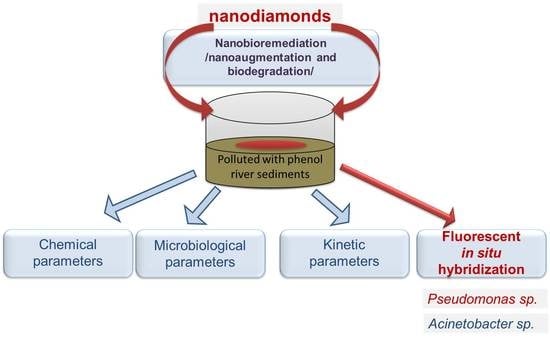
Application of Nanodiamonds in Modelled Bioremediation of Phenol Pollution in River Sediments
Abstract
1. Introduction
2. Materials and Methods
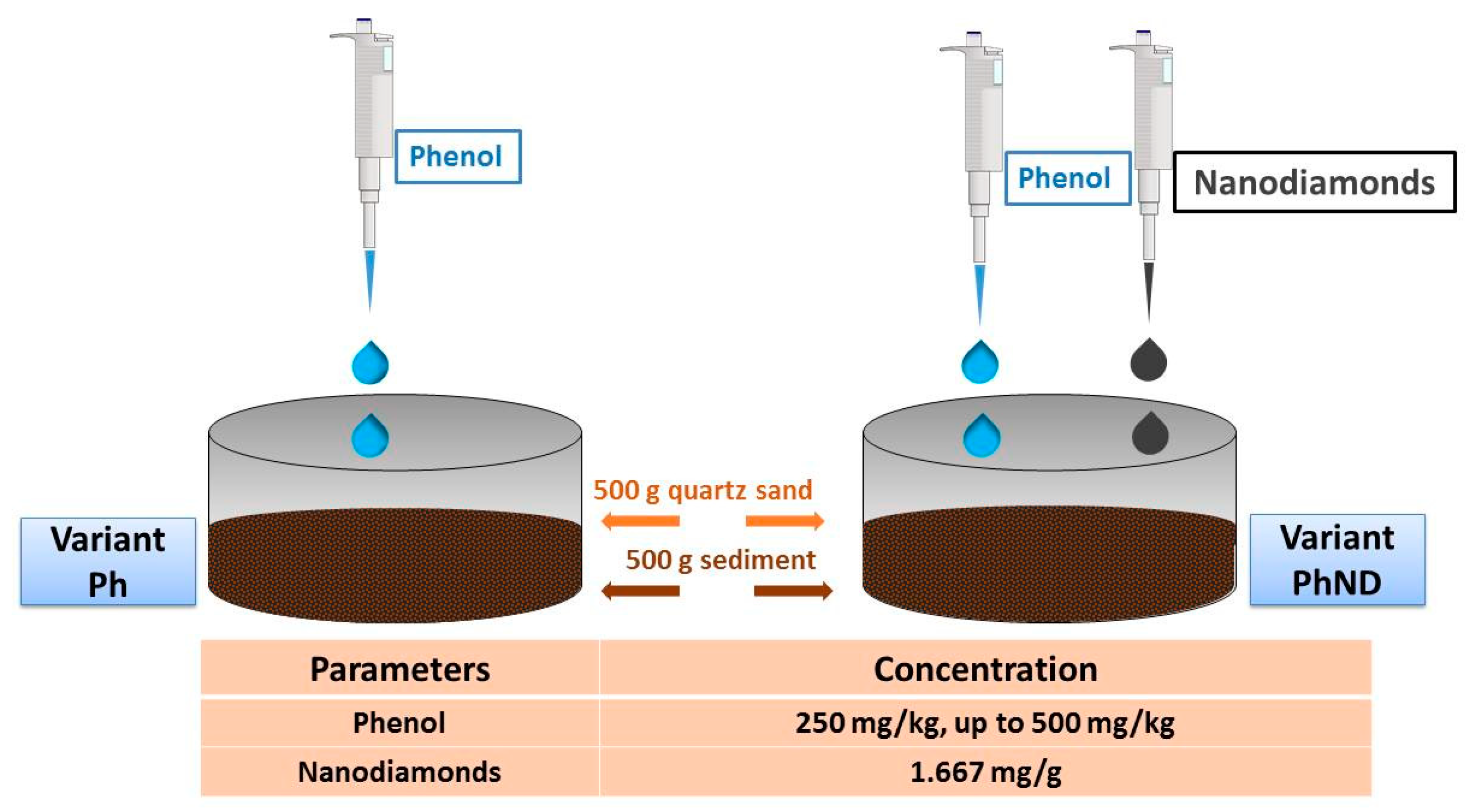
2.1. Experimental Design
- Pollution with a comparatively low concentration of phenol: from the 2nd–48th hours;
- Simulation of a critical situation with the entering of a high concentration of phenol: at the 49th and 72nd hours;
- Study of the system after a long adaptation and entering into the phase of system recovery after the explosive phenol loading: from the 144th–214th hours.
2.2. Analytical Methods and Indicators
2.3. Microbiological Analysis
2.4. FISH Analysis
2.5. Application of Nanodiamonds
3. Results and Discussion
3.1. Residual Concentration of Phenol
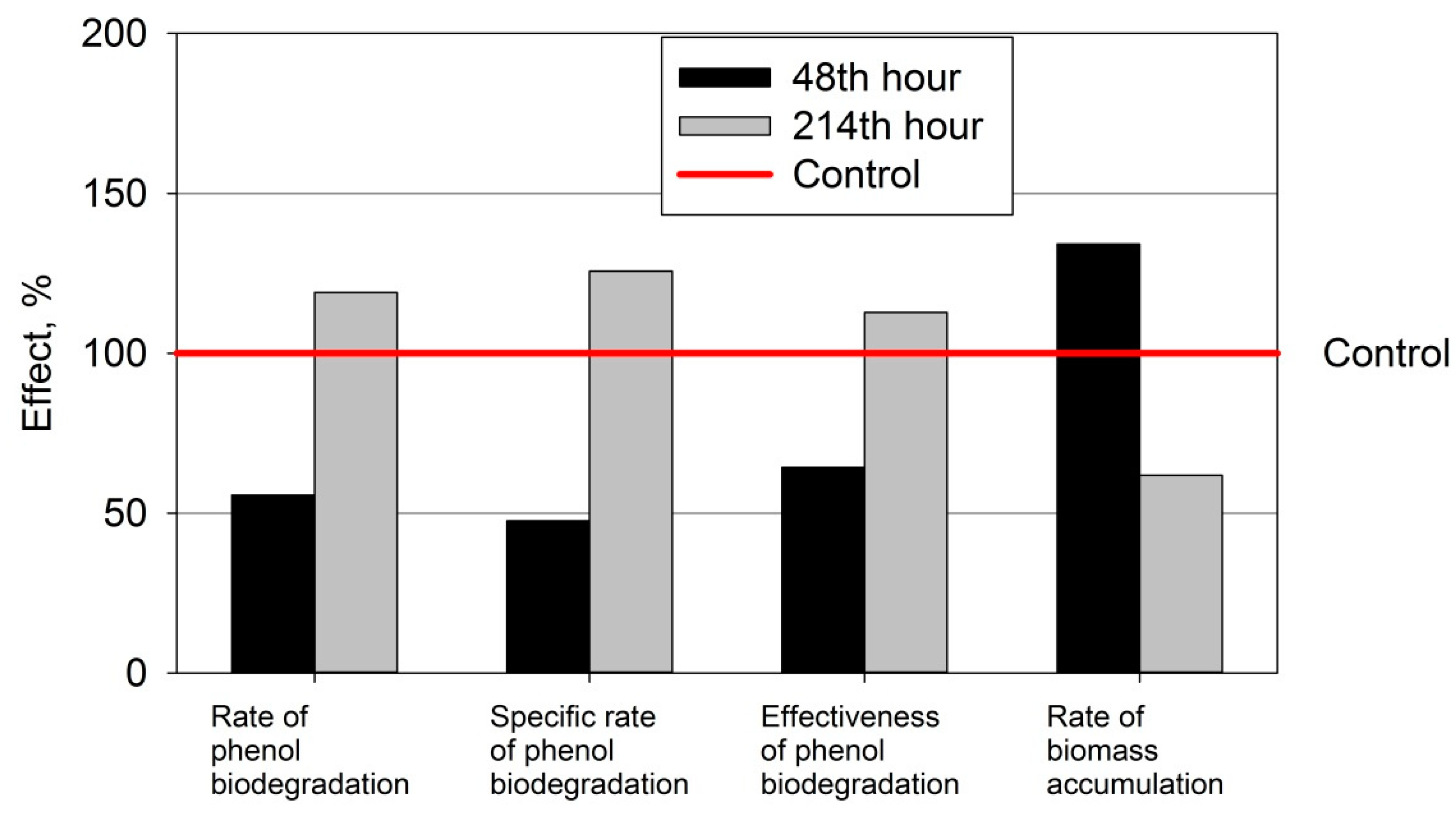
3.2. Kinetic Parameters of Biodetoxification Processes
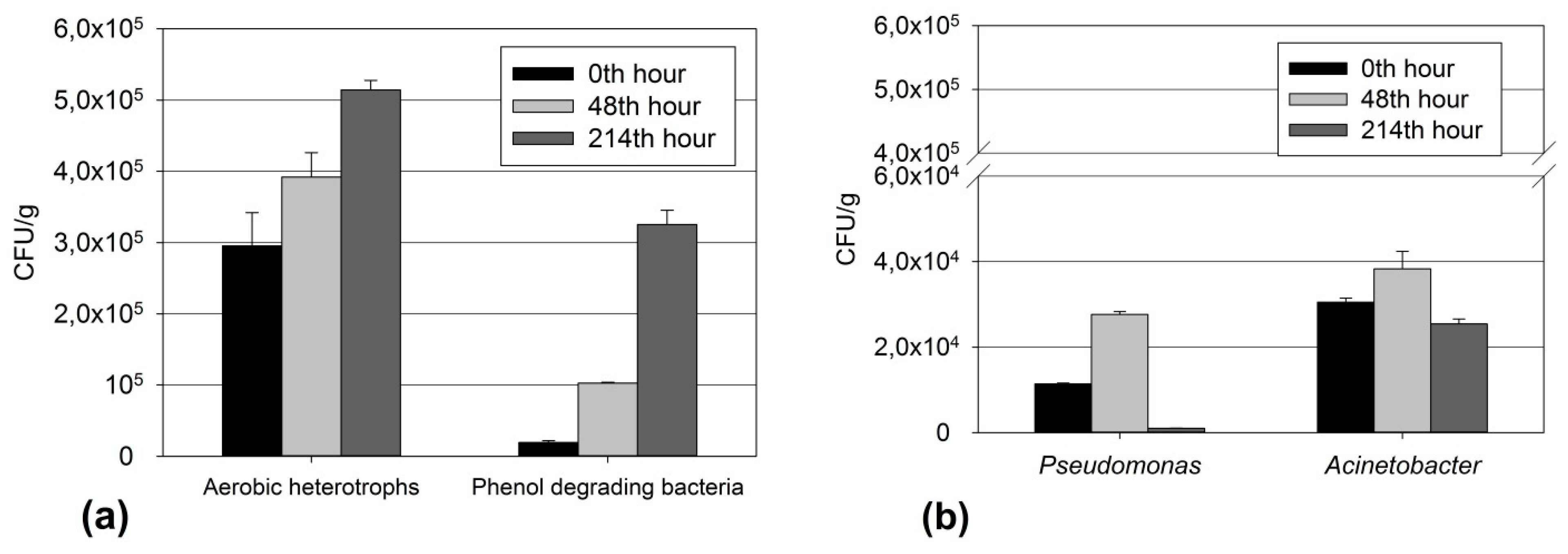
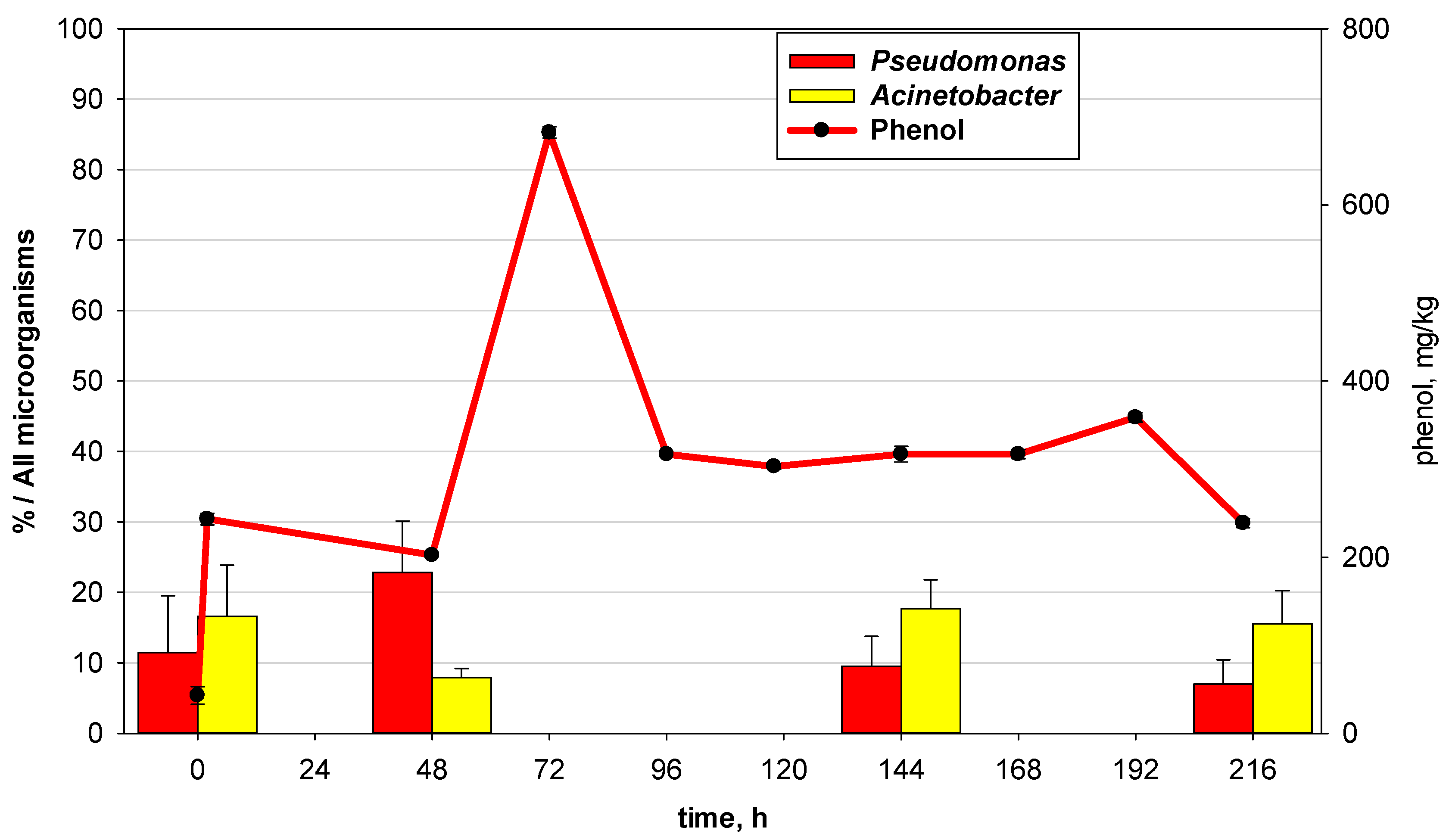
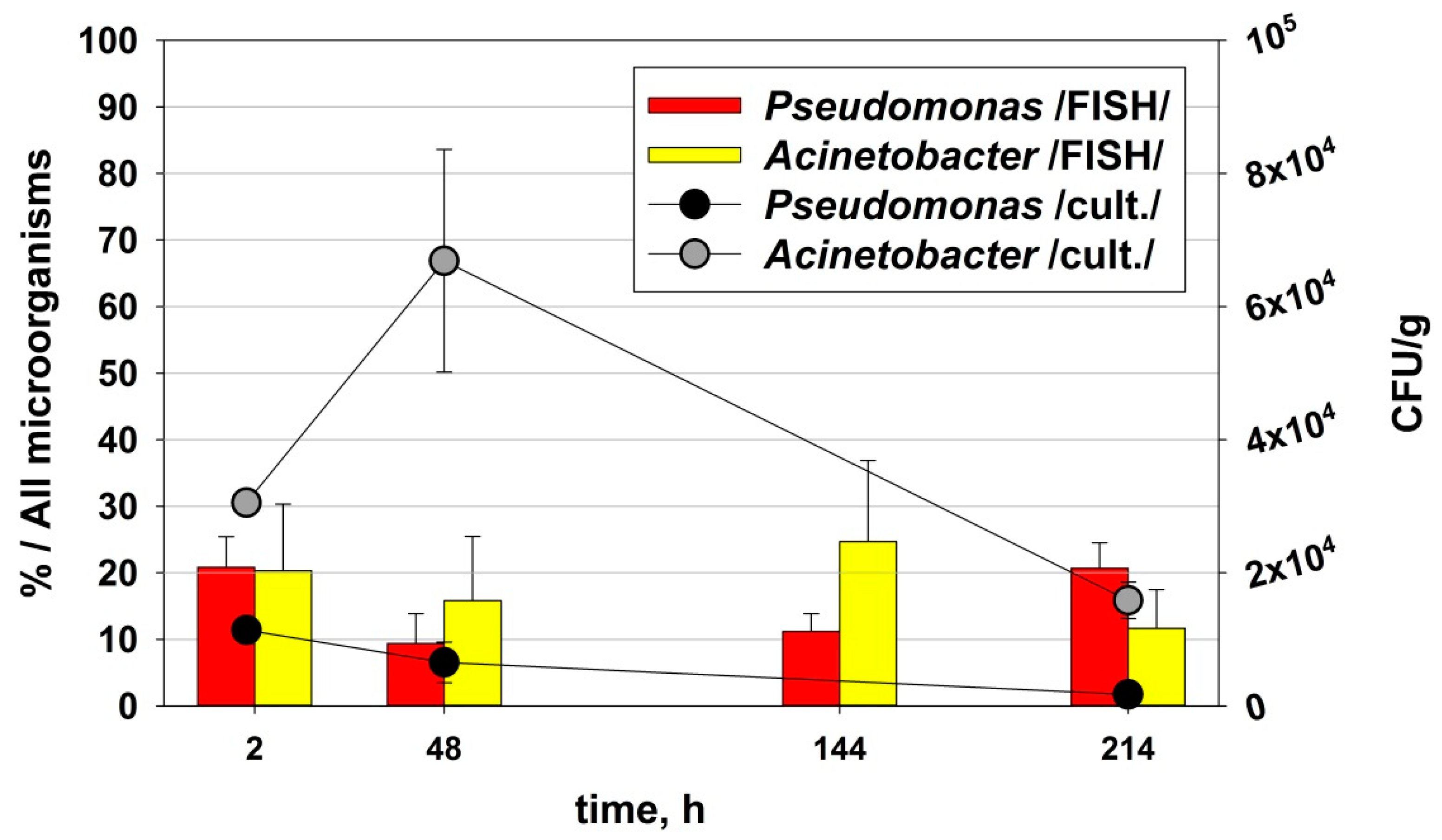
3.3. Relationship of Culturable and Non-Culturable Microorganisms during Adaptation without ND
- (1)
- In the early phase (at the 48th hour—in the biodegradation process while at phenol concentrations that are close to critical) of adaptation to low phenol concentrations, microbial communities respond by increasing the quantity of microorganisms from the key groups for the biodegradation process.
- (2)
- In the late phase (after the intoxication shock and the start of the readaptation process) after prolonged adaptation to high concentrations of xenobiotics that are close to a critical level, the adaptive response is more complex and highly specialized. Synergistic relationships and zones are formed in the communities with high oxygenase enzyme activity, formation of mature biofilm with zones with high biodegradation activity and other similar mechanisms. In this case, the quantity of culturable microorganisms is not the only adaptive response. For the genera Pseudomonas and Acinetobacter, the mechanisms that are associated with increased activity to degrade the xenobiotic are valid.
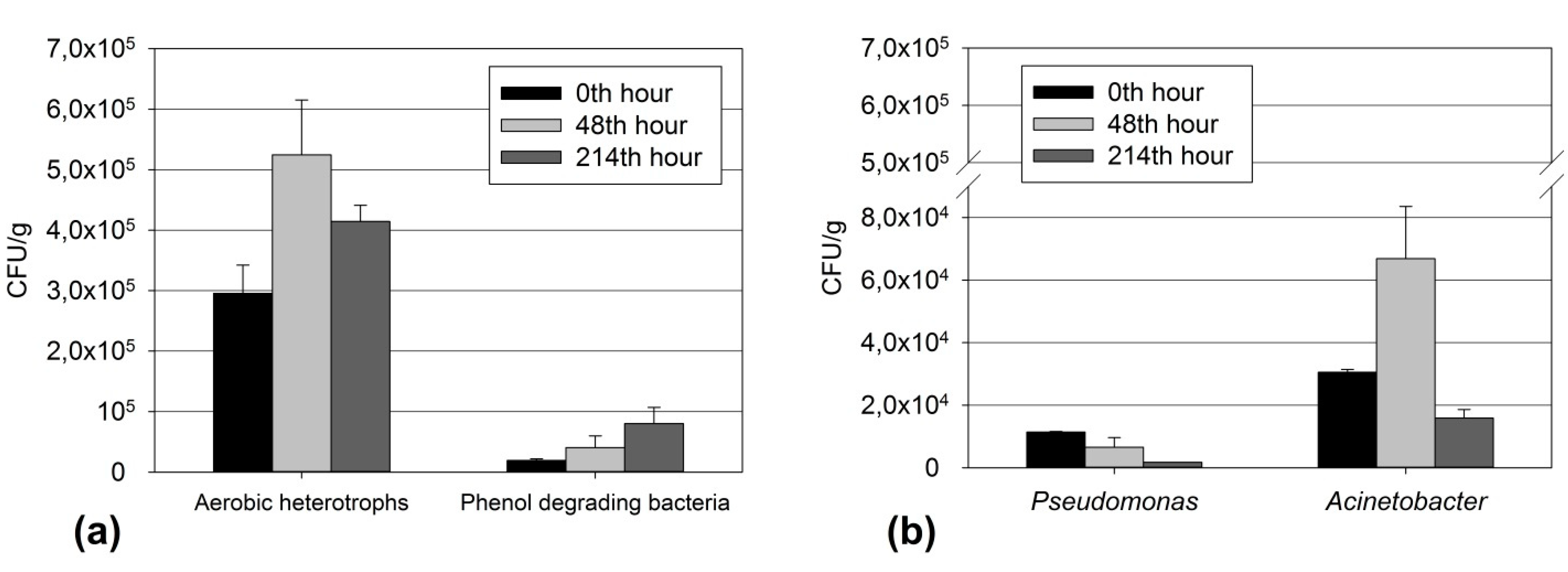
3.4. Relationship of Culturable and Non-Culturable Microorganisms during Adaptation with the Participation of ND
- The genus Pseudomonas reduced their quantity. In this standard method of studying microbial cenosis, ND inhibited the growth and adaptation of culturable bacteria of this genus within 48 h.
- At the 48th hour, the other two physiological and taxonomic groups increased their populations of culturable bacteria, namely Acinetobacter and the phenol-degrading microorganisms. Bacteria of the genus Acinetobacter played a leading role in the bioremediation of the phenol-contaminated sediments. These bacteria have occupied this ecological niche and there was an increase in their population compared to the observed decline in the quantity of bacteria of the genus Pseudomonas.
3.5. Effect of ND on the Restructuring of the Microbial Community
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carere, M.; Marcheggiani, S.; Miniero, R.; Pilozzi, A.; Mancini, L. Risk Assessment Elements for the Management of Contaminated Sediments. Preface. Ann. Del’istituto Super. Di Sanità 2008, 44, 217. [Google Scholar]
- De Groot, R.S. Functions of Nature: Evaluation of Nature in Environmental Planning, Management and Decision-Making; Wolters Noordhoff BV: Groningen, The Neatherlands, 1992. [Google Scholar]
- Heise, S. Risk management of sediments. Ann. Ist. Super. Sanità 2008, 44, 224–232. [Google Scholar] [PubMed]
- Heise, S.; Apitz, S.E.; Babut, M.; Bergmann, H.; Besten, P.D.; Ellen, G.J.; Joziasse, J.; Katsiri, A.; Maaß, V.; Oen, A.; et al. Risk management of sediments and communication. Synthesis of the SedNet working group 5 outcomes. J. Soils Sediments 2004, 4, 233–235. [Google Scholar] [CrossRef]
- Salomons, W.; Brils, J. Contaminated Sediments in European River Basins; European Sediment Research Network SedNet. EC Contract No. EVKI-CT-2001-20002, Key Action 1.4. 1 Abatement of Water Pollution from Contaminated Land, Landfills and Sediments; TNO: Den Helder, The Netherlands, 2004. [Google Scholar]
- Marcheggiani, C.; Mancini, L. Microbiological Quality of River Sediments and Primary Prevention. In Ecosystems Biodiversity; Grillo, O., Venora, G., Eds.; IntechOpen: London, UK, 2011. [Google Scholar]
- Jones, J.G. Some differences in the microbiology of profundal and littoral lake sediments. Microbiology 1980, 117, 285–292. [Google Scholar] [CrossRef][Green Version]
- Kuznezow, S.I. Die Rolle der Mikroorganismen im Stoffkreislauf der Seen; Deutscher Verlag d. Wissenschaften: Berlin, Germany, 1959; p. 301. [Google Scholar]
- Rheinheimer, G. Aquatic Microbiology, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 1992; p. 362. [Google Scholar]
- Lincheva, S.; Todorova, Y.; Topalova, Y. Spatial and seasonal fluctuations in microbial segment of Iskar River in the part of cascade of mini-hydroelectric power stations “Middle Iskar”. In Proceedings of the Youth Scientific Conference “Klimentovi Dni, Sofia Bulgaria, 22–23 November 2010; Volume 1, pp. 46–50. [Google Scholar]
- Yotinov, I.; Lincheva, S.; Kenderov, L.; Schneider, I.; Topalova, Y. Functional Structure of Microbial Communities—Mechanism for Management of Biotransformation Processes in the Sediments. In Proceedings of the International Conference Ecology—Interdisciplinary Science and Practice, Sofia, Bulgaria, 25–26 October 2012; Volume 1, pp. 100–107. [Google Scholar]
- Lincheva, S. Algorithms for Biocontrol of the Transformation Processes in the Sediments of the Iskar River in the Area of the Cascade of Middle Middle Iskar. Ph.D. Disertation, Sofia University “St. Kliment Ohridski”, Sofia, Bulgaria, 2015; p. 181. [Google Scholar]
- Yotinov, I.; Lincheva, S.; Kenderov, L.; Schneider, I.; Topalova, Y. Evaluation of the self-purification in the waters of the micro-dams in the small hydroelectric power plants (hepps) Lakatnik and Svrazhen: Potential of the bioalgorithms. Bulg. J. Agric. Sci. 2013, 19, 135–138. [Google Scholar]
- Paisio, C.E.; Talano, M.A.; González, P.S.; Pajuelo-Domínguez, E.; Agostini, E. Characterization of a phenol-degrading bacterium isolated from an industrial effluent and its potential application for bioremediation. Environ. Technol. 2013, 34, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Krastanov, A.; Alexieva, Z.; Yemendzhiev, H. Microbial degradation of phenol and phenolic derivatives. Eng. Life Sci. 2013, 13, 76–87. [Google Scholar] [CrossRef]
- Qiao, M.; Wang, C.; Huang, S.; Wang, D.; Wang, Z. Composition, sources, and potential toxicological significance of PAHs in the surface sediments of the Meiliang Bay, Taihu Lake, China. Environ. Int. 2006, 32, 28–33. [Google Scholar] [CrossRef]
- WHO. International Programme on Chemical Safety (IPCS); Environmental Health Criteria 161 Phenol; World Health Organization (WHO): Geneva, Switzerland, 1994. [Google Scholar]
- Topalova, Y.; Dimkov, R. Biodegradation of Xenobiotics; Faculty of Biology, Sofia University “St. Kliment Ohridski”: Sofia, Bulgaria, 2003; p. 120. [Google Scholar]
- Asenova, M.; Topalova, Y.; Dimkov, R. Validation of TTC-dehydrogenase activity method for measurement of bioremediation potencial of plankton community in oil contaminated ponds. Biotechnol. Biotechnol. Equip. 2003, 17, 16–26. [Google Scholar]
- Nair, C.I.; Jayachandran, K.; Shashidhar, S. Biodegradation of phenol. Afr. J. Biotechnol. 2008, 7, 25. [Google Scholar]
- Haack, S.K.; Bekins, B.A. Microbial populations in contaminant plumes. Hydrogeol. J. 2000, 8, 63. [Google Scholar] [CrossRef]
- Madsen, E.L. Nucleic-acid characterization of the identity and activity of subsurface microorganisms. Hydrogeol. J. 2000, 8, 112. [Google Scholar] [CrossRef]
- Amann, R.; Ludwig, W. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 2000, 24, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, B.J. Molecular approaches to the study of aquatic microbial communities. Curr. Opin. Biotechnol. 1999, 10, 220–224. [Google Scholar] [CrossRef]
- Magrez, A.; Kasas, S.; Salicio, V.; Pasquier, N.; Seo, J.W.; Celio, M.; Forró, L. Cellular toxicity of carbon-based nanomaterials. Nano Lett. 2006, 6, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 1998, 73, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 1999, 2, 317–322. [Google Scholar] [CrossRef]
- Aktan, S.; Salih, B.A. Fluorescent in situ hybridization (FISH) for the detection of bacterial community in activated sludge from textile factories. Environ. Technol. 2006, 27, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Belouhova, M.; Schneider, I.; Chakarov, S.; Ivanova, I.; Topalova, Y. Microbial community development of biofilm in Amaranth decolourization technology analysed by FISH. Biotechnol. Biotechnol. Equip. 2014, 28, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Daims, H.; Lemmer, H. FISH Handbook for Biological Wastewater Treatment: Identification and Quantification of Microorganisms in Activated Sludge and Biofilms by FISH; IWA Publishing: London, UK, 2009; p. 200. ISBN 9781843392316. [Google Scholar]
- Amann, R.I.; Krumholz, L.; Stahl, D.A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 1990, 172, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Breuker, A.; Köweker, G.; Blazejak, A.; Schippers, A. The deep biosphere in terrestrial sediments in the Chesapeake Bay area, Virginia, USA. Front. Microbiol. 2011, 2, 156. [Google Scholar] [CrossRef] [PubMed]
- Tischer, K.; Zeder, M.; Klug, R.; Pernthaler, J.; Schattenhofer, M.; Harms, H.; Wendeberg, A. Fluorescence in situ hybridization (CARD-FISH) of microorganisms in hydrocarbon contaminated aquifer sediment samples. Syst. Appl. Microbiol. 2012, 35, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Langendijk, P.S.; Schut, F.; Jansen, G.J.; Raangs, G.C.; Kamphuis, G.R.; Wilkinson, M.H.; Welling, G.W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 1995, 61, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Suárez, A.; López-López, A.; Tovar-Sánchez, A.; Yarza, P.; Orfila, A.; Terrados, J.; Rosselló-Móra, R. Response of sulfate-reducing bacteria to an artificial oil-spill in a coastal marine sediment. Environ. Microbiol. 2011, 13, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Gibello, A.; Caracciolo, A.B.; Fajardo, C.; Nande, M.; Vargas, R.; Martín, M. A new fluorescent oligonucleotide probe for in situ detection of s-triazine-degrading Rhodococcus wratislaviensis in contaminated groundwater and soil samples. Water Res. 2009, 43, 2999–3008. [Google Scholar] [CrossRef] [PubMed]
- Kleikemper, J.; Pombo, S.A.; Schroth, M.H.; Sigler, W.V.; Pesaro, M.; Zeyer, J. Activity and diversity of methanogens in a petroleum hydrocarbon-contaminated aquifer. Appl. Environ. Microbiol. 2005, 71, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Pombo, S.A.; Kleikemper, J.; Schroth, M.H.; Zeyer, J. Field-scale isotopic labeling of phospholipid fatty acids from acetate-degrading sulfate-reducing bacteria. FEMS Microbiol. Ecol. 2005, 51, 197–207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rogers, S.W.; Ong, S.K.; Moorman, T.B. Mineralization of PAHs in coal–tar impacted aquifer sediments and associated microbial community structure investigated with FISH. Chemosphere 2007, 69, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Ficker, M.; Krastel, K.; Orlicky, S.; Edwards, E. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 1999, 65, 5576–5585. [Google Scholar] [CrossRef]
- Kleikemper, J.; Pelz, O.; Schroth, M.H.; Zeyer, J. Sulfate-reducing bacterial community response to carbon source amendments in contaminated aquifer microcosms. FEMS Microbiol. Ecol. 2002, 42, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Pombo, S.A.; Pelz, O.; Schroth, M.H.; Zeyer, J. Field-scale 13C-labeling of phospholipid fatty acids (PLFA) and dissolved inorganic carbon: Tracing acetate assimilation and mineralization in a petroleum hydrocarbon-contaminated aquifer. FEMS Microbiol. Ecol. 2002, 41, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Anneser, B.; Pilloni, G.; Bayer, A.; Lueders, T.; Griebler, C.; Einsiedl, F.; Richters, L. High resolution analysis of contaminated aquifer sediments and groundwater—what can be learned in terms of natural attenuation? Geomicrobiol. J. 2010, 27, 130–142. [Google Scholar] [CrossRef]
- Griebler, C.; Mindl, B.; Slezak, D.; Geiger-Kaiser, M. Distribution patterns of attached and suspended bacteria in pristine and contaminated shallow aquifers studied with an in situ sediment exposure microcosm. Aquat. Microb. Ecol. 2002, 28, 117–129. [Google Scholar] [CrossRef]
- Albrechtsen, H.J.; Winding, A. Microbial biomass and activity in subsurface sediments from Vejen, Denmark. Microb. Ecol. 1992, 23, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Fera, M.T.; Gugliandolo, C.; Lentini, V.; Favaloro, A.; Bonanno, D.; La Camera, E.; Maugeri, T.L. Specific detection of Arcobacter spp. in estuarine waters of Southern Italy by PCR and fluorescent in situ hybridization. Lett. Appl. Microbiol. 2010, 50, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Holm, P.E.; Nielsen, P.H.; Albrechtsen, H.J.; Christensen, T.H. Importance of unattached bacteria and bacteria attached to sediment in determining potentials for degradation of xenobiