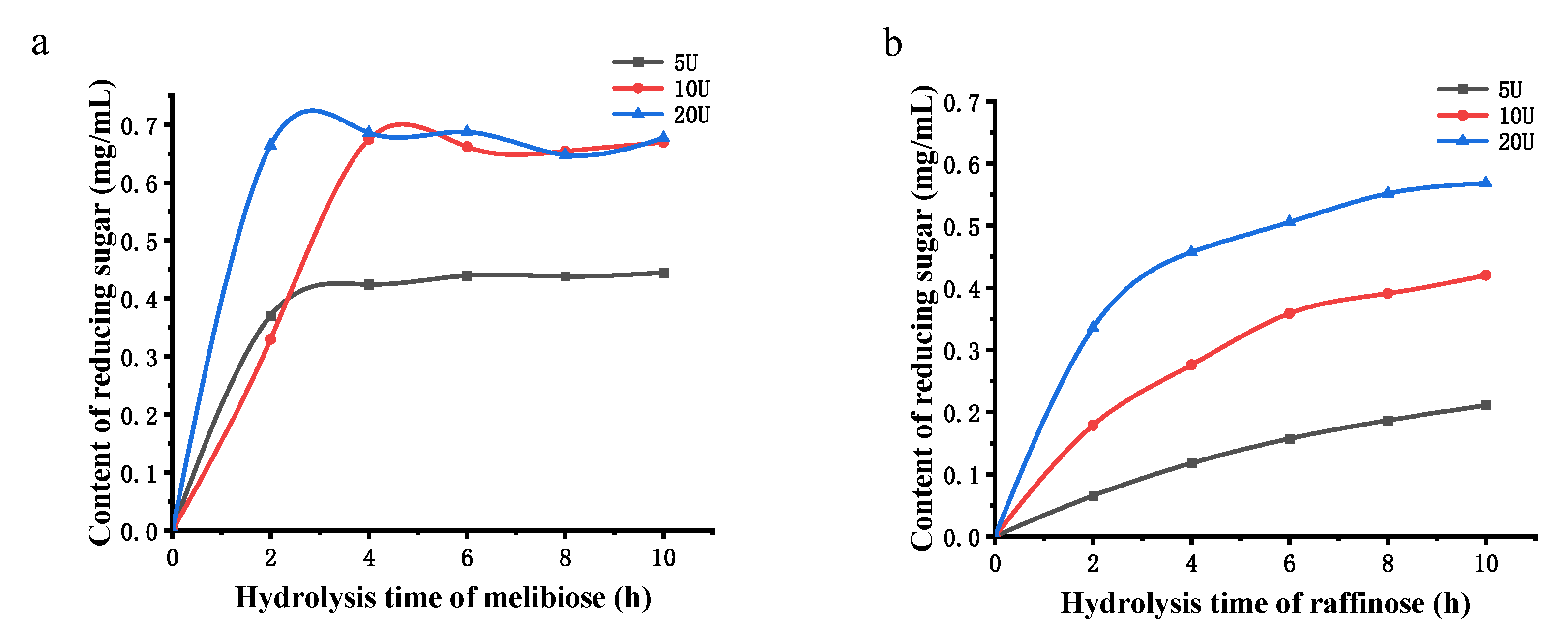
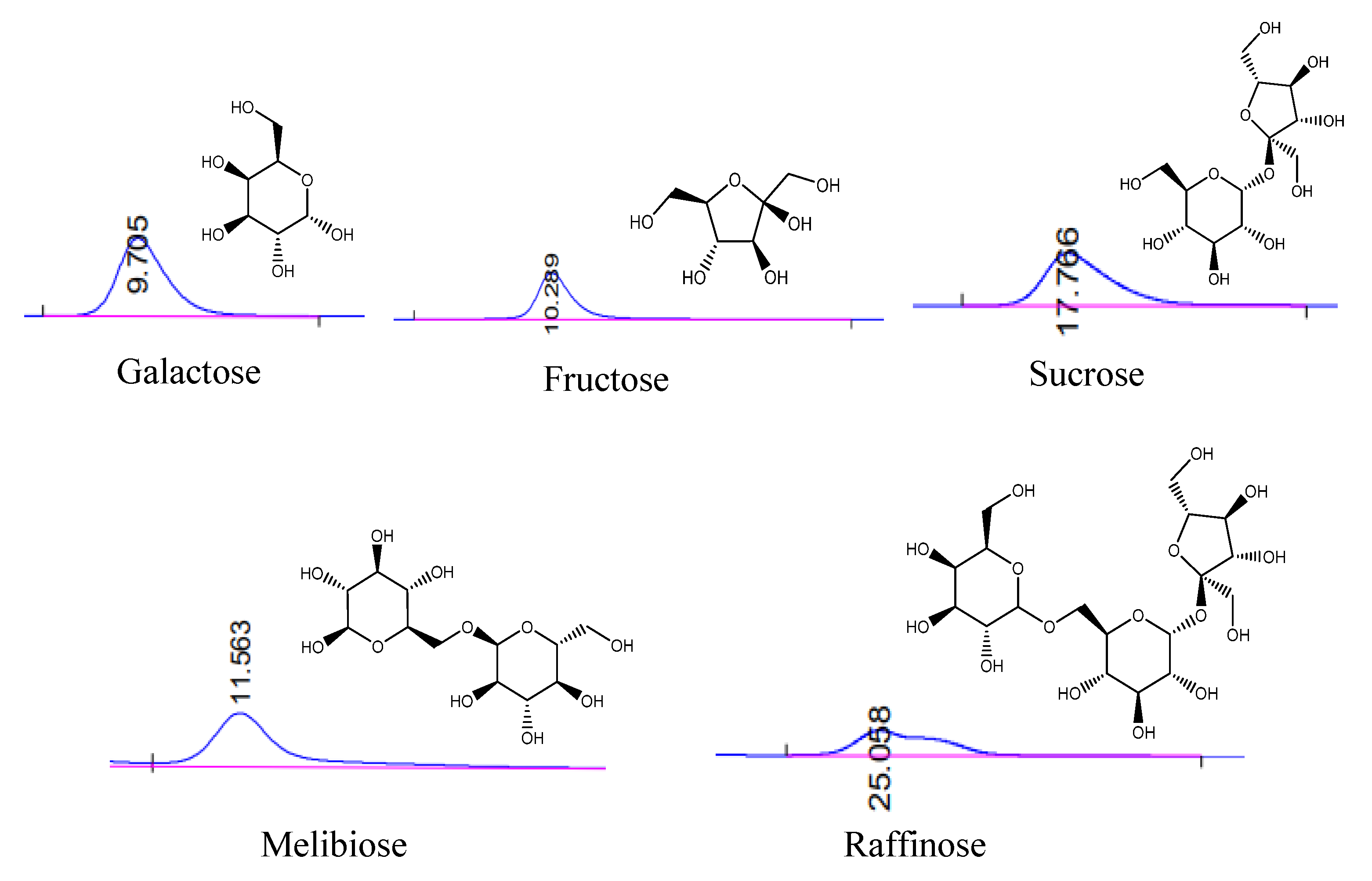
2.9. Enzymatic Hydrolysis of Melibiose and Raffinose
3. Results and Discussion
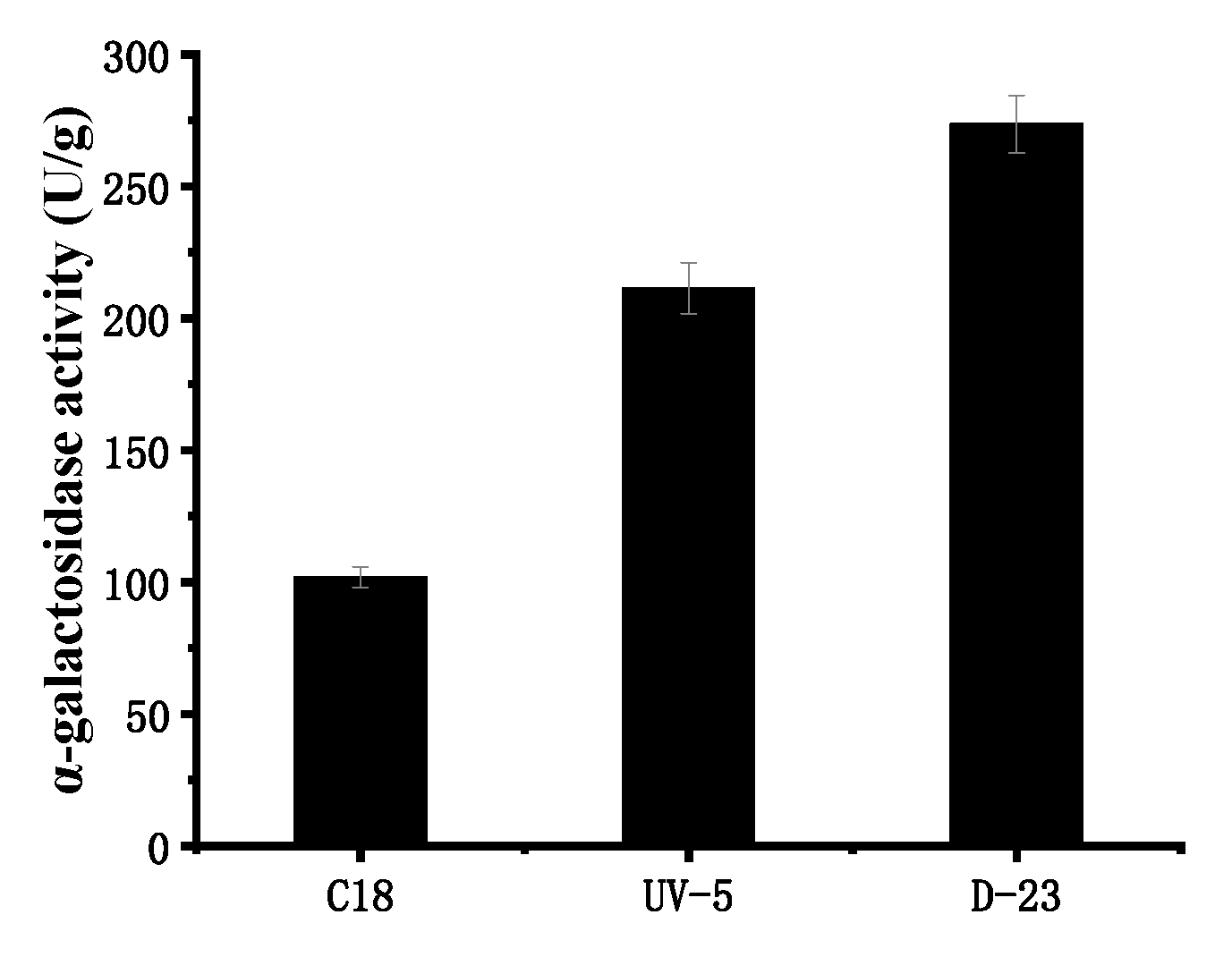
3.1. Compound Mutagenesis of Ultraviolet and Diethyl Sulfate
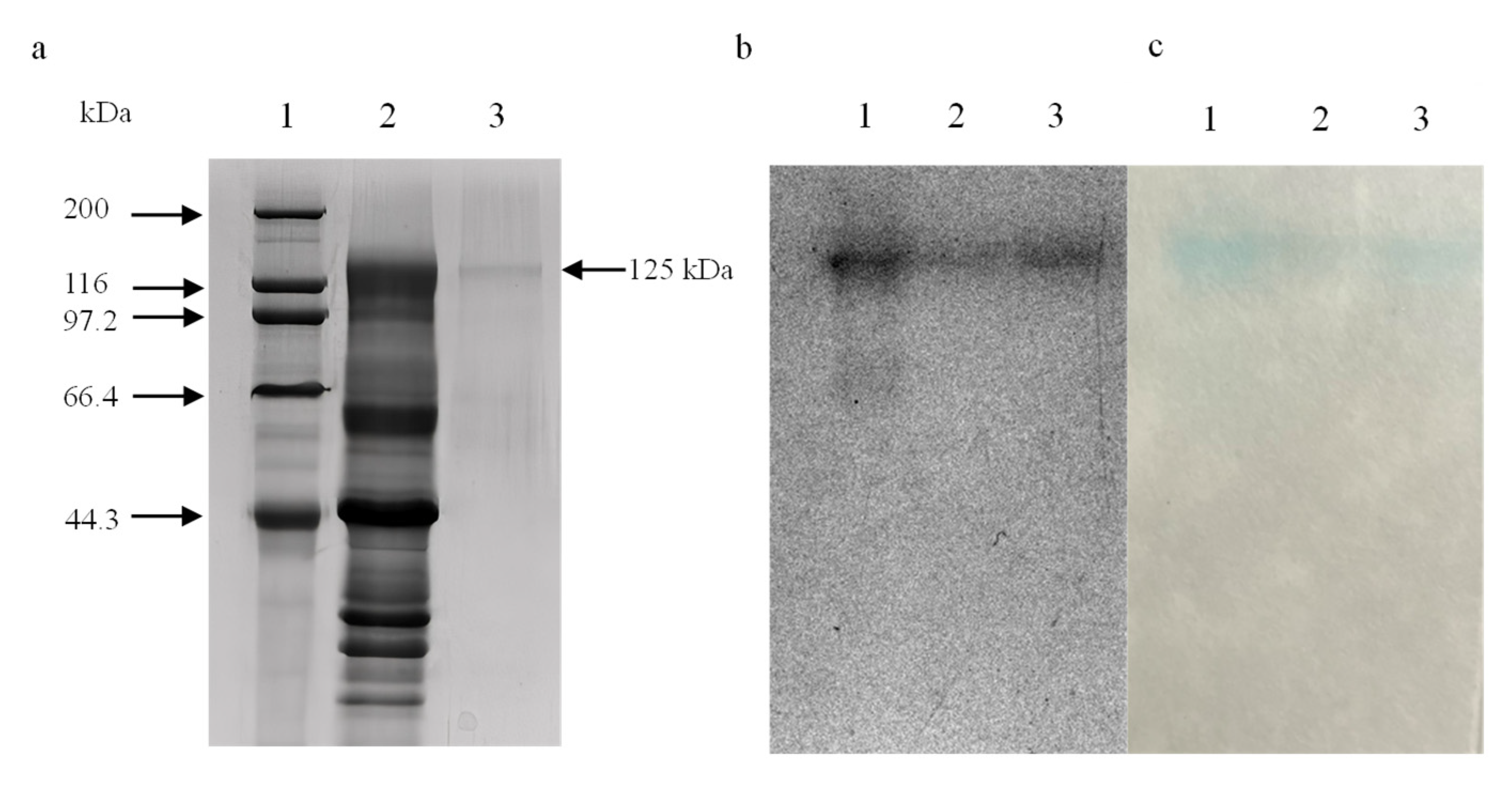
3.2. Purification of α-Galactosidase
3.3. Homogeneity and Molecular Mass of α-Galactosidase
3.4. Biochemical Characterization of α-Galactosidase
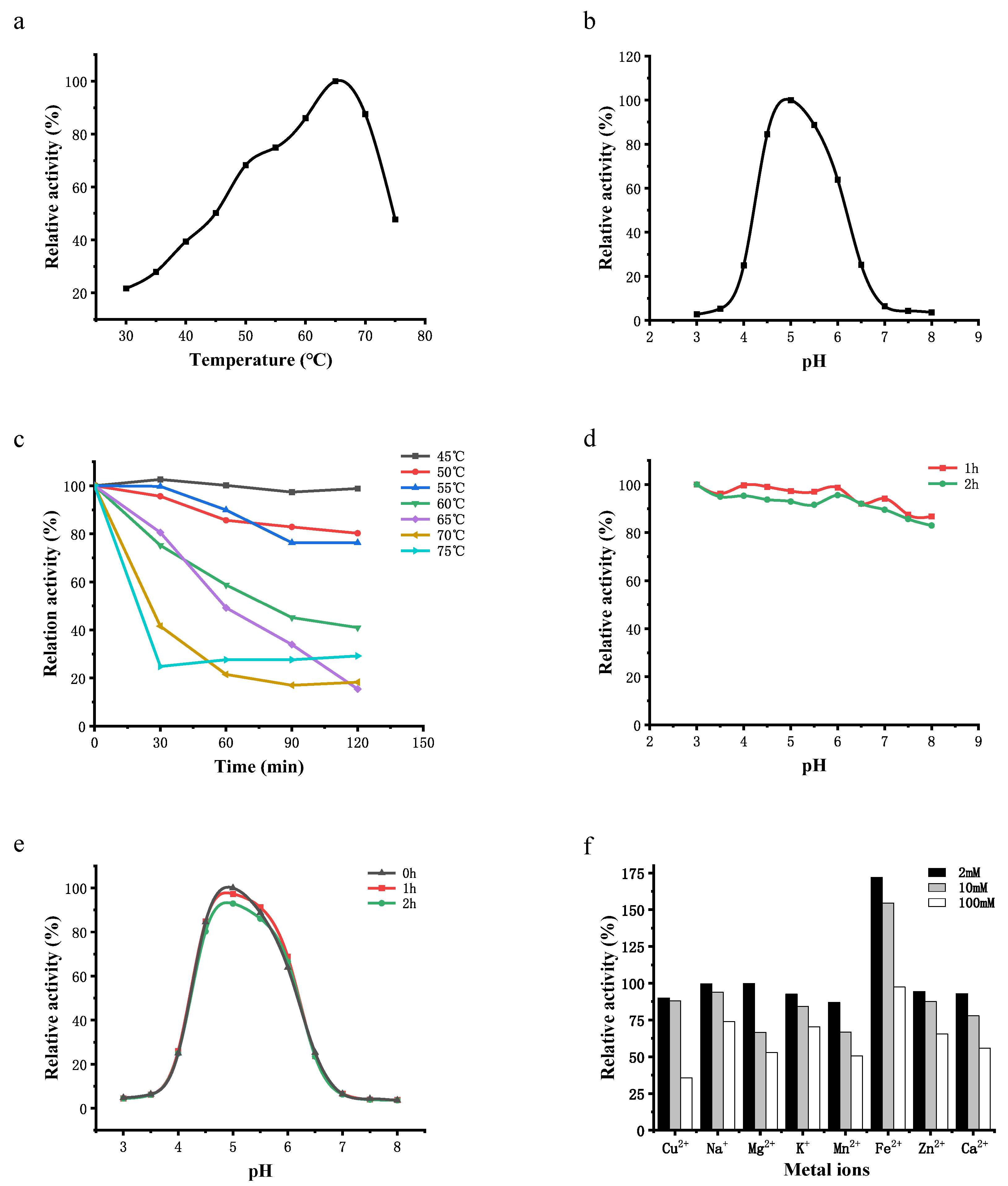
3.4.1. Temperature and pH Optima and Stability
3.4.2. Effect of Metal Ions
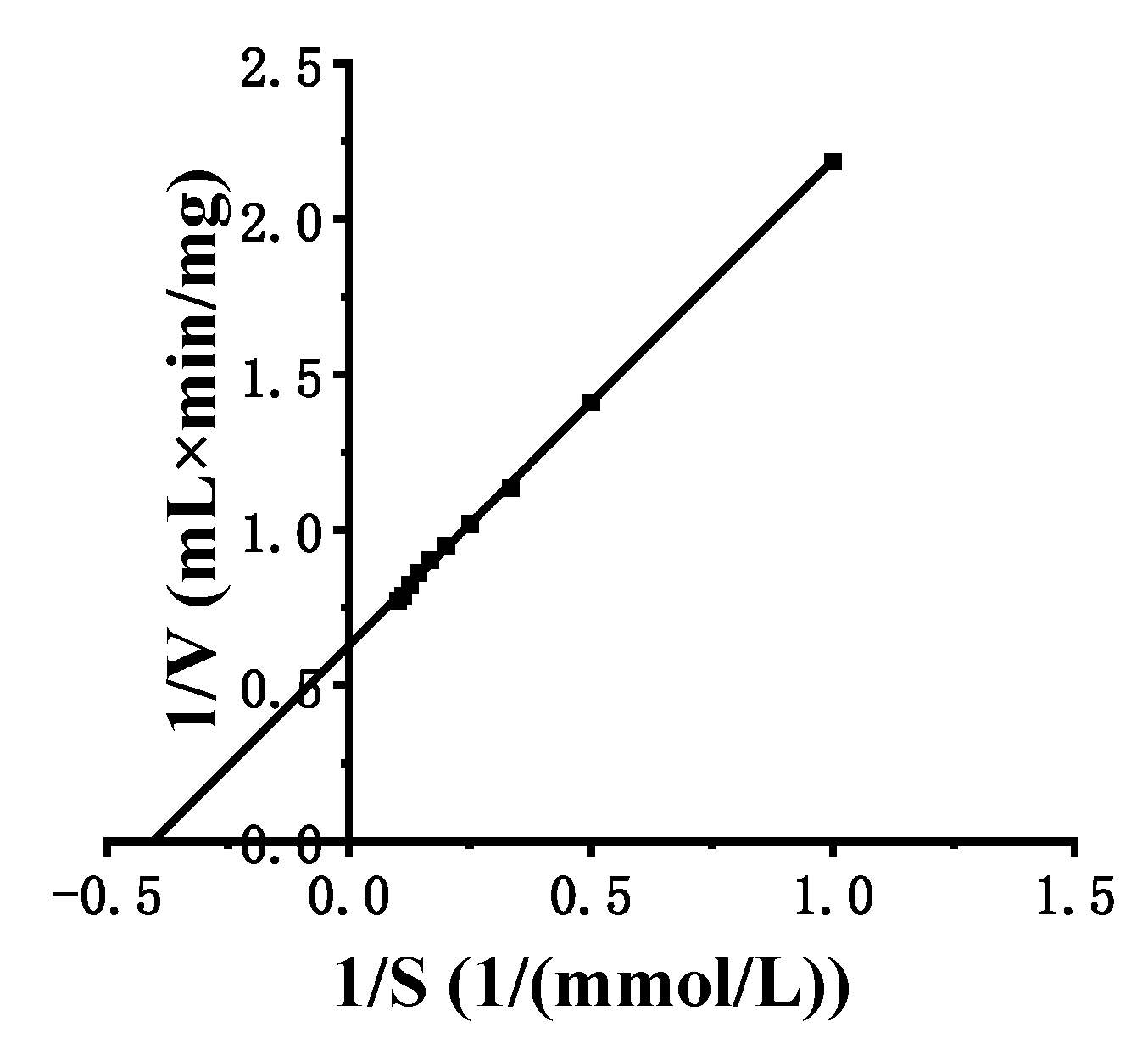
3.5. Kinetic Characterization and Substrate Specificity
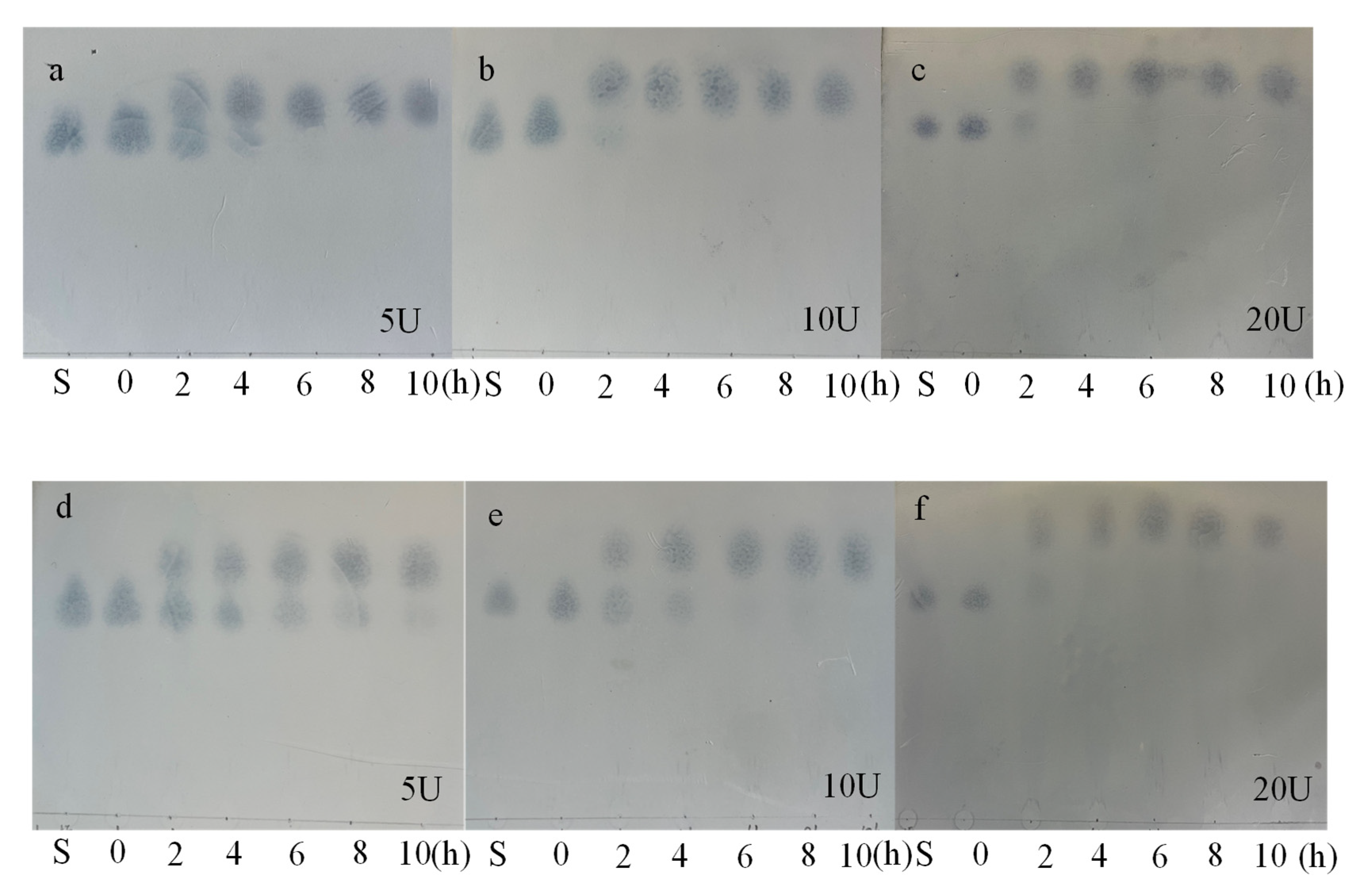
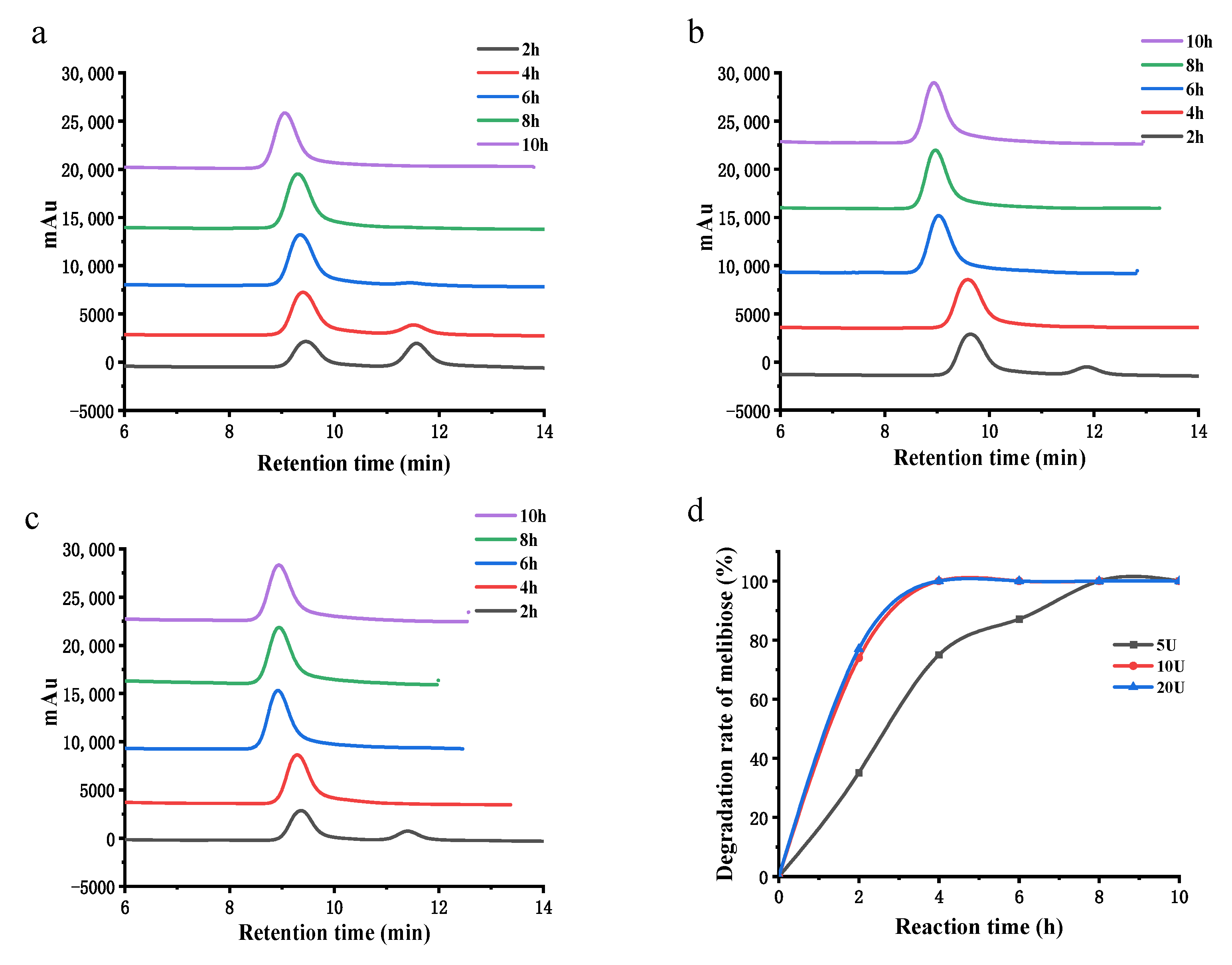
3.6. Hydrolysis of Melibiose and Raffinose by α-Galactosidase
3.6.1. TLC Analysis of the Hydrolysate
3.6.2. HPLC Analysis of the Hydrolysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Anisha, G.S. 16-α-Galactosidases. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Negi, S., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 369–394. [Google Scholar]
- Anisha, G.S. Microbial alpha-galactosidases: Efficient biocatalysts for bioprocess technology. Bioresour. Technol. 2022, 344, 126293. [Google Scholar] [CrossRef] [PubMed]
