Abstract
Proteases that can remain active under extreme conditions such as high temperature, pH, and salt concentration are widely applicable in the commercial sector. The majority of the proteases are rendered useless under harsh conditions in industries. Therefore, there is a need to search for new proteases that can tolerate and function in harsh conditions, thus improving their commercial value. In this study, 142 bacterial isolates were isolated from diverse alkaline soil habitats. The two highest protease-producing bacterial isolates were identified as Bacillus subtilis S1 and Bacillus amyloliquefaciens KSM12, respectively, based on 16S rRNA sequencing. Optimal protease production was detected at pH 8, 37 °C, 48 h, 5% (w/v) NaCl for Bacillus subtilis S1 (99.8 U/mL) and pH 9, 37 °C, 72 h, 10% (w/v) NaCl for Bacillus amyloliquefaciens KSM12 (94.6 U/mL). The molecular weight of these partially purified proteases was then assessed on SDS-PAGE (17 kDa for Bacillus subtilis S1 and 65 kDa for Bacillus amyloliquefaciens KSM12), respectively. The maximum protease activity for Bacillus subtilis S1 was detected at pH 8, 40 °C, and for Bacillus amyloliquefaciens KSM12 at pH 9, 60 °C. These results suggest that the proteases secreted by Bacillus subtilis S1 and Bacillus amyloliquefaciens KSM12 are suitable for industries working in a highly alkaline environment.
1. Introduction
Proteases hold great commercial value and are involved in the breakdown of large proteins into smaller peptide fragments and amino acids [1]. Microbial proteases are an important family of proteases and have an advantage over animal and plant proteases, as microbes can be cultured on a large scale in less time, and the growth conditions can easily be optimized in lab conditions. Therefore, high yields with low cost can be achieved consistently. These microbes can also withstand extreme environmental conditions, thus making them a suitable tool for producing enzymes with high stability and activity under extreme conditions [2].
Many proteases, especially the worldwide sales of alkaline proteases, are growing tremendously. Nearly 60% of the global protease market consists of alkaline proteases. Alkaline proteinases show optimal enzyme activity at pH 7–11 and are comprised of metalloproteases and serine proteases [3]. Most proteases available in the market today are secreted by the genus Bacillus. Other bacterial strains, such as Escherichia coli, Lactococci, Streptomyces, Pseudomonas, and Vibrio spp. can produce proteases, but Bacillus is preferred due to its heterotrophic nature, high yield, and ease in growth on a variety of substrates. Alkaline proteases produced by Bacillus spp. possess great biochemical diversity and stability, therefore increasing its commercial value. These proteases are also used for various applications, including food, medicine, detergents, silver recovery, waste treatment, etc. Few of the important Bacillus spp. used for protease production, are B. subtilis and B. licheniformis [4]. A recent study isolated alkaline protease-producing bacterial strains of Bacillus cereus species from leather industrial effluent [5]. The optimization of microbial growth media has a significant role in improving product yield. It can affect cultural growth, which can, in turn, affect the target protein production.
The remarkable industrial and commercial value of high tolerance proteases leads to the continuous search for new microbial sources with protease production. Furthermore, the physicochemical characterization of an enzyme indicates its performance and potential applications in different industries. With this in view, we isolated an alkali-thermotolerant, B. subtilis S1 and B. amyloliquefaciens KSM12, from dump soil (Islamabad) and salt mines (Karak), respectively. Furthermore, we optimized the protease production and activity under different physicochemical factors like pH, temperature, and salinity for possible industrial applications.
2. Material and Methods
2.1. Sampling and Strain Isolation
Soil samples were collected from Bahadur Khel Salt Mines, Karak (33.1057° N, 70.5715° E), and sector I-12 dump area, Islamabad (33.6279° N, 72.9991° E), at a depth of 5–10 cm. Various physiological parameters such as temperature and pH were recorded, and samples were processed on the same day. The samples were serially diluted up to 10−10, and 200 µL solution from each dilution was dispensed onto nutrient agar plates. The plates were incubated at 37 °C for 48 h, and bacterial isolates were purified using a repeated subculturing technique [6].
2.2. Screening for Protease Production
Preliminary screening for protease production by the bacterial isolates was performed on casein agar plates under sterile conditions. The purified isolates were then streaked on skim milk agar plates and incubated at 37 °C for 24–48 h; finally, zones of hydrolysis were measured. The isolates producing larger zones were subsequently subjected to quantitative assay.
2.3. Quantitative Assay for Protease Activity
Overnight culture of selected protease-producing bacterial isolates were transferred to a 500 mL nutrient broth flask, supplemented with casein (1% w/v), at pH 7. The culture was incubated for 24–48 h at 37 °C with continuous shaking at 120 rpm. Afterward, the bacterial culture was centrifuged for 15 min at 10,000 rpm, and 4 °C. The cell-free supernatant was collected, taken as crude proteinase extract, and examined for proteinase activity [7].
