Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of an Adnectin-Derived Phage Display Library
2.3. Biopaning of Adnectins against PPM1D430 Using Adnectin-Derived Phage Display Library
2.4. Expression and Purification of Recombinant PPM1D-Specific Adnectins
2.5. Binding Analysis of Adnectin-Derived Phages
2.6. Binding Analysis of Recombinant Adnectins to Ser/The Protein Phosphatases
2.7. Biolayer Interferometry BLItz System Assay
2.8. Binding Analysis of Peptide-Conjugated Bacterial Alkaline Phosphatase
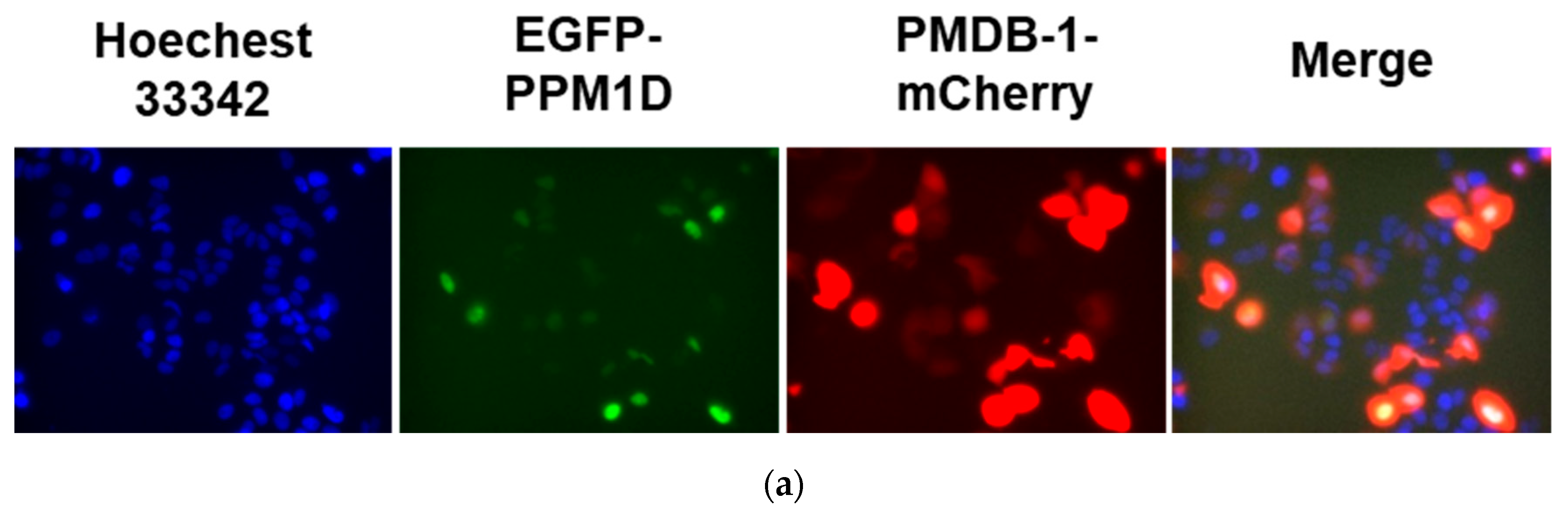
2.9. Subcellular Localization Analysis in Living Cells
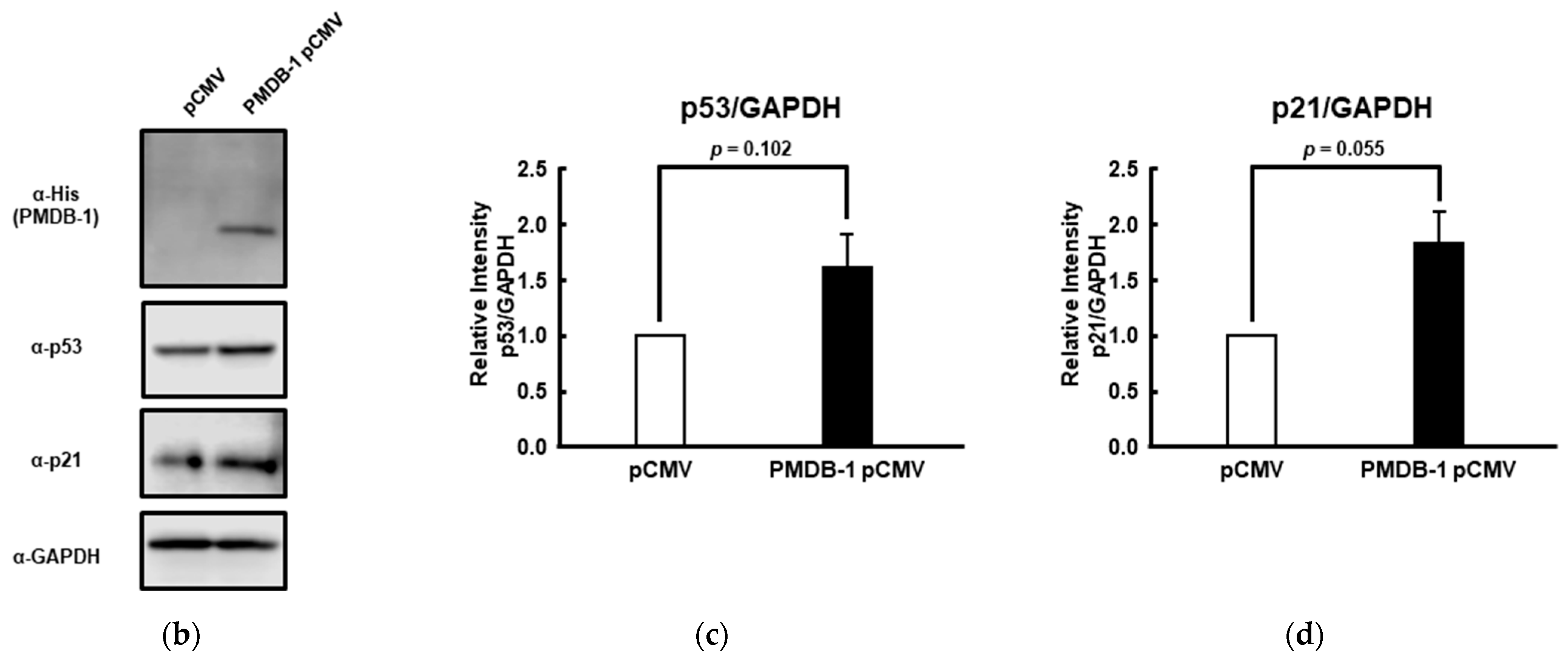
2.10. Antibodies and Western Blotting
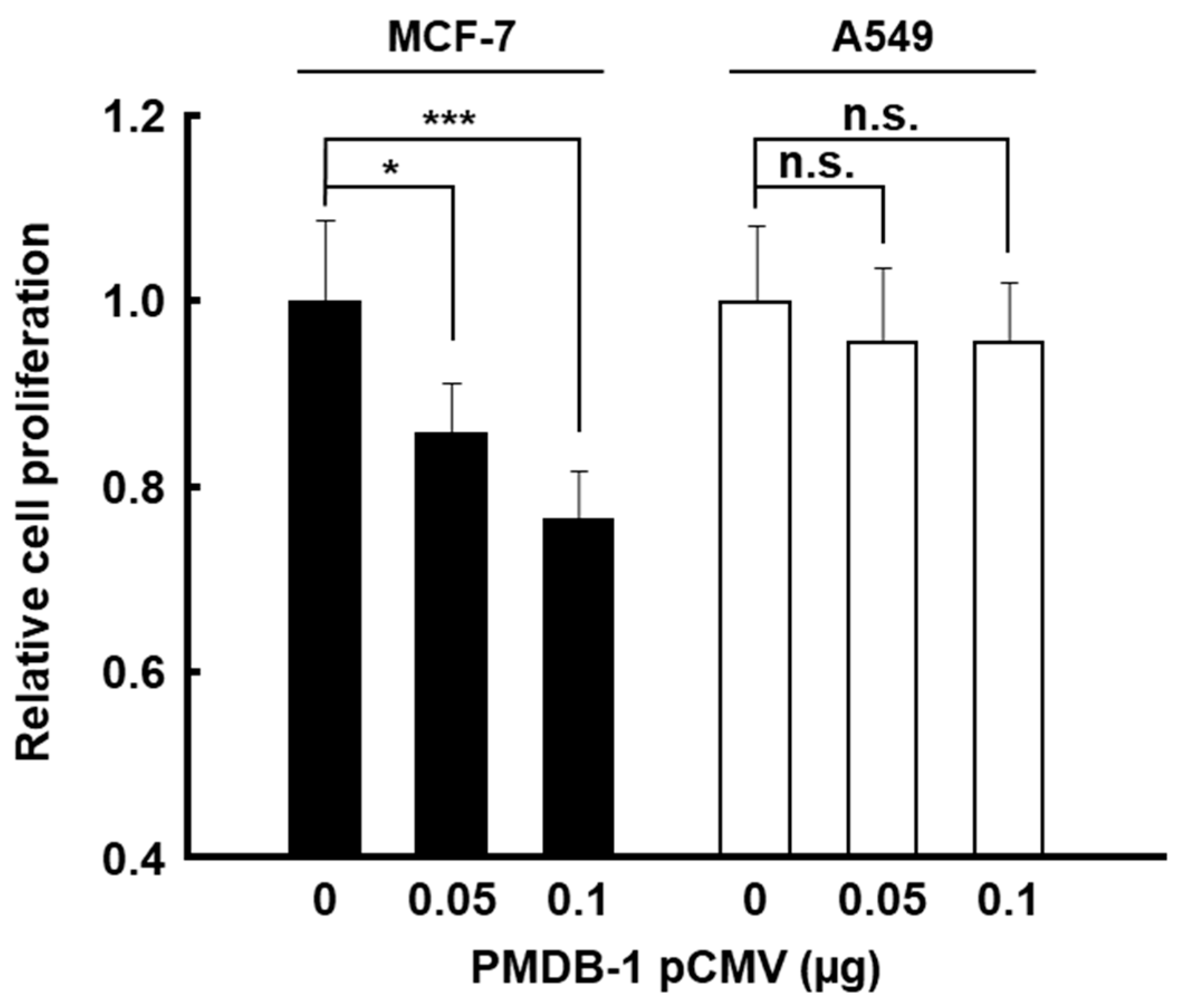
2.11. Cell Proliferation Assay
3. Results
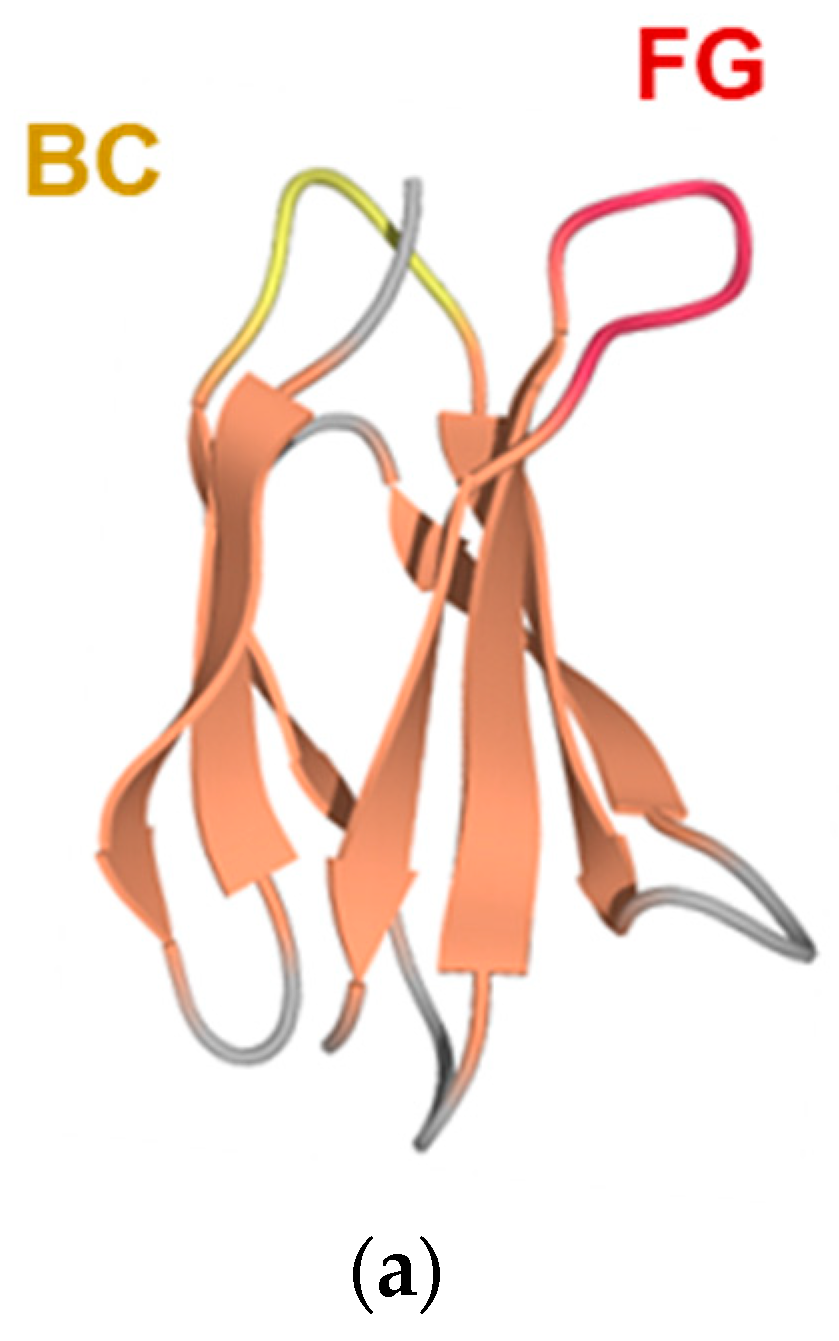
3.1. Construction of an Adnectin-Derived Phage Display Library with Randomized BC and FG Loops
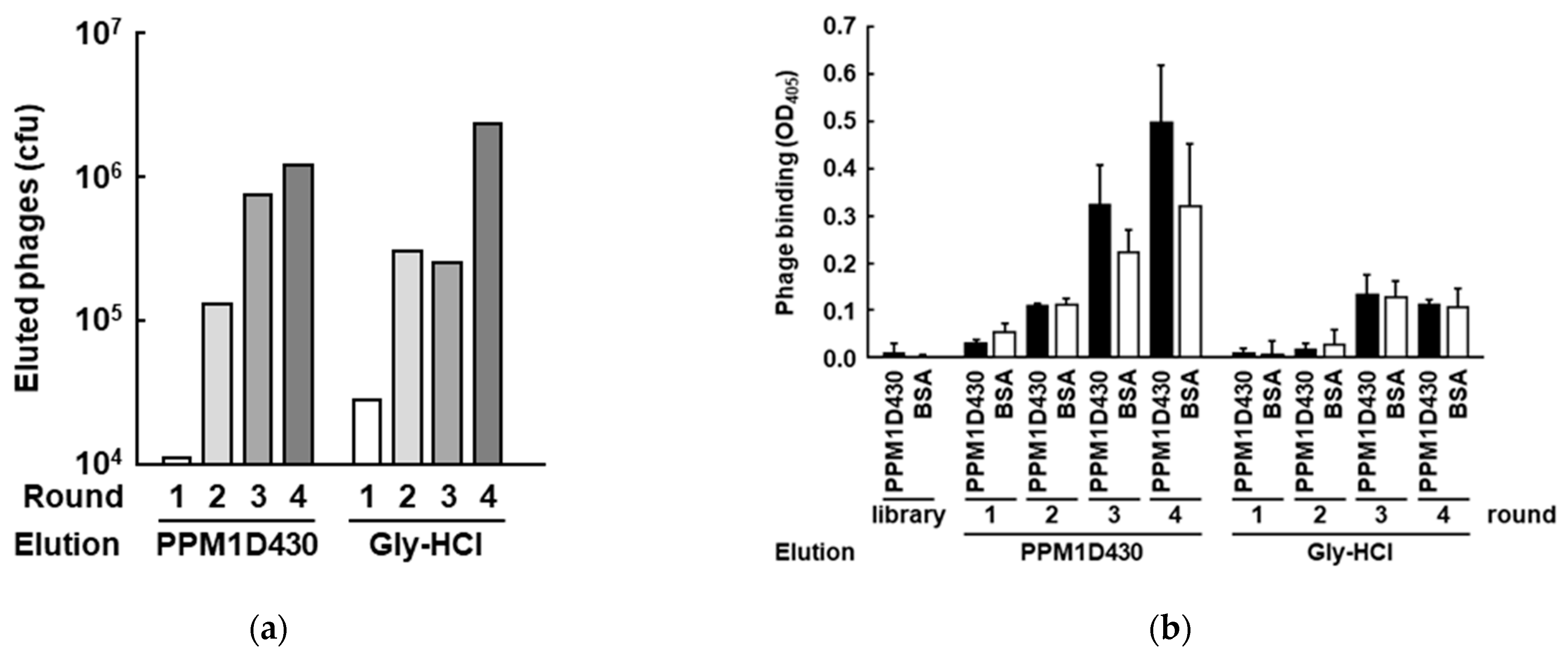
3.2. Screening of PPM1D-Specific Adnectins from the Constructed Phage Display Library
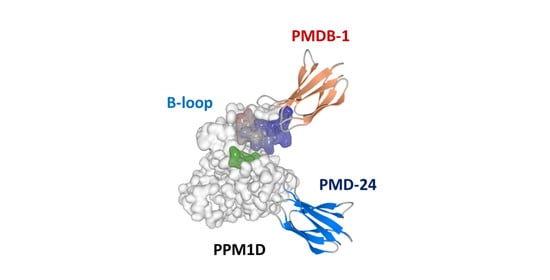
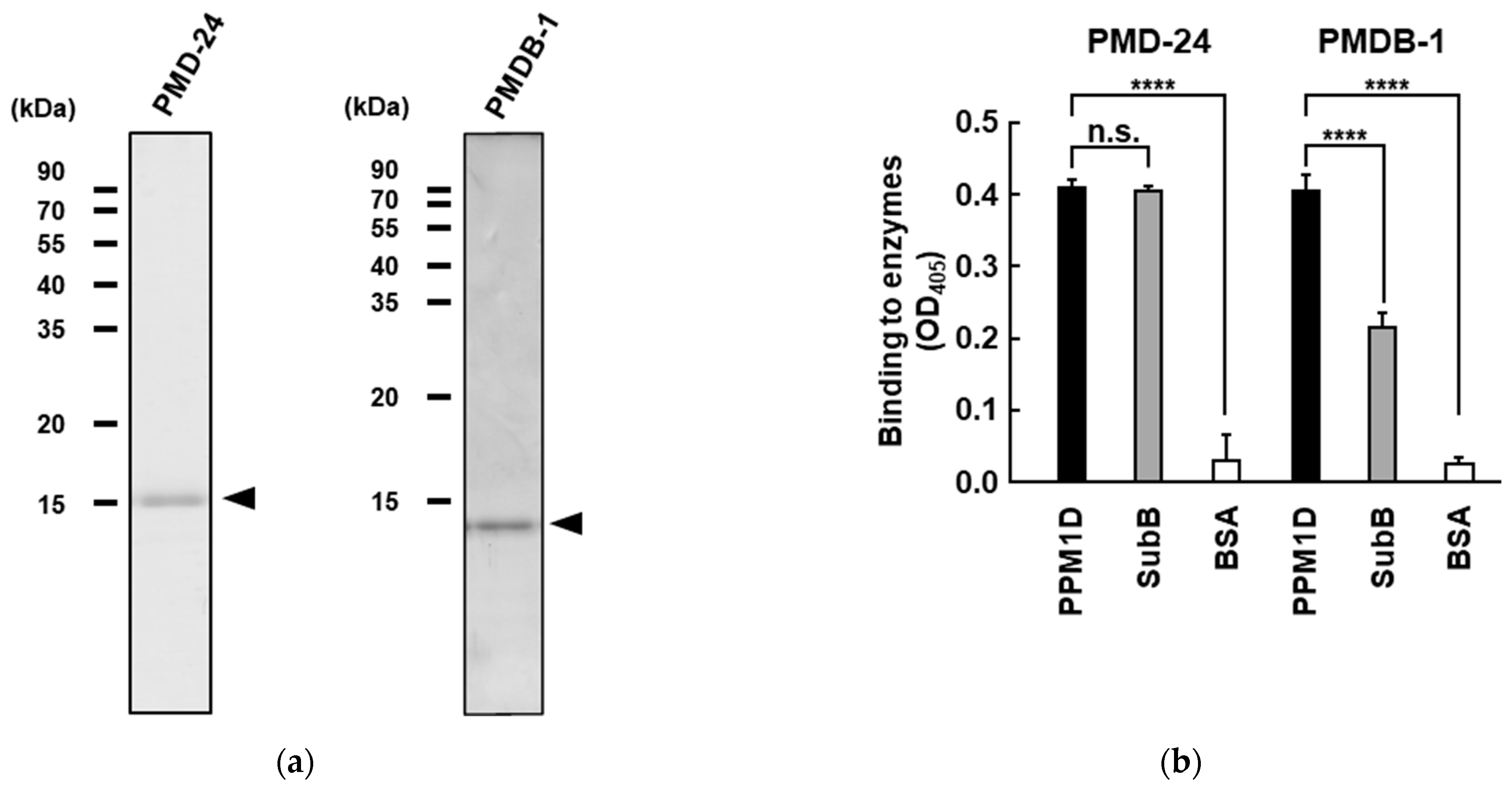
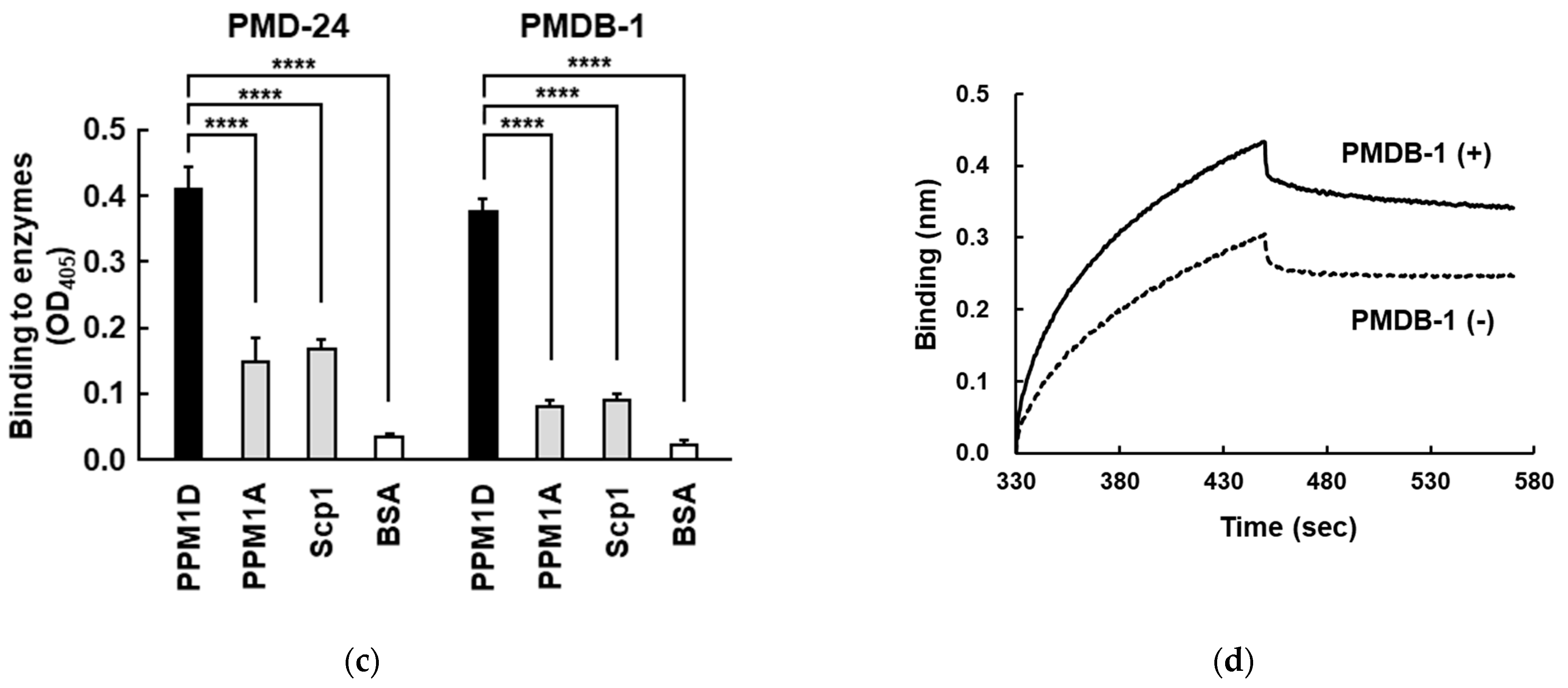
3.3. Analysis of Recombinant PPM1D-Specific Adnectin Binding
3.4. Inhibitory Activity of PMDB-1 Adnectin in Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Barford, D.; Das, A.K.; Egloff, M.P. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys Biomol. Struct. 1998, 27, 133–164. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, G.B.; Trinkle-Mulcahy, L.; Ulke-Lemée, A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007, 8, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Shreeram, S.; Demidov, O.N.; Hee, W.K.; Yamaguchi, H.; Onishi, N.; Kek, C.; Timofeev, O.N.; Dudgeon, C.; Fornace, A.J.; Anderson, C.W.; et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol. Cell 2006, 23, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Horejsí, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehesdted, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Takekawa, M.; Adachi, M.; Nakahata, A.; Nakayama, I.; Itoh, F.; Tsukuda, H.; Taya, Y.; Imai, K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000, 19, 6517–6526. [Google Scholar] [CrossRef]
- Lu, X.; Nannenga, B.; Donehower, L.A. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005, 19, 1162–1174. [Google Scholar] [CrossRef]
- Fujimoto, H.; Onishi, N.; Kato, N.; Takekawa, M.; Xu, X.Z.; Kosugi, A.; Kondo, T.; Imamura, M.; Oishi, I.; Yoda, A.; et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006, 13, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Hjermind Justesen, E.; Grønbæk, K. Protein phosphatase, Mg2+/Mn2+-dependent 1D (PPM1D) mutations in haematological cancer. Br. J. Haematol. 2021, 192, 697–705. [Google Scholar] [CrossRef]
- Fiscella, M.; Zhang, H.; Fan, S.; Sakaguchi, K.; Shen, S.; Mercer, W.E.; Vande Woude, G.F.; O’Connor, P.M.; Appella, E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc. Natl. Acad. Sci. USA 1997, 94, 6048–6053. [Google Scholar] [CrossRef]
- Lu, X.; Nguyen, T.A.; Moon, S.H.; Darlington, Y.; Sommer, M.; Donehower, L.A. The type 2C phosphatase Wip1: An oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008, 27, 123–135. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Chuman, Y.; Yagi, H.; Fukuda, T.; Nomura, T.; Matsukizono, M.; Shimohigashi, Y.; Sakaguchi, K. Characterization of the active site and a unique uncompetitive inhibitor of the PPM1-type protein phosphatase PPM1D. Protein Pept. Lett. 2008, 15, 938–948. [Google Scholar] [CrossRef]
- Gudipaty, S.A.; McNamara, R.P.; Morton, E.L.; D’Orso, I. PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation. Mol. Cell Biol. 2015, 35, 3810–3828. [Google Scholar] [CrossRef]
