1. Introduction
In the three-dimensional culture method, it is possible to culture cells in a state close to that of an actual living body. Therefore, the results of 3D culture are closer to in vivo than those of 2D culture, such as supply of medium components, cell–cell interaction, cell differentiation potential, proliferation, survival rate, toxicity and metabolism of drugs, and stimulus response [
1,
2,
3,
4,
5]. One of the three-dimensional culture methods is to culture a cell aggregate. Cell aggregates are formed by cell–cell interactions with E-cadherin and cell–extracellular matrix with integrins [
5]. In addition, there is a concentration gradient of oxygen and nutrients supplied to cells inside the cell aggregate; so, the same physiological environment as in the living body can be reproduced [
6]. The natural shape of the cell is maintained within the cell morphology. As a characteristic of the cell aggregate, high gene expression and metabolic activity have been confirmed because the cell–cell interaction is enhanced as compared with the cells cultivated two-dimensionally on the same culture medium. Furthermore, it has been reported that the diffusion limit of oxygen and nutrients supplied to the cell aggregate and waste products inside the cell aggregate is 150 to 200 µm [
5]. Therefore, cell aggregates with controlled size are applied in various fields such as regenerative medicine and drug toxicity testing [
7,
8,
9,
10].
Until now, various cell aggregation methods have been reported and utilized for cell aggregate culture. The mainstream method is to coat the surface of the base material with a super hydrophilic polymer or the like. This method takes advantage of the fact that cells show nonadhesiveness on highly hydrophilic and hydrophobic surfaces [
9], and the cells are aggregated at the bottom of the substrate. Another method of culturing cell aggregates is conducted on a positively charged substrate surface. Cell aggregates cultured on these substrates have been reported to enhance cell function compared with monolayer cells [
8]. However, with these methods, it is problematic that the cells peel off from the adhesion surface, the aggregate size is nonuniform, and that strong positive charges show cytotoxicity [
7].
We have previously reported that KP24 is a peptide that induces uniform-sized cell aggregates only in cell suspension [
11]. By immobilizing this peptide on the substrate, we thought that it could be applied to a three-dimensional culture substrate that can induce cell aggregates with controlled size. Therefore, in this study, mouse fibroblasts (L929) and human Mesenchymal Stem Cells (hMSC) were seeded on a glass substrate on which KP24 was immobilized, and the cell behavior was observed. These results can contribute to the development of regenerative medicine such as cell transplantation substrates and microarrays.
KP24 peptide induces uniform-sized cell aggregates only in cell suspension. However, it is unclear as to the mechanism of KP24 that induces cell aggregation. In this study, KP24 is chemically fixed on the surface of the substrate; so, it is possible to trace the mechanism of interaction between KP24 and the cells surface. With this fact, this experiment may help in the analysis of the mechanism of action of KP24. It can be determined whether or not the KP24 peptide is taken up into the cell for induction of cell aggregates. For mechanism analysis of the cell aggregation ability of KP24, we investigated how KP24 acts on the cell surface without being taken up into cells.
KP24, a 12-cycle repeating sequence consisting of Lys and Pro, was synthesized by the Fmoc solid-phase synthesis method. By immobilizing the synthesized KP24 from the end, two types of KP24-immobilized base materials having different surface functional groups were prepared [
11]. Immobilization of KP24 was evaluated by analyzing the wettability of the substrate surface and surface elements. In the cell experiment, L929 was seeded on the KP24-immobilized substrate to observe whether the cell aggregate could be induced on the substrate. Furthermore, the viability of cells involved in cell assembly formation was evaluated by Live Dead staining. A cell mass microscope (Mil-Cell) was used to observe the three-dimensional shape of the cell aggregate induced on the P24-immobilized substrate. Based on the observation results of Mil-Cell, the three-dimensional shape and sphericity of each cell aggregate were compared [
11].
2. Materials and Methods
2.1. Materials
All 9-fluorenylmethyloxycarbonyl (Fmoc) amino acid derivatives and solid-phase peptide synthesis resin were purchased from Watanabe Chemical Industries, Ltd. (Hiroshima, Japan). All other reagents including organic solvents were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan).
Dulbecco’s modified Eagle’s medium (DMEM) and Eagle’s Minimal Essential Medium (EMEM) for cell culture were purchased from Nissui Pharmaceutical Co., Ltd., Tokyo, Japan. Fetal bovine serum (FBS) for cell culture was from HyClone, Cytiva, Sheffield, UK. A total 100 U/mL penicillin and 100 μg/mL streptomycin were obtained from Thermo Fisher Scientific, MA, USA. Other cell culture additives were obtained from Invitrogen (USA) and FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan).
2.2. Peptide Synthesis
KP24 peptides that induce the cell aggregate were synthesized by the solid-phase peptide synthesis procedure in previous paper [
11]. The KP24 was synthesized on Alko–PEG resin using the handmade standard manual Fmoc-protocol with a 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) activation procedure. The Fmoc group deprotection and condensation reactions were repeated to synthesize the Pro-Lys(Boc)
12-TrtA-PEG-Resin. Finally, peptide protecting group deprotection and resin cleavage were conducted under acidic conditions. The KP24 peptide was dialyzed with a membrane with a molecular weight cutoff range of 100–500 Da (Spectra/Pro Dialysis Membrane Biotech CE Tubing, MWCO: 100–500 D). Finally, all peptides were purified by high-performance liquid chromatography (HPLC8020 System; Tosoh Corp., Tokyo, Japan; Column: TSKgel-ODS-100V 5 mm) with a gradient of water/acetonitrile containing 0.1% TFA. All peptides were identified by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF-MS; Microflex LRF System, Bruker Corp., Billerica, MA, USA) using a-cyano-4-hydroxycinnamic acid as a matrix [
11]..
2.3. Cells and Culture
Mouse fibroblasts (L929) (RIKEN Biosource Research Center, Ibaraki, Japan) were cultured in DMEM containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cells were maintained at 37 ℃ in a humidified 5% CO2/95% air atmosphere. EMEM powder, 2.82 g was dissolved in 261 mL of ultrapure water, sterilized in an autoclave at 120 ℃ for 15 min, and then filtered and sterilized. To prepare 10% (w/v) FBS EMEM medium, 7.50% sodium hydrogen carbonate (NaHCO3) aqueous solution 6.00 mL, 3% L-glutamine aqueous solution 3.00 mL, and 30 mL of FBS were mixed. Next, 2.82 g of Eagle’s Minimal Essential Medium (EMEM) powder was dissolved in 291 mL of ultrapure water, sterilized in an autoclave at 120 ℃ for 15 min, and then filtered and sterilized. To prepare serum-free EMEM medium, 7.5% NaHCO3 aqueous solution 6.00 mL and 3.0% L-glutamine 3.00 mL of aqueous solution were mixed.
Human Mesenchymal Stem Cells (hMSC, RCB1451) (RIKEN Biosource Research Center, Ibaraki, Japan) were cultured in DMEM containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 g/mL streptomycin. All cells were maintained at 37 ℃ in a humidified 5% CO2/95% air atmosphere.
L929 cells were selected because they were used in the previous report [
11]. L929 cells are commonly used to evaluate cytotoxicity and other activities [
12,
13,
14]. The toxicity evaluation of KP24 and induction of cell aggregates was performed in a previous report [
11] and was used again in this study. On the other hand, hMSC were used because of their potential for developing into regenerative medicine such as cell transplantation.