2.4. Protease Specific Activity Assay
Protease-specific activity assay was performed according to the method of Cupp-Enyard and Aldrich [8]. Briefly, 5 mL of 0.65% casein dissolved in 50 mM potassium phosphate buffer (pH 7.5) was added to both sample and blank tubes and incubated at 37 °C for 5 min. Subsequently, 1 mL of crude protease extract was added to only the sample tube and incubated for 10 min at 37 °C. The reaction was stopped with the addition of 5 mL of 110 mM trichloroacetic acid (TCA) solution in both the sample and blank tubes. After the incubation was completed, protease extract was also added to the blank tube, and the mixture was filtered using a 0.45 µm syringe filter. Finally, 5 mL of 500 mM sodium carbonate was added to both the tubes, followed by the addition of 1 mL of 0.5 mM Folin–Ciolcaltea Reagent. The mixture was then incubated for 30 min at 37 °C. UV-Vis spectrophotometer (UV-6100A) was used to determine the absorbance value at 660 nm. The standard curve was generated, and enzyme activity in units/mL was calculated. The unit of enzyme activity is defined as “the amount of enzyme that catalyzes the conversion of one micromole substrate per minute under the specified conditions of the assay method” [9].
Total protein estimation: The crude extract was measured for the total protein content using Bradford’s method [10]. For this purpose, standard solutions were prepared using bovine serum albumin (BSA), ranging from 0–1 mg/mL.
2.5. Identification of Highest Protease Producers
The highest protease-producing bacteria (n = 2) were identified based on 16S rRNA gene sequence analysis. The 16S rRNA sequencing reaction was performed by Macrogen Inc. Seoul, South Korea (https://dna.macrogen.com/ accessed on 12 December 2021), using universal primers: 785F (GGATTAGATACCCTGGTA) and 907R (CCGTCAATTCMTTTRAGTTT). The sequenced gene was analyzed via BioEdit software v. 7.2 [11], and chimera sequences were removed using DECIPHER software [12]. A gene homology search was performed on the BLASTn search engine [13], and sequence data of closely related strains were retrieved from the NCBI database. The alignment was performed by the Clustal X program [14] and the phylogenetic tree was constructed by the neighbor-joining method, based on the Kimura 2-parameter model [15] using the MEGA 7 software [16]. All positions containing gaps and missing data were eliminated, and there were a total of 1371 positions in the final dataset. To validate the reproducibility of the branching pattern, a bootstrap value of 1000 was taken and shown on each branch.
2.6. Optimization of Protease Production
Optimum physicochemical conditions required for maximum bacterial growth and protease secretion by both strains were determined by subjecting them to different conditions, such as pH, incubation time, incubation temperature, and salt (NaCl) concentration, in the presence of casein modified nutrient broth (casein (1% w/v) at pH 7). The pH ranged from 5 to 11, with an interval of 1 unit. The temperature varied from 22 to 47 °C with an interval of 5 °C. The optimal incubation time was determined by inoculating bacterial cultures for 24–96 h with 24 h intervals. Salt (NaCl) concentration was optimized by inoculating the bacteria in casein-modified nutrient broth with 5–40% NaCl. All other conditions were kept constant. After incubation, OD600 was measured for bacterial growth, and protease secretion was estimated using the assay described earlier.
2.7. Partial Purification of Proteases
The crude protease obtained was then subjected to partial purification through ammonium sulfate precipitation [17]. To obtain cell-free supernatant, the bacterial culture was centrifuged at 15,000 rpm for 10 min at 4 °C. Ammonium sulfate was added slowly to the chilled cell-free proteinase extract along with gentle stirring. Ammonium sulfate fractions were then collected at 0–80% saturation and 4 °C for 2 h. The precipitate was obtained through centrifugation at 15,000 rpm for 15 min at 4 °C. The protein precipitate was re-suspended into 20 mM Tris-HCl at pH 8.
2.8. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
SDS-PAGE (12%) was used to determine the purity and molecular weight of the partially purified proteases under denaturing conditions as described by Laemmli [18]. A ladder of 10–180 kDa (PageRulerPrestained Protein Ladder) was used as a standard marker, and the gel was allowed to run at 80 V constantly for 2–2.5 h. The gel was then washed and stained with Coomassie Brilliant Blue staining solution and kept overnight at room temperature. The gel was finally de-stained using a de-staining solution (10% glacial acetic acid, 40% methanol, 50% distilled water) until bands were visualized on a transparent background.