- Kamada, R.; Kudoh, F.; Ito, S.; Tani, I.; Janairo, J.I.B.; Omichinski, J.G.; Sakaguchi, K. Metal-dependent Ser/Thr protein phosphatase PPM family: Evolution, structures, diseases and inhibitors. Pharmacol. Ther. 2020, 215, 107622. [Google Scholar] [CrossRef] [PubMed]
- Chuman, Y.; Kurihashi, W.; Mizukami, Y.; Nashimoto, T.; Yagi, H.; Sakaguchi, K. PPM1D430, a novel alternative splicing variant of the human PPM1D, can dephosphorylate p53 and exhibits specific tissue expression. J. Biochem. 2009, 145, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kleiblova, P.; Shaltiel, I.A.; Benada, J.; Ševčík, J.; Pecháčková, S.; Pohlreich, P.; Voest, E.E.; Dundr, P.; Bartek, J.; Kleibl, Z.; et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J. Cell Biol. 2013, 201, 511–521. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.H.; Wan, H.; Yang, R.; Wang, Z.; Feng, J.; Yang, S.; Jones, S.; Wang, S.; Zhou, W.; et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat. Genet. 2014, 46, 726–730. [Google Scholar] [CrossRef]
- Ruark, E.; Snape, K.; Humburg, P.; Loveday, C.; Bajrami, I.; Brough, R.; Rodrigues, D.N.; Renwick, A.; Seal, S.; Ramsay, E.; et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013, 493, 406–410. [Google Scholar] [CrossRef]
- Catherine, C.C.; Zehir, A.; Devlin, S.M.; Kishtagari, A.; Syed, A.; Jonsson, P.; Hyman, D.M.; Solit, D.B.; Robson, M.E.; Baselga, J.; et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017, 21, 374–382. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, J.R.; Kwon, O.S.; Kim, H.; Fornace, A.J., Jr.; Cha, H.J. Off-target response of a Wip1 chemical inhibitor in skin keratinocytes. J. Dermatol. Sci. 2014, 73, 125–134. [Google Scholar] [CrossRef]
- Yagi, H.; Chuman, Y.; Kozakai, Y.; Imagawa, T.; Takahashi, Y.; Yoshimura, F.; Tanino, K.; Sakaguchi, K. A small molecule inhibitor of p53-inducible protein phosphatase PPM1D. Bioorg. Med. Chem. Lett. 2012, 22, 729–732. [Google Scholar] [CrossRef]
- Ogasawara, S.; Kiyota, Y.; Chuman, Y.; Kowata, A.; Yoshimura, F.; Tanino, K.; Kamada, R.; Sakaguchi, K. Novel inhibitors targeting PPM1D phosphatase potently suppress cancer cell proliferation. Bioorg. Med. Chem. 2015, 23, 6246–6249. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Durell, S.R.; Feng, H.; Bai, Y.; Anderson, C.W.; Appella, E. Development of a substrate-based cyclic phosphopeptide inhibitor of protein phosphatase 2Cdelta, Wip1. Biochemistry 2006, 45, 13193–13202. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R.; Tanoue, K.; Durell, S.R.; Chatterjee, D.K.; Jenkins, L.M.; Appella, D.H.; Appella, E. Optimization of a cyclic peptide inhibitor of Ser/Thr phosphatase PPM1D (Wip1). Biochemistry 2011, 50, 4537–4549. [Google Scholar] [CrossRef] [PubMed]
- Gilmartin, A.G.; Faitg, T.H.; Richter, M.; Groy, A.; Seefeld, M.A.; Darcy, M.G.; Peng, X.; Minthorn, E.; Yang, J.; Zhang, S.Y.; et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat. Chem. Biol. 2014, 10, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Yokoo, T.; Tanabe, A.; Yoshida, Y.; Caaveiro, J.M.M.; Nakakido, M.; Ikeda, Y.; Fujimura, Y.; Matsumoto, M.; Entzminger, K.; Maruyama, T.; et al. Antibody recognition of complement factor H reveals a flexible loop involved in atypical hemolytic uremic syndrome pathogenesis. J. Biol. Chem. 2022, 298, 101962. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Eggenstein, E.; Skerra, A. Anticalins: Exploiting a non-Ig scaffold with hypervariable loops for the engineering of binding proteins. FEBS Lett. 2014, 588, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Helma, J.; Cardoso, M.C.; Muyldermans, S.; Leonhardt, H. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 2015, 209, 633–644. [Google Scholar] [CrossRef]
- Yu, X.; Yang, Y.P.; Dikici, E.; Deo, S.K.; Daunert, S. Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem. 2017, 10, 293–320. [Google Scholar] [CrossRef]
- Gebauer, M.; Skerra, A. Engineered Protein Scaffolds as Next-Generation Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 391–415. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, L.; Jin, D.; Liu, Y. Nanobody-A versatile tool for cancer diagnosis and therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1697. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, S.; Gräslund, T.; Eriksson Karlström, A.; Frejd, F.Y.; Nygren, P.Å.; Löfblom, J. Affibody Molecules in Biotechnological and Medical Applications. Trends Biotechnol. 2017, 35, 691–712. [Google Scholar] [CrossRef] [PubMed]
- Plückthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 489–511. [Google Scholar] [CrossRef]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef]
- Park, S.H.; Park, S.; Kim, D.Y.; Pyo, A.; Kimura, R.H.; Sathirachinda, A.; Choy, H.E.; Min, J.J.; Gambhir, S.S.; Hong, Y. Isolation and Characterization of a Monobody with a Fibronectin Domain III Scaffold That Specifically Binds EphA2. PLoS ONE 2015, 10, e0132976. [Google Scholar] [CrossRef]
- Bloom, L.; Calabro, V. FN3: A new protein scaffold reaches the clinic. Drug Discov. Today 2009, 14, 949–955. [Google Scholar] [CrossRef]
- Wojcik, J.; Lamontanara, A.J.; Grabe, G.; Koide, A.; Akin, L.; Gerig, B.; Hantschel, O.; Koide, S. Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface. J. Biol. Chem. 2016, 291, 8836–8847. [Google Scholar] [CrossRef]
- Mitchell, T.; Chao, G.; Sitkoff, D.; Lo, F.; Monshizadegan, H.; Meyers, D.; Low, S.; Russo, K.; DiBella, R.; Denhez, F.; et al. Pharmacologic Profile of the Adnectin BMS-962476, a Small Protein Biologic Alternative to PCSK9 Antibodies for Low-Density Lipoprotein Lowering. J. Pharmacol. Exp. Ther. 2014, 350, 412–424. [Google Scholar] [CrossRef]
- Kondo, T.; Iwatani, Y.; Matsuoka, K.; Fujino, T.; Umemoto, S.; Yokomaku, Y.; Ishizaki, K.; Kito, S.; Sezaki, T.; Hayashi, G.; et al. Antibody-like proteins that capture and neutralize SARS-CoV-2. Sci. Adv. 2020, 6, eabd3916. [Google Scholar] [CrossRef]
- Sha, F.; Salzman, G.; Gupta, A.; Koide, S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017, 26, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Wojcik, J.; Gilbreth, R.N.; Hoey, R.J.; Koide, S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 2012, 415, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Verdine, G.L.; Walensky, L.D. The challenge of drugging undruggable targets in cancer: Lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 2007, 13, 7264–7270. [Google Scholar] [CrossRef] [PubMed]
- Mamluk, R.; Carvajal, I.M.; Morse, B.A.; Wong, H.; Abramowitz, J.; Aslanian, S.; Lim, A.C.; Gokemeijer, J.; Storek, M.J.; Lee, J.; et al. Anti-tumor effect of CT-322 as an adnectin inhibitor of vascular endothelial growth factor receptor-2. MAbs 2010, 2, 199–208. [Google Scholar] [CrossRef]
- Schiff, D.; Kesari, S.; de Groot, J.; Mikkelsen, T.; Drappatz, J.; Coyle, T.; Fichtel, L.; Silver, B.; Walters, I.; Reardon, D. Phase 2 study of CT-322, a targeted biologic inhibitor of VEGFR-2 based on a domain of human fibronectin, in recurrent glioblastoma. Investig. New Drugs 2015, 33, 247–253. [Google Scholar] [CrossRef]
- Yamabhai, M.; Kay, B.K. Examining the specificity of Src homology 3 domain-ligand interactions with alkaline phosphatase fusion proteins. Anal. Biochem. 1997, 247, 143–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaneko, A.; Watari, M.; Mizunuma, M.; Saito, H.; Furukawa, K.; Chuman, Y. Development of Specific Inhibitors for Oncogenic Phosphatase PPM1D by Using Ion-Responsive DNA Aptamer Library. Catalysts 2020, 10, 1153. [Google Scholar] [CrossRef]
- Rayter, S.; Elliott, R.; Travers, J.; Rowlands, M.G.; Richardson, T.B.; Boxall, K.; Jones, K.; Linardopoulos, S.; Workman, P.; Aherne, W.; et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene 2008, 27, 1036–1044. [Google Scholar] [CrossRef]
- Deng, W.; Li, J.; Dorrah, K.; Jimenez-Tapia, D.; Arriaga, B.; Hao, Q.; Cao, W.; Gao, Z.; Vadgama, J.; Wu, Y. The role of PPM1D in cancer and advances in studies of its inhibitors. Biomed. Pharmacother. 2020, 125, 109956. [Google Scholar] [CrossRef]
- Batori, V.; Koide, A.; Koide, S. Exploring the potential of the monobody scaffold: Effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002, 15, 1015–1020. [Google Scholar] [CrossRef]
- Chuman, Y.; Uren, A.; Cahill, J.; Regan, C.; Wolf, V.; Kay, B.K.; Rubin, J.S. Identification of a peptide binding motif for secreted frizzled-related protein-1. Peptides 2004, 25, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Helps, N.R.; Cohen, P.T.; Barford, D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996, 15, 6798–6809. [Google Scholar] [CrossRef] [PubMed]
- Almo, S.C.; Bonanno, J.B.; Sauder, J.M.; Emtage, S.; Dilorenzo, T.P.; Malashkevich, V.; Wasserman, S.R.; Swaminathan, S.; Eswaramoorthy, S.; Agarwal, R.; et al. Structural genomics of protein phosphatases. J. Struct. Funct. Genom. 2007, 8, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Waschbüsch, D.; Berndsen, K.; Lis, P.; Knebel, A.; Lam, Y.P.; Alessi, D.R.; Khan, A.R. Structural basis for the specificity of PPM1H phosphatase for Rab GTPases. EMBO Rep. 2021, 22, e52675. [Google Scholar] [CrossRef] [PubMed]
- Nagarathinam, K.; Nakada-Nakura, Y.; Parthier, C.; Terada, T.; Juge, N.; Jaenecke, F.; Liu, K.; Hotta, Y.; Miyaji, T.; Omote, H.; et al. Outward open conformation of a Major Facilitator Superfamily multidrug/H+ antiporter provides insights into switching mechanism. Nat. Commun. 2018, 9, 4005. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Ongusaha, P.; Lee, S.W.; Liou, Y.C. Loss of Wip1 Sensitizes Cells to Stress- and DNA Damage-induced Apoptosis. J. Biol. Chem. 2009, 284, 17428–17437. [Google Scholar] [CrossRef]


| Targets | Name | Sequence | Frequency | |
|---|---|---|---|---|
| BC | FG | |||
| PPM1D430 | PMD-24 | AREQAIY | GQGWKML | 12 |
| PMD-42 | SWTHQAT | ELHSWGS | 1 | |
| PMD-3 | MLLDNPK | RGNGAQP | 1 | |
| Total | 14 | |||
| B-loop | PMDB-1 | NMGWSQG | KGLDFLC | 9(43 *) |
| PMDB-2 | NESLRTL | NQRSIYF | 1 | |
| Other | (28 *) | |||
| Total | 10(71 *) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeura, M.; Tashiro, H.; Yamagata, Y.; Saito, H.; Kobayashi, T.; Mizunuma, M.; Yamazaki, K.; Baba, K.; Furukawa, K.; Chuman, Y. Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D. Processes 2022, 10, 1501. https://doi.org/10.3390/pr10081501
Ikeura M, Tashiro H, Yamagata Y, Saito H, Kobayashi T, Mizunuma M, Yamazaki K, Baba K, Furukawa K, Chuman Y. Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D. Processes. 2022; 10(8):1501. https://doi.org/10.3390/pr10081501
Chicago/Turabian StyleIkeura, Megumi, Hiroto Tashiro, Yuka Yamagata, Hikaru Saito, Tamaki Kobayashi, Masataka Mizunuma, Kazuki Yamazaki, Keisuke Baba, Kazuhiro Furukawa, and Yoshiro Chuman. 2022. "Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D" Processes 10, no. 8: 1501. https://doi.org/10.3390/pr10081501
APA StyleIkeura, M., Tashiro, H., Yamagata, Y., Saito, H., Kobayashi, T., Mizunuma, M., Yamazaki, K., Baba, K., Furukawa, K., & Chuman, Y. (2022). Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D. Processes, 10(8), 1501. https://doi.org/10.3390/pr10081501