2.4. Preparation of KP24-Immobilized Glass
A schematic description for preparation of KP24-immobilized glass is shown in
Figure 1. A cover glass (Matsunami Glass Industry Co., Ltd. Kishiwada, Japan) with a diameter of 15 mm was used for the KP24-immobilized substrate. Hydrogen peroxide (3.0 mL) and concentrated sulfuric acid (7.0 mL) were mixed in a petri dish, and the cover glass was immersed for 2 h for piranha treatment. The piranha-treated cover glass was immersed in a 2.0% APTES solution diluted with toluene for 6 h to prepare an APTES substrate. KP24 (1.0 mg, 0.37 µmol) and excess amounts of WSCD/HCl and NHS were added to a 2.0 mL tube and dissolved in 1.0 mL of PBS. The mixture was stirred at room temperature for 4 h and the aqueous peptide solution was transferred to a 12-well plate. KP24 was immobilized by immersing the APTES substrate in the 12-well plate at 4 °C for 24 h.
CMETS was modified on the bar glass. In order to hydrolyze the methoxy group of the CMETS substrate, it was immersed in 35% concentrated hydrochloric acid for 24 h. WSCD/HCl and NHS were dissolved in PBS and the aqueous solution was added to a 12-well plate. The CMETS (COOH) substrate was immersed at room temperature for 4 h to activate the carboxy group on the substrate surface. KP24 (1.0 mg, 0.37 µmol) was dissolved in PBS, the aqueous solution was added onto an activated CMETS (COOH) substrate, and the mixture was shaken at room temperature for 4 h to immobilize KP24.
2.5. Characterization of KP24-Immobilized Glass
The water contact angle of the KP24-immobilized substrate surface was measured. The amount of water dropped onto the cover glass surface was kept constant at 5.0 μL (n = 6) and used to calculate the average value. Surface elements analysis of the KP24-immobilized substrate was achieved by XPS. Other elements analyzed were carbon, oxygen, nitrogen, and silicon. Elements present in the range of 0 eV to 1100 eV on the substrate surface were also analyzed.
2.6. L929 Cell Aggregate Induced on KP24-Immobilized Glass
KP24-immobilized substrate was washed with 70 % ethanol and PBS and placed in a 24-well plate (Iwaki, AGC Techno Glass Co., Ltd. Shizuoka, Japan). Serum-containing EMEM medium in the well in which L929 was cultured was removed and washed three times with PBS. The supernatant was removed and resuspended with serum-free EMEM. The L929 cells in the cell suspension were seeded at 1.5 × 105 cells each on KP24-immobilized substrate and incubated at 5% CO2, 95%HR at 37 °C, for 7 days. During the incubation period, the volume of medium in each well was set at 1 mL and medium was changed at 500 µL/day.
Live/Dead assay was carried out for cell aggregate on KP24-immobilized substrate. L929 cell aggregates on KP24-immobilized substrate were detached by pipetting and seeded into triple wells (Iwaki, AGC Techno Glass Co., Ltd. Shizuoka, Japan). As it was anticipated that the cell aggregates would detach during staining, a small amount of EMEM medium containing FBS was added and incubated for 24 h under humidified conditions with 5% CO2, at 37 °C. For other substrates, they were cracked with tweezers at the time of staining and placed in a triple well. Cells on the substrates were washed once with PBS and the prepared staining solution was added; incubated under humidified conditions with 5% CO2, at 37 °C, for 45 min; and observed with a confocal laser microscope.
2.7. hMSCs Cell Aggregate on KP24-Immobilized glass and Differentiation to Osteoblast
As with L929 cells, hMSCs [
15,
16,
17,
18,
19] were seeded at 1.5 × 10
5 cells each on KP24-immobilized substrate and incubated under humidified conditions with 5% CO
2, at 37 °C, and humidified conditions for 7 days.
Osteoblast-induced differentiation medium was prepared by adding 1.0% ascorbic acid, 2.0% β-glycerophosphate, and 0.20% hydrocortisone to DMEM medium containing serum [
18,
19].
The KP24 immobilized substrate was seeded with 1.5 × 105 cells of hMSCs and cultured under humidified conditions with 5% CO2, at 37 °C, for 7 days. 1.0 mL of DMEM medium was removed from 24 well and 1.0 mL of osteoblast differentiation inducing medium was added. They were cultured under humidified conditions with 5% CO2, at 37 °C.
ALP activity was measured by Laboratory assay ALP (Fujifilm Wako Pure Chemical Industries, Ltd.). hMSCs on KP24-immobilized substrate cultured in osteoblast differentiation induction medium were washed once with saline. A total 1.0 mL of 1.0 % (v/v) Triton X-100/saline was added to each well of a 24-well plate and incubated for 1 h under 5% CO2, at 37 °C, with humidified conditions. The cell membrane solubilized solution from each well was transferred to a 2 mL tube and the supernatant was collected by centrifugation at 4 °C, 1500 rpm, for 15 min. A total 100 µL of the substrate buffer supplied with the lab assay ALP and 20 µL of the collected supernatant were added to a 96-well plate (Iwaki, AGC Techno Glass Co., Ltd. Shizuoka, Japan) The 96-well plate was incubated in humidified conditions with 5% CO2 at 37 °C for 15 min. The absorbance was measured with a microplate reader (Tecan Inc. Ltd. Männedorf, Switzerland) at an absorption wavelength of 405 nm with a reference wavelength of 620 nm.
2.8. Measuring the Size of Cell Aggregates and Sphericity
Three-dimensional analysis of cell aggregates was performed using Mil-Cell (Sumitomo Electric Industries, Ltd., Osaka, Japan) and 3D analysis imaging software [
11].
The size of the cell aggregate was measured under a microscope observation, and analysis was performed using OLYMPUS imaging software cellSens (Olympus Corp., Tokyo, Japan) [
11].
3. Results and Discussion
3.1. Peptide Synthesis
KP24 peptide was synthesized manually by solid-phase peptide synthesis. This peptide was characterized by HPLC, MALDI-TOF-MS, and amino-acid analysis. All peptides were purified by RP-HPLC to a single peak. Results of MALDI-TOF-MS spectra m/z of [M+H]+, [M+Na]+, and [M+K]+ were 2724.6, 2746.1, 2762.7, respectively. This result suggests that KP24 was synthesized, as intended in the molecular design.
3.2. Characterization of KP24-Immobilized Substrates
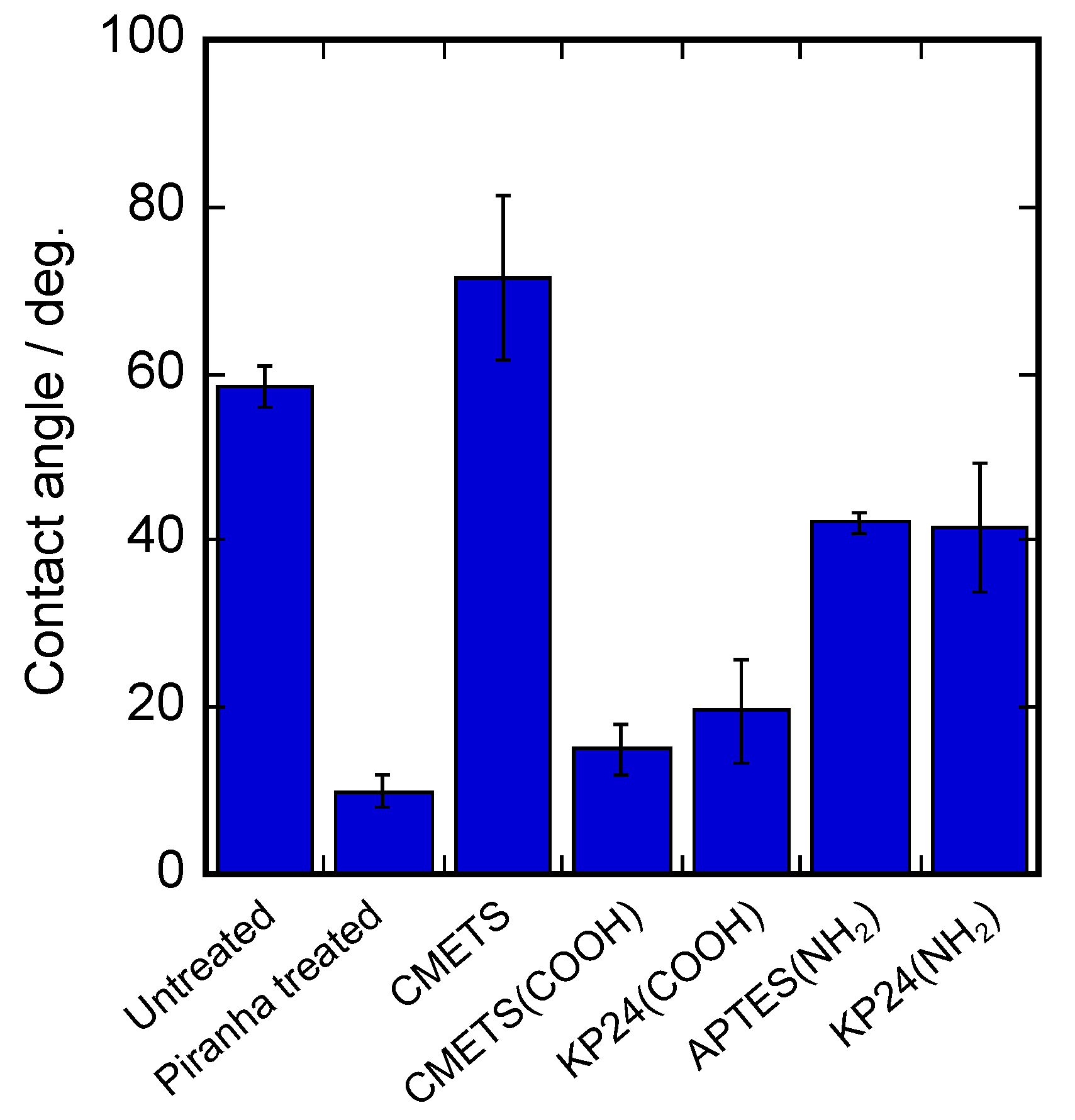
The results of water contact angle measurements of the prepared KP24-immobilized substrates are shown in
Figure 2. It shows that the water contact angle was about 60° for the untreated condition, but it decreased to about 10° after the piranha treatment. On the CMETS substrate surface, the contact angle increased to about 70° after piranha treatment. In the CMETS (COOH) substrate, the substrate surface changed to a carboxyl group by hydrolysis. In the case of KP24 (COOH), there was no significant change in the contact angle because the carboxy groups of KP24 were exposed. For the APTES substrate, the contact angle increased to about 50° after piranha treatment, suggesting that the modification of APTES changed the surface of the substrate to amino groups. The contact angle of the KP24 (NH
2) substrate was similar to that of the APTES substrate, suggesting that the amino groups of KP24 were exposed because KP24 was immobilized from the carboxyl groups on the APTES substrate. Surface elements were analyzed by XPS. The results of surface element analysis of the prepared KP24-immobilized substrates are shown in
Figure 3. These results show the nitrogen peaks on the glass substrate surface. In the case of KP24 (COOH), the nitrogen peak was detected at around 401 eV due to the presence of nitrogen in KP24. In the KP24 (NH
2) substrate, the nitrogen peak shifted to the higher energy side from the nitrogen peak of the APTES substrate and was detected at around 401 eV as in the KP24 (COOH) substrate. The surface element ratios show that the percentage of nitrogen on the surface of the KP24 (NH
2) substrate increased compared with that of the APTES substrate. Therefore, it was concluded that the KP24 (NH
2) substrate was immobilized.