2.9. Zymography
Zymography was performed as described by Lantz and Ciborowski [19]. Separating gel (8%) was prepared according to the standard procedure. Gelatin (1 mg/mL) was added as a substrate in the separating gel. Sample buffer (2X) was added and mixed well with the samples and kept at room temperature for 10 min without heating. The samples were then loaded on the gel under standard conditions, and the gel was allowed to run at 80 V constantly for 2–2.5 h. The gel was finally stained and de-stained until clear bands were visualized against a dark background, indicating proteolytic activity.
The standard marker of 10–180 kDa (PageRuler Prestained Protein Ladder) was used, and molecular weights of the protein bands were determined using a logarithmic curve of Rf value versus the molecular weight.
2.10. Characterization of Partially Purified Protease
A pH range of 6–11 was selected to study the effect of pH on partially purified protease activity, using different buffers such as phosphate buffer (pH 6–7), Tris buffer (pH 8–9), Glycine-NaOH buffer (pH 9–10), and dilute NaOH for pH 11. The buffers and proteases were mixed in an equal ratio. pH values were adjusted at 25 °C. A temperature range of 20 °C to 70 °C was used to study the effect of temperature on protease activity. The effect of surfactants (1%) on proteinase activity was observed using ionic (SDS) and non-ionic (Tween 80 and Triton X-100) surfactants. The previously described assay was used to determine the protease-specific activity and relative activity. The formula used was:
[Relative Activity of Enzyme (%) = (activity of sample of interest/activity of control) × 100]
2.11. Statistical Analysis
All reactions were performed in triplicates, and their means and standard deviations were calculated by Microsoft Excel (2016). Statistical analysis was performed by one-way analysis of variance (ANOVA).
3. Results
3.1. Geographical Location and Physiological Parameter of Soil Samples
Bacterial strains (n = 142) were isolated from four different sites in Islamabad and Karak (Pakistan). The pH and temperature at the dump area (I-12, Islamabad, Pakistan) were 8.3 and 22 °C and at the salt mine (Karak, Khyber Pakhtunkhwa, Pakistan) (Figure 1) was 8.7 and 24 °C.
Figure 1.
Map of Karak showing the location of Bahadur Khel Salt Mines (red circle).
3.2. Preliminary Screening for Protease Production
Out of 142 isolated strains, 80 (56.3%) exhibited halo zones around bacterial colonies on skim milk agar plates. Out of these 80 bacterial isolates, B. subtilis S1 and B. amyloliquefaciens KSM12 produced larger zones of hydrolysis, i.e., 22 mm and 19 mm, respectively (Figure 2).
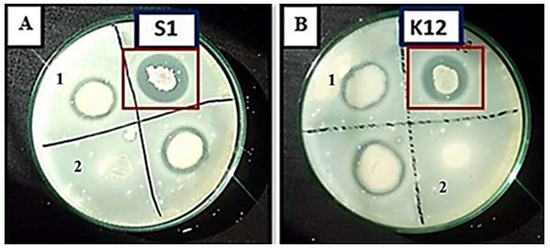
Figure 2.
Zones of hydrolysis formed on skim milk agar plates by (A) B. subtilis S1, and (B) B. amyloliquefaciens KSM12. Representative halo zone produced by the rest protease producers (1). Bacterial strains with no protease production (2).
3.3. Protease Assay
In preliminary screening, 14 best protease-producing bacterial isolates were selected based on the halo zones produced on skim milk agar plates. These selected isolates were then subjected to protease assay, which was performed after 72 h of incubation with casein as a substrate.
The results show varying levels of protease activity (Table 1). Two strains were finally selected based on maximum proteinase production, i.e., B. subtilis S1 (34.3 U/mL) and B. amyloliquefaciens KSM12 (27.9 U/mL). Proteases produced from Bacillus spp. are known to be widely used in the enzyme industry.

Table 1.
Quantitative assay for protease production by the 14 selected isolates. The highest protease-producing strains (bold) were selected for further analysis.
3.4. Bacterial Strains Identification
Molecular identification of the two highest protease producers was carried out by comparing their 16S rRNA gene sequences with other closely related 16S rRNA sequences on NCBI BLASTn. Strain S1 was identified as B. subtilis S1 (accession no. MN963814) with the sequence similarity of 100% with B. subtilis strain RS10 [18] (accession no. CP046860.1), and strain KSM12 was identified as B. amyloliquefaciens KSM12 (accession no. MN391010) with the sequence similarity of 99.93% with B. amyloliquefaciens strain BCRC 11601 (accession no. NR116022). The phylogenetic tree of both B. subtilis S1 and B. amyloliquefaciens KSM12 shows that both bacterial strains belong to the genus Bacillus and are closely related to B. subtilis and B. amyloliquefaciens, respectively (Figure 3).