- Gajdhane, S.B.; Bhagwat, P.K.; Dandge, P.B. Response surface methodology-based optimization of production media and purification of alpha-galactosidase in solid-state fermentation by Fusarium moniliforme NCIM 1099. 3 Biotech 2016, 6, 260. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.-K.; Loman, A.A.; Islam, S.M. α-Galactosidase and Its Applications in Food Processing. In Encyclopedia of Food Chemistry; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Rättö, M.; Siika-Aho, M.; Buchert, J.; Valkeajävi, A.; Viikari, L. Enzymatic hydrolosis of isolated and fibre-bound galactoglucomannans from pine-wood and pine kraft pulp. Appl. Microbiol. Biotechnol. 1993, 40, 449–454. [Google Scholar] [CrossRef]
- Linden, J.C. Immobilized α-d-galactosidase in the sugar beet industry. Enzym. Microb. Technol. 1982, 4, 130–136. [Google Scholar] [CrossRef]
- Kizhner, T.; Azulay, Y.; Hainrichson, M.; Tekoah, Y.; Arvatz, G.; Shulman, A.; Ruderfer, I.; Aviezer, D.; Shaaltiel, Y. Characterization of a chemically modified plant cell culture expressed human α-Galactosidase-A enzyme for treatment of Fabry disease. Mol. Genet. Metab. 2015, 114, 259–267. [Google Scholar] [CrossRef]
- Bhatia, S.; Singh, A.; Batra, N.; Singh, J. Microbial production and biotechnological applications of alpha-galactosidase. Int. J. Biol. Macromol. 2020, 150, 1294–1313. [Google Scholar] [CrossRef]