3.3. Induction of L929 Cell Aggregates on KP24-Immobilized Substrate
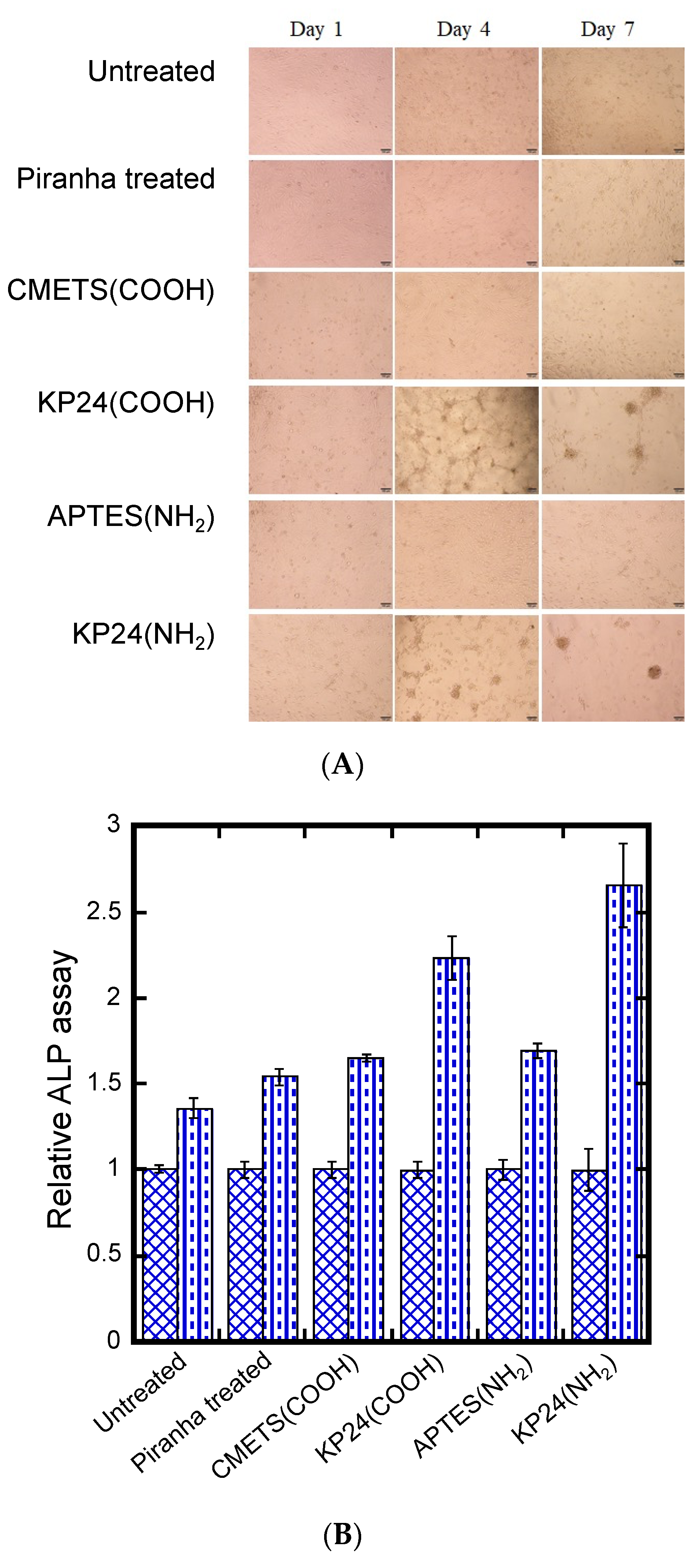
The results of phase contrast microscopy of L929 seeded on the prepared KP24-immobilized substrate are shown in
Figure 4A. On the first day after seeding, L929 was adherent and extended over all substrate’s surfaces, and no changes were observed between substrates. However, on day 4 after seeding, cell aggregates were induced on two KP24-immobilized substrates, KP24 (COOH) and KP24 (NH
2) substrate. Observation on day 7 after seeding showed that induction of cell aggregates was complete. The sequence of KP24 contains 12 residues of lysine, a basic amino acid with a positive charge. This positive charge was thought to induce the cell through electrostatic interaction with the negative charge of the cell membrane. However, the amino groups on the main and side chains of KP24 interacted with the carboxyl groups on the CMTES (COOH) substrate surface, weakening the positive charge and inhibiting the induction of cell assembly (
Figure 4C). It is speculated that the reason for the formation of cell aggregates on the KP24 (COOH) substrate is that some amino groups of KP24 do not interact with the carboxyl groups on the substrate (
Figure 4C).
The sequence of KP24 contains 12 residues of lysine, a basic amino acid with a positive charge. This positive charge was thought to induce the cell through electrostatic interaction with the negative charge of the cell membrane. However, the amino group of KP24 and substrate of carboxyl groups on the CMTES (COOH) interacted by electrostatic interaction before KP24 immobilization. The amino groups on the main and side chains of KP24 interacted with the carboxyl groups on the CMTES (COOH) substrate surface, weakening the positive charge and inhibiting the induction of cell assembly. It is speculated that the reason for the formation of cell aggregates on the KP24 (COOH) substrate is that some amino groups of KP24 do not interact with the carboxyl groups on the substrate.
Most of the cell aggregates induced on the KP24 (COOH) substrate were less than 100 µm in diameter. In contrast, many cell aggregates larger than 100 µm in diameter were induced on the KP24 (NH
2) substrate. The average diameter of L929 cell aggregates induced on KP24 (COOH) substrate was 86.6 µm. On the KP24 (NH
2) substrate, cell aggregates with an average diameter of 122.9µm were induced. Therefore, it is assumed that some of the amino groups of Lys that do not interact with the substrate surface interact with the cells surface. In the KP24 (NH
2) substrate, the amino groups of KP24 do not interact with the substrate surface because KP24 is immobilized on the substrate with exposed amino groups, see
Figure 4D. In this fact, it is likely that a large number of amino groups of KP24 interacted with the cells. These results suggest that the induced cell aggregates depend on the number of amino groups of KP24 that interact, see
Figure 4C,D.
Live/Dead staining of cell aggregates on KP24-immobilized substrates by calcein AM and ethidium homodimer I is shown in
Figure 4B. On the two types of KP24-immobilized substrates, the cells surrounding the cell assemblies remained elongated, but these cells were cells originated from the cell assemblies. Therefore, cell–cell interaction was expected, and it may have led to the retention of cell morphology. Live/Dead staining results showed a small number of dead cells in all substrates as well as inside the cell aggregates. The outer cells of the cell assembly are in direct contact with the culture medium and are easily supplied with factors such as nutrients and oxygen. However, the inner cells have difficulty receiving these factors. Therefore, a small number of dead cells were observed inside the cell aggregates. Since these results were also observed in cell aggregates formed by the hanging drop method, they are not likely due to the effect of KP24 [
20].
This fact suggests that the KP24 peptide is not taken up into the cell, but interacts with the cell surface to induce cell aggregates. It is considered that KP24 also interacts with the cell surface to induce cell aggregates when added to the cell culture medium.
3.4. Induction of hMSC Cell Aggregates on KP24-Immobilized Substrate
The results of hMSC seeding on KP24-immobilized substrates are shown
Figure 5A. These results show that hMSCs seeded on KP24-immobilized substrates adhered and expanded on all substrates on the first day of seeding. However, as with the case of L929, cell aggregation was not observed on KP24 (COOH) and KP24 (NH
2) substrates until around day 4 of seeding. Furthermore, we confirmed that cell aggregation was completed on the seventh day of seeding. The observation showed that there was a size difference in the cell aggregates induced on KP24(COOH) and KP24(NH
2) substrates in the same way as L929 cell aggregates.
We measured the size of the induced hMSC cell assemblies. The average diameter of hMSC cell aggregates induced on KP24 (COOH) substrate was 127.6 µm. On the KP24(NH
2) substrate, it was 169.2 µm. The size difference of the cell aggregates between the two substrates may depend on the number of amino groups in KP24 that are involved in aggregation, which is similar to what happened to L929 cell aggregates. To characterize hMSC cell aggregates induced on KP24-immobilized substrate, we induced differentiation into osteoblasts and measured ALP activity. The results are shown in
Figure 5B.
These results suggest that ALP activity of hMSCs on all substrates increased in proportion to the duration of culture in differentiation medium. The percentage of increase in activity value was higher on KP24 (COOH) and KP24 (NH2) substrates. This may be due to the induction of hMSC cell aggregates on the KP24-immobilized substrate. Unlike cells in a monolayer, cell aggregates interact with each other in three dimensions, which led to high values of ALP activity. In addition, we also stained ALP expressed in hMSC cell aggregates. ALP staining was performed on day 1 and day 7 of differentiation. No difference in staining was observed between monolayer cells and cell aggregates on day 1. However, on day 7, we observed more staining in the cell aggregates induced on the KP24-immobilized substrate. This is similar to the ALP activity assay and may be due to enhanced ALP expression in the cell aggregates due to three-dimensional cell–cell interactions.
3.5. Measuring the Size of Cell Aggregates and Sphericity
The microphotograph of Mil-Cell analysis indicated that the cell aggregates of L929 and hMSC certainly had a three-dimensional structure (
Figure 6A). The average of sphericity of the L929 cell aggregate induced by KP24 was about 70–80% on KP24 (COOH) and KP24(NH
2) substrate as calculated by the Mil-Cell 3D imaging analysis software (
Figure 6B). However, hMSC cell aggregates were more distorted and less spherical than L929 cell aggregate. The cell aggregates induced by KP24 are also expected to have a different shape depending on the cell type.
4. Conclusions
Based on the water contact angle and XPS results, APTES and CMETS modified surfaces were constructed. In addition, KP24-modified surfaces could be created. In experiments with L929 seeded on the prepared substrates, the formation of cell aggregates was observed on the KP24 (COOH) and KP24 (NH2) substrates.
Cell aggregates on the two types of KP24-immobilized substrates were found to be smaller on the KP24 (COOH) substrate than on the KP24 (NH2) substrate. However, all aggregates on the KP24 (NH2) substrate were larger than 100 μm. When hMSC cells were seeded onto the KP24-immobilized substrate, cell aggregation was observed around day 4, as in the case of L929, and aggregation was observed to be complete by day 7. The induced hMSC cell aggregates were differentiated using osteoblast differentiation induction medium, and ALP activity and ALP staining results were evaluated. The results showed that the substrate on which hMSC cell aggregates were induced showed high ALP activity values. The sizes of the induced hMSC cell aggregates were similar to that of the L929 cell aggregates, but was different for the two types of KP24-immobilized substrates.
The amino groups on the main and side chains of KP24 interacted with the carboxyl groups on the CMTES (COOH) substrate surface, weakening the positive charge and inhibiting the induction of cell assembly. The Mil-Cell analysis indicated that the cell aggregates of L929 and hMSC certainly had a three-dimensional structure. The average of sphericity of the L929 cell aggregate induced by KP24 was about 70–80% on the KP24 (COOH) and KP24(NH2) substrate as calculated by the Mil-Cell 3D imaging analysis software. hMSC cell aggregates were more distorted and less spherical than L929 cell aggregate. The cell aggregates induced by KP24 are also expected to have different shapes depending on the cell type.
These results are expected to be applied as a new culture substrate that can induce cell aggregates from 2D culture. We propose a concept for a new cell culture technology [
21,
22,
23,
24].