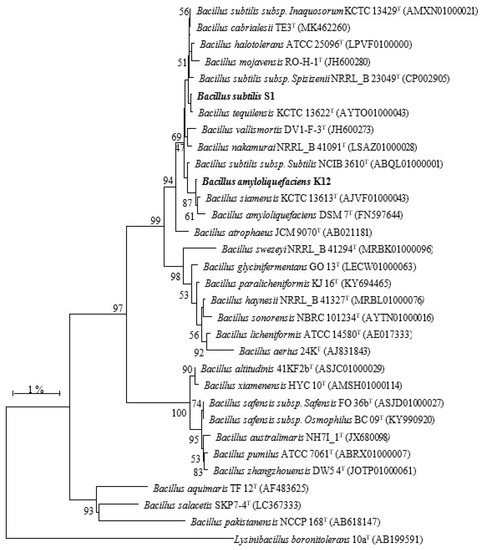
Figure 3.
Phylogenetic analysis of strains S1 and KSM12 inferred from 16S rRNA gene sequence using Neighbor-Joining method based on the Kimura 2-parameter model. The type strain of Lysinibacillus boronitolerans was used as an outgroup.
3.5. Optimization of Protease Production
Protease production of both B. subtilis S1 and B. amyloliquefaciens KSM12 was observed maximum in the neutral to the alkaline range (Figure 4A). The optimal protease production for B. amyloliquefaciens KSM12 can be seen at pH 9 (27.9 U/mL protease activity) while for B. subtilis S1 at pH 8 (34.3 U/mL protease activity).
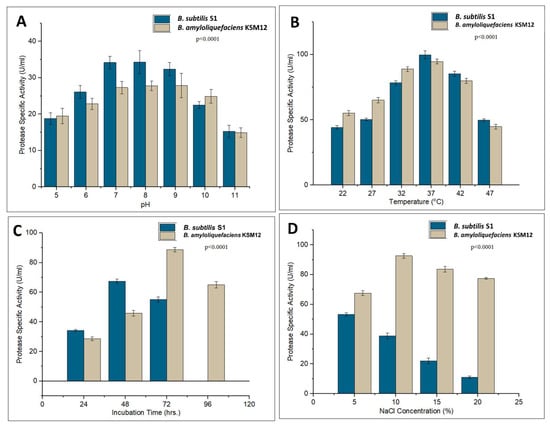
Figure 4.
Optimization for protease production by B. subtilis S1 and B. amyloliquefaciens KSM12. (A) pH optimization (F = 20.03, p < 0.0001), (B) temperature optimization (F = 237.6, p < 0.0001), (C) incubation time optimization (F = 338.72, p < 0.0001), and (D) NaCl optimization (F = 73.75, p < 0.0001).
Both strains, B. subtilis S1 and B. amyloliquefaciens KSM12 showed maximum protease production at 37 °C (Figure 4B). At this temperature, B. subtilis S1 exhibited 99.8 U/mL of protease activity, and B. amyloliquefaciens KSM12 exhibited 94.6 U/mL of protease activity. After 37 °C, protease production gradually started declining. More than 80% activity was retained at 42 °C, but less than 50% activity at 47 °C.
Optimization of incubation time indicated that B. amyloliquefaciens KSM12 was able to produce maximum protease (88.9 U/mL) after 72 h, and B. subtilis S1 produced maximum protease (67.5 U/mL) after 48 h (Figure 4C).
Optimum protease production (94.2 U/mL) was observed at 10% (w/v) NaCl by B. amyloliquefaciens KSM12, and (53.3 U/mL) at 5% (w/v) NaCl by B. subtilis S1, after that it started to decline rapidly (Figure 4D).
3.6. SDS-PAGE and Zymogram Analysis
Partially purified proteases produced a single band conforming to a molecular weight of 17 kDa and 65 KDa on an SDS-PAGE gel for B. subtilis S1 and B. amyloliquefaciens KSM12, respectively (Figure 5). The zymogram activities for both the proteases also showed a single band corresponding to the band in SDS-PAGE.
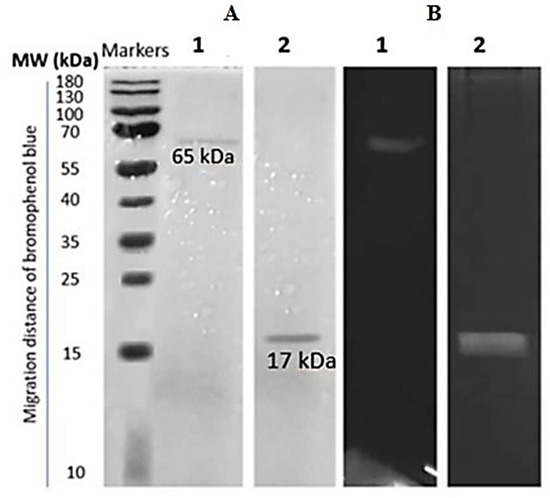
Figure 5.
The 12% SDS-PAGE gel (A) and zymogram analysis (B) of partially purified protease from B. sp. Lane 1: B. subtilis strain S1 and Lane 2: B. amyloliquefaciens strain KSM12.
3.7. Physicochemical Characterization of Partially Purified Protease
Alkaline proteases generally exhibit optimum activity between pH 9–11. The proteases purified from both B. subtilis S1 and B. amyloliquefaciens KSM12 showed significant activity in the pH range of 6–11. Optimal protease activity for B. amyloliquefaciens KSM12 was observed at pH 9, while 98% of activity was retained at pH 10, 72% at pH 11, and 86% at pH 6. Optimal protease activity for B. subtilis S1 was observed at pH 8, while it retained 82% activity at pH 10 and 63% at pH 11 (Figure 6A).
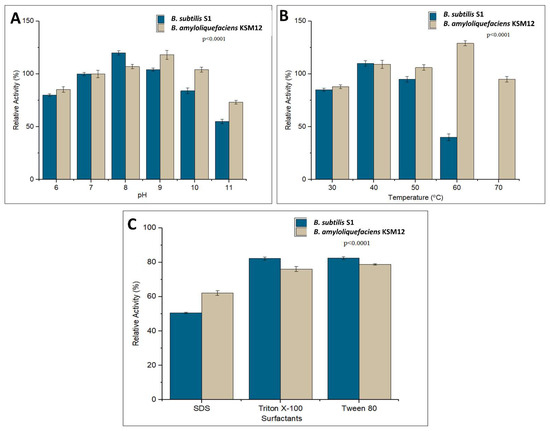
Figure 6.
Physicochemical characterization of partially purified proteases of B. subtilis S1 and B. amyloliquefaciens KSM12. (A) Effect of pH (F = 112.97, p < 0.0001), (B) effect of temperature (F = 177.37, p < 0.0001), and (C) effect of surfactants (F = 378.01, p < 0.0001).
Proteases from both B. subtilis S1 and B. amyloliquefaciens KSM12 showed significantly higher activity at 37 °C and 60 °C, respectively (Figure 6B). Both proteases were active in the range of 30–60 °C. The protease from B. subtilis S1 retained 40% activity at 60 °C and the protease from B. amyloliquefaciens KSM12 retained almost 95% activity at 70 °C.
The detergent industry requires enzymes to be compatible with surfactants and detergents. Both these proteases were not only active under ionic surfactants such as SDS, but also non-ionic surfactants such as Tween 80 and Triton X-100. B. subtilis S1 and B. amyloliquefaciens KSM12 proteases retained activity of 50% and 60%, respectively, in the presence of SDS (Figure 6C). While these proteases retained activity of 80% and 75%, respectively, in the presence of Triton X-100. Inconsistent with the results of Triton X-100 for Tween 80, B. subtilis S1 and B. amyloliquefaciens KSM12 proteases retained activity of 80% and 78%, respectively (Figure 6C).
4. Discussion
Many industries employ hostile environmental conditions such as extremes of pH, temperature, and salinity, thus limiting the wide applicability of many commercially available proteases. In the present study, two alkaline soil bacteria, i.e., B. subtilis S1 and B. amyloliquefaciens KSM12, were isolated from an alkaline dump and sodic soil, respectively. The increase in pH of waste or dumpsites might be due to the presence of high organic matter in the soil [20], which in turn increases the rate of mineralization and exchangeable cations [21,22]. The high pH of salt mine soil might be due to the high amount of exchangeable sodium (cation) in sodic soils [23]. Microorganisms surviving in both dump and sodic soils have been reported to adapt to harsh environmental conditions. This might be due to biochemical modifications at cellular and sub-cellular levels [24,25]. Bacillus spp. from soil has been reported to produce alkali-thermostable proteases [26].
Preliminary screening showed halo zones produced on skim milk agar plates, indicating proteolytic reaction performed by extracellular proteases secreted by these bacterial strains.