- Çalcı, E.; Önal, S. Comparative affinity immobilization of α-galactosidase on chitosan functionalized with Concanavalin A and its useability for the hydrolysis of raffinose. React. Funct. Polym. 2022, 172, 105181. [Google Scholar] [CrossRef]
- Jang, J.M.; Yang, Y.; Wang, R.; Bao, H.; Yuan, H.; Yang, J. Characterization of a high performance α-galactosidase from Irpex lacteus and its usage in removal of raffinose family oligosaccharides from soymilk. Int. J. Biol. Macromol. 2019, 131, 1138–1146. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Yang, Y.; Liu, Y.; Piao, X.; Cao, Y. Characterization of a protease-resistant alpha-galactosidase from Aspergillus oryzae YZ1 and its application in hydrolysis of raffinose family oligosaccharides from soymilk. Int. J. Biol. Macromol. 2020, 158, 708–720. [Google Scholar] [CrossRef]
- Wang, H.; Shi, P.; Luo, H.; Huang, H.; Yang, P.; Yao, B. A thermophilic α-galactosidase from Neosartorya fischeri P1 with high specific activity, broad substrate specificity and significant hydrolysis ability of soymilk. Bioresour. Technol. 2014, 153, 361–364. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, Q.; Guan, L.; Jiang, Z.; Yang, S. Biochemical characterization of a novel protease-resistant α-galactosidase from Paecilomyces thermophila suitable for raffinose family oligosaccharides degradation. Process Biochem. 2020, 94, 370–379. [Google Scholar] [CrossRef]
- Cao, Y.; Xiong, E.M.; True, A.D.; Xiong, Y.L. The pH-dependent protection of α-galactosidase activity by proteins against degradative enzymes during soymilk in vitro digestion. LWT Food Sci. Technol. 2016, 69, 244–250. [Google Scholar] [CrossRef]
- Mi, S.; Meng, K.; Wang, Y.; Bai, Y.; Yuan, T.; Luo, H.; Yao, B. Molecular cloning and characterization of a novel α-galactosidase gene from Penicillium sp. F63 CGMCC 1669 and expression in Pichia pastoris. Enzym. Microb. Technol. 2007, 40, 1373–1380. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, Y.; Li, Z.; Lu, W.; Cao, Y. Cloning and functional expression of α-galactosidase cDNA from Penicillium janczewskii zaleski. Biologia 2011, 66, 205–212. [Google Scholar] [CrossRef]
- Rezessy-Szabó, J.M.; Nguyen, Q.D.; Hoschke, Á.; Braet, C.; Hajós, G.; Claeyssens, M. A novel thermostable α-galactosidase from the thermophilic fungus Thermomyces lanuginosus CBS 395.62/b: Purification and characterization. Biochim. Biophys. Acta Gen. Subj. 2007, 1770, 55–62. [Google Scholar] [CrossRef]
- Katrolia, P.; Jia, H.; Yan, Q.; Song, S.; Jiang, Z.; Xu, H. Characterization of a protease-resistant α-galactosidase from the thermophilic fungus Rhizomucor miehei and its application in removal of raffinose family oligosaccharides. Bioresour. Technol. 2012, 110, 578–586. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Ye, F.; Geng, X.-R.; Xu, L.-J.; Chang, M.-C.; Feng, C.-P.; Meng, J.-L. Purification and characterization of a novel protease-resistant GH27 α-galactosidase from Hericium erinaceus. Int. J. Biol. Macromol. 2018, 120, 2165–2174. [Google Scholar] [CrossRef]
- Albertson, P.L.; Grof, C.P. Application of high performance anion exchange-pulsed amperometric detection to measure the activity of key sucrose metabolising enzymes in sugarcane. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 845, 151–156. [Google Scholar] [CrossRef]
- Vidya, C.H.; Gnanesh Kumar, B.S.; Chinmayee, C.V.; Singh, S.A. Purification, characterization and specificity of a new GH family 35 galactosidase from Aspergillus awamori. Int. J. Biol. Macromol. 2020, 156, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Fan, J.; Xu, L.; Wang, H.; Ng, T.B. Hydrolysis of oligosaccharides by a fungal α-galactosidase from fruiting bodies of a wild mushroom Leucopaxillus tricolor. J. Basic Microbiol. 2018, 58, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Shivam, K.; Mishra, S.K. Purification and characterization of a thermostable α-galactosidase with transglycosylation activity from Aspergillus parasiticus MTCC-2796. Process Biochem. 2010, 45, 1088–1093. [Google Scholar] [CrossRef]
- Xie, J.; Wang, B.; He, Z.; Pan, L. A thermophilic fungal GH36 alpha-galactosidase from Lichtheimia ramosa and its synergistic hydrolysis of locust bean gum. Carbohydr. Res. 2020, 491, 107911. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, H.; Ma, R.; Shi, P.; Niu, C.; Luo, H.; Yang, P.; Yao, B. Biochemical characterization of a novel thermophilic α-galactosidase from Talaromyces leycettanus JCM12802 with significant transglycosylation activity. J. Biosci. Bioeng. 2016, 121, 7–12. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, P.; Shi, P.; Wang, Y.; Luo, H.; Meng, K.; Zhang, Z.; Wu, N.; Yao, B.; Fan, Y. Purification and characterization of a novel protease-resistant α-galactosidase from Rhizopus sp. F78 ACCC 30795. Enzym. Microb. Technol. 2007, 41, 835–841. [Google Scholar] [CrossRef]
- Sakharayapatna Ranganatha, K.; Venugopal, A.; Chinthapalli, D.K.; Subramanyam, R.; Nadimpalli, S.K. Purification, biochemical and biophysical characterization of an acidic alpha-galactosidase from the seeds of Annona squamosa (custard apple). Int. J. Biol. Macromol. 2021, 175, 558–571. [Google Scholar] [CrossRef]
- Blöchl, A.; Peterbauer, T.; Hofmann, J.; Richter, A. Enzymatic breakdown of raffinose oligosaccharides in pea seeds. Planta 2008, 228, 99–110. [Google Scholar] [CrossRef]
- Geng, X.; Yang, D.; Zhang, Q.; Chang, M.; Xu, L.; Cheng, Y.; Wang, H.; Meng, J. Good hydrolysis activity on raffinose family oligosaccharides by a novel α-galactosidase from Tremella aurantialba. Int. J. Biol. Macromol. 2020, 150, 1249–1257. [Google Scholar] [CrossRef]
- Singh, N.; Kayastha, A.M. Purification and characterization of α-galactosidase from white chickpea (Cicer arietinum). J. Agric. Food Chem. 2012, 60, 3253–3259. [Google Scholar] [CrossRef]
- Hu, Y.; Tian, G.; Geng, X.; Zhang, W.; Zhao, L.; Wang, H.; Ng, T.B. A protease-resistant α-galactosidase from Pleurotus citrinopileatus with broad substrate specificity and good hydrolytic activity on raffinose family oligosaccharides. Process Biochem. 2016, 51, 491–499. [Google Scholar] [CrossRef]
- Chen, Z.; Yan, Q.; Jiang, Z.; Liu, Y.; Li, Y. High-level expression of a novel α-galactosidase gene from Rhizomucor miehei in Pichia pastoris and characterization of the recombinant enyzme. Protein Expr. Purif. 2015, 110, 107–114. [Google Scholar] [CrossRef]
- Hu, Y.; Tian, G.; Zhao, L.; Wang, H.; Ng, T.B. A protease-resistant α-galactosidase from Pleurotus djamor with broad pH stability and good hydrolytic activity toward raffinose family oligosaccharides. Int. J. Biol. Macromol. 2017, 94, 122–130. [Google Scholar] [CrossRef]
- Ferreira, J.G.; Reis, A.P.; Guimaraes, V.M.; Falkoski, D.L.; Fialho Lda, S.; De Rezende, S.T. Purification and characterization of Aspergillus terreus alpha-galactosidases and their use for hydrolysis of soymilk oligosaccharides. Appl. Biochem. Biotechnol. 2011, 164, 1111–1125. [Google Scholar] [CrossRef]

| Procedures | Total Activity (U) | Protein (mg) | Specific Activity (U/mg) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|
| Crude enzyme | 390.00 | 1.78 | 219.45 | 100.00 | 1.00 |
| Q | 251.34 | 0.14 | 1682.70 | 64.46 | 7.67 |
| Substrate | Concentration (mmol/L) | Relative Activity (%) |
|---|---|---|
| p-Nitrophenyl α-D-galactopyranoside (p-NPG) | 8 | 100.00 |
| Melibiose | 50 | 17.63 |
| Raffinose | 50 | 10.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Zhang, J.; Liu, X.; Zhang, P.; Yue, W.; Cai, Z. High Molecular Weight α-Galactosidase from the Novel Strain Aspergillus sp. D-23 and Its Hydrolysis Performance. Processes 2023, 11, 255. https://doi.org/10.3390/pr11010255
Chen K, Zhang J, Liu X, Zhang P, Yue W, Cai Z. High Molecular Weight α-Galactosidase from the Novel Strain Aspergillus sp. D-23 and Its Hydrolysis Performance. Processes. 2023; 11(1):255. https://doi.org/10.3390/pr11010255
Chicago/Turabian StyleChen, Ke, Jingyun Zhang, Xing Liu, Peiyuan Zhang, Wenlong Yue, and Zhiqiang Cai. 2023. "High Molecular Weight α-Galactosidase from the Novel Strain Aspergillus sp. D-23 and Its Hydrolysis Performance" Processes 11, no. 1: 255. https://doi.org/10.3390/pr11010255
APA StyleChen, K., Zhang, J., Liu, X., Zhang, P., Yue, W., & Cai, Z. (2023). High Molecular Weight α-Galactosidase from the Novel Strain Aspergillus sp. D-23 and Its Hydrolysis Performance. Processes, 11(1), 255. https://doi.org/10.3390/pr11010255