Protease production by both bacterial strains was observed maximum in the neutral to alkaline pH range. The optimum protease activity for both B. subtilis S1 and B. amyloliquefaciens KSM12 was observed at pH 8 and 9, respectively, which suggests that both strains might be producing alkaline proteases. These results conform with several earlier reports involving maximal protease production at pH 8 [27] and 9 [28,29] by Bacillus spp. Temperature optimization showed that both the bacterial strains, B. subtilis S1 and B. amyloliquefaciens KSM12, produced maximum protease at 37 °C suggesting their mesophilic nature. Further increase in temperature decreased protease production, suggesting that an increase in temperature eventually disrupts the molecular structure of bacteria and thus stops protease production. Other studies on protease production by Bacillus subtilis have also reported 37 °C to be the optimal temperature for protease production [30]. The optimal incubation time for B. subtilis S1 and B. amyloliquefaciens KSM12 was after 48 h and 72 h of incubation, respectively. Bajaj and Sharma [31] observed maximum protease production by Streptomyces ambofaciens after 48 h. Lakshmi et al. also observed maximal proteinase secretion by B. licheniformis after 72 h [28]. Maximum production of extracellular proteases occurs in the late logarithmic and early stationary phase when bacteria start secreting proteases to digest complex nutrients as cultural media starts to get deprived of the primary sources of carbon and nitrogen [32]. The decline in protease activity of B. subtilis S1 and B. amyloliquefaciens KSM12 after 48 h and 72 h, respectively, may be due to the accumulation of toxic metabolites produced by bacteria in a culture medium. The optimization of salt concentration for B. amyloliquefaciens KSM12 showed maximal proteinase secretion at 10% NaCl (w/v), after which it rapidly started decreasing. Proteases from Bacillus species are characterized by a molecular weight range from 27 to 71 kDa, optimal pH range of 6–10, temperature of 37–60 °C, and stability values over a wide range of pH and temperature [33]. After optimization, protease production increased by 2.9-fold for B. subtilis S1, i.e., 99.8 U/mL from 34.3 U/mL, and 3.4-fold for B. amyloliquefaciens, i.e., 94.6 U/mL from 27.9 U/mL. These results showed that these bacteria might be able to produce highly halotolerant proteases. The decrease in protease production afterward may be due to high ionic strength, which disrupts the enzymatic structures of this bacterium. This indicates that the protease identified in the current study has high halo-tolerance and can be used in several industries requiring high salt conditions.
The most imperative characteristic of partially purified proteases from both the strains was their tolerance to a wide range of pH (8–11), which shows their high alkali-tolerance nature. The optimal activity of protease from B. subtilis S1 was observed at pH 8 and for B. amyloliquefaciens KSM12 at pH 9. The maximum activity of many commercially important proteases is reported in the pH range of 8–11 [1]. High protease activity at such a wide range of pH and optimal activity at higher pH makes this enzyme suitable for the detergent industry, as many commercially important proteases used in the detergent industry operate at a pH of 7–11. It can also be used for dehairing and bating processes in the leather industry, preparation of protein hydrolysates, and as an additive in cleaning and dishwashing detergents [34].
The maximum protease activity for B. subtilis S1 and B. amyloliquefaciens KSM12 was observed at 40 °C and 60 °C, respectively, which indicates the thermo-tolerant nature of these proteases. This can also be supported by a study that reports optimal protease activity up to 60 °C [35]. These results show that the proteases are not only alkaline but can also remain active at a wide range of temperatures. Optimal temperature ranges up to 60 °C show the thermostable behavior of proteases, thus making them an ideal candidate for the detergent industry. Furthermore, such thermostable proteases can be used to avoid the risk of contamination by running processes not suitable for mesophilic microorganisms [36].
The stability of proteases in surfactants suggests that these proteases might be suitable for the detergent industry. Both these proteases were not only active under ionic surfactants such as SDS but also under non-ionic surfactants such as Tween 80 and Triton X-100. The stability of these proteases towards SDS is a very crucial factor for the application in the detergent industry as SDS is a very common additive in detergents. Also, many proteases produced from Bacillus spp. are reported to be SDS-stable [36]. Alkaline proteases produced by microbes are also extensively used in the leather industry. These proteases are especially useful for the removal of unwanted pigments during the dehairing process [37]. Apart from this, Bacillus spp. are also reported to be suitable for other industries. Table 2 shows the current research progress and the utilization of different Bacillus strains and their applications in various industries.

Table 2.
Current research trends and industrial applications of proteases secreted from Bacillus spp.
These results show that the extracellular alkaline proteinases secreted by B. subtilis S1 and B. amyloliquefaciens KSM12 might serve as a potential candidate for different industrial applications such as detergents, food, leather, textiles, and pharmaceuticals. Among proteases, alkaline bacterial proteases play a vital role in different industries, and their future use is likely to be increased.
5. Conclusions
In the present study, two bacterial strains Bacillus subtilis S1 and Bacillus amyloliquefaciens KSM12 were isolated from the soil for their ability to produce proteases. The best conditions for protease production were optimized at pH 8, 37 °C, 48 h, 5% (w/v) NaCl for Bacillus subtilis S1 and pH 9, 37 °C, 72 h, 10% (w/v) NaCl for Bacillus amyloliquefaciens KSM12. The predicted maximum protease activity was 99.8 U/mL and 94.6 U/mL for B. subtilis S1 and B. amyloliquefaciens KSM12, respectively. Maximum activity and stability of protease enzyme were retained from B. subtilis S1 at pH 8, 40 °C and B. amyloliquefaciens KSM12 at pH 9, 60 °C. The proteases were also stable against surfactants.
The characteristics exhibited by these extracellular proteases might help them to be utilized as a potential additive in detergent formulations and as a catalyst in the textile, food, and pharmaceutical industry with improved yield and characteristics. Other applications include protein hydrolysis, thermal tolerance, substrate specificity, processing of photographic films, and production of zein hydrolysates.
The thermostable nature of these enzymes makes them suitable for industries employing higher temperatures along with shorter reaction times and a low risk of contamination. Specifically, the tolerance towards alkaline pH makes these bacterial isolates more prominent to act as biocatalysts and makes them useful for the local industrial sector.
This study concludes that produced surfactant stable compatible, thermotolerant, alkaline protease has the potential for industrial applications.
Author Contributions
S.H. and S.I. designed the project, S.H. performed experiments under the supervision of H.A.J., S.I. was involved in bacterial isolation from soil samples, and I.A. performed phylogenetic analysis of bacterial isolates. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The GenBank accession number for the 16S rRNA gene sequence of strain B. subtilis S1 is MN963814, and for strain KSM12 identified as B. amyloliquefaciens is MN391010.
Acknowledgments
The authors are thankful to ASAB-NUST for scientific and administrative support and for MS Student Research Fund.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. Biomed. Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Beg, Q.; Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002, 59, 15–32. [Google Scholar] [PubMed]
- Degering, C.; Eggert, T.; Puls, M.; Bongaerts, J.; Evers, S.; Maurer, K.-H.; Jaeger, K.-E. Optimization of Protease Secretion in Bacillus subtilis and Bacillus licheniformis by Screening of Homologous and Heterologous Signal Peptides. Appl. Environ. Microbiol. 2010, 76, 6370–6376. [Google Scholar] [CrossRef] [Green Version]
- Masi, C.; Gemechu, G.; Tafesse, M. Isolation, screening, characterization, and identification of alkaline protease-producing bacteria from leather industry effluent. Ann. Microbiol. 2021, 71, 24. [Google Scholar] [CrossRef]
- Bishop, M.L.; Fody, E.P.; Schoeff, L.E. Clinical Chemistry: Techniques, Principles, and Correlations; Wolters Kluwer: Alphen aan den Rijn, The Netherlands, 2018. [Google Scholar]
- Ahmed, M.; Rehman, R.; Siddique, A.; Hasan, F.; Naeem, A.; Hameed, A. Production, purification and characterization of detergent-stable, halotolerant alkaline protease for eco-friendly application in detergents’ industry. Int. J. Biosci. 2016, 8, 47–65. [Google Scholar]
- Cupp-Enyard, C. Sigma’s non-specific protease activity assay-casein as a substrate. J. Vis. Exp. 2008, 19, e899. [Google Scholar] [CrossRef]
- Labuda, J.; Bowater, R.; Fojta, M.; Gauglitz, G.; Glatz, Z.; Hapala, I.; Havliš, J.; Kilar, F.; Kilar, A.; Malinovská, L.; et al. Terminology of bioanalytical methods (IUPAC Recommendations 2018). Pure Appl. Chem. 2018, 90, 1121–1198. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Wright, E.S.; Yilmaz, L.S.; Noguera, D.R. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 2012, 78, 717–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Englard, S.; Seifter, S. Precipitation techniques. Methods Enzymol. 1990, 182, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lantz, M.S.; Ciborowski, P. Zymographic techniques for detection and characterization of microbial proteases. Methods Enzymol. 1994, 235, 563–594. [Google Scholar] [CrossRef]
- Anikwe, M.A.N. Amelioration of a heavy clay loam soil with rice husk dust and its effect on soil physical properties and maize yield. Bioresour. Technol. 2000, 74, 169–173. [Google Scholar] [CrossRef]
- Anikwe, M.; Nwobodo, K. Long term effect of municipal waste disposal on soil properties and productivity of sites used for urban agriculture in Abakaliki, Nigeria. Bioresour. Technol. 2002, 83, 241–250. [Google Scholar] [CrossRef]
- Woomer, P.L.; Martin, A.; Albrecht, A.; Resck, D.V.S.; Scharpenseel, H.W. The importance and management of soil organic matter in the tropics. Biol. Manag. Trop. Soils Fertil. 1994. [Google Scholar]
- Aderoju, D.O.; Festus, A.G. Influence of salinity on soil chemical properties and surrounding vegetation of Awe salt mining site, Nasarawa State, Nigeria. Afr. J. Environ. Sci. Technol. 2013, 7, 1070–1075. [Google Scholar]
- Sayaniya, A.V.; Patel, P. Isolation and Characterization of Detergent Compatible Alkaline Protease Producing Bacillus Subtilis APO-1. Biomed. J. Sci. Tech. Res. 2021, 35, 27949–27955. [Google Scholar]
- Otlewska, A.; Migliore, M.; Dybka-Stępień, K.; Manfredini, A.; Struszczyk-Świta, K.; Napoli, R.; Pinzari, F. When salt meddles between plant, soil, and microorganisms. Front. Plant Sci. 2020, 11, 1429. [Google Scholar] [CrossRef] [PubMed]
- Gaur, R.; Tiwari, S.; Sharma, A. Protease of Bacillus sp. P-02. Am. J. Food Technol. 2014, 9, 246–256. [Google Scholar] [CrossRef]
- Nascimento, W.C.A.D.; Martins, M.L.L. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol. 2004, 35, 91–96. [Google Scholar] [CrossRef]
- Lakshmi, B.K.M.; Sri, P.R.; Devi, K.A.; Hemalatha, K.P.J. Media optimization of protease production by Bacillus licheniformis and partial characterization of Alkaline protease. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 650–659. [Google Scholar]
- Swamy, M.K.; Kashyap, S.S.N.; Vijay, R.; Tiwari, R.; Anuradha, M. Production and optimization of extracellular protease from Bacillus sp. isolated from soil. Int. J. Adv. Biotechnol. Res. 2012, 3, 564–569. [Google Scholar]
- Abusham, R.A.; Rahman, R.N.; Salleh, A.B.; Basri, M. Optimization of physical factors affecting the production of thermo-stable organic solvent-tolerant protease from a newly isolated halo tolerant Bacillus subtilis strain Rand. Microb. Cell Factories 2009, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, B.K.; Sharma, P. An alkali-thermotolerant extracellular protease from a newly isolated Streptomyces sp. DP2. N. Biotechnology 2011, 28, 725–732. [Google Scholar] [CrossRef]
- Ward, O.P. Proteolytic enzymes. Compr. Biotechnol. 1985, 3, 789–818. [Google Scholar]
- Karray, A.; Alonazi, M.; Horchani, H.; Ben Bacha, A. A novel thermostable and alkaline protease produced from Bacillus stearothermphilus isolated from olive oil mill sols suitable to industrial Biotechnology. Molecules 2021, 26, 1139. [Google Scholar] [CrossRef] [PubMed]
- Neklyudov, A.D.; Ivankin, A.N.; Berdutina, A.V. Properties and uses of protein hydrolysates. Appl. Biochem. Microbiol. 2000, 36, 452–459. [Google Scholar] [CrossRef]
- Tang, X.Y.; Wu, B.; Ying, H.J.; He, B.F. Biochemical Properties and Potential Applications of a Solvent-Stable Protease from the High-Yield Protease Producer Pseudomonas aeruginosa PT121. Appl. Biochem. Biotechnol. 2009, 160, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, A.; Sivakumar, N.; Gujarathi, A.M.; Victor, R. Production of thermotolerant, detergent stable alkaline protease using the gut waste of Sardinella longiceps as a substrate: Optimization and characterization. Sci. Rep. 2018, 8, 12442. [Google Scholar] [CrossRef]
- Brandelli, A.; Daroit, D.; Riffel, A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010, 85, 1735–1750. [Google Scholar] [CrossRef]
- Maurer, K.H. Detergent proteases. Curr. Opin. Biotechnol. 2004, 15, 330–334. [Google Scholar] [CrossRef]
- Hammami, A.; Fakhfakh, N.; Abdelhedi, O.; Nasri, M.; Bayoudh, A. Proteolytic and amylolytic enzymes from a newly isolated Bacillus mojavensis S.A.: Characterization and applications as laundry detergent additive and in leather processing. Int. J. Biol. Macromol. 2018, 108, 56–68. [Google Scholar] [CrossRef]
- Plotnikov, M.B.; Dygai, A.M.; Aliev, O.I.; Chernyshova, G.A.; Smol’yakova, V.I.; Vasil’ev, A.S.; Markov, V.A.; Vyshlov, E.V.; Vereschagin, E.I.; Kinsht, D.N. Antithrombotic and Thrombolytic Effects of a New Proteolytic Preparation Trombovazim (Russia). Bull. Exp. Biol. Med. 2009, 147, 438–440. [Google Scholar] [CrossRef]
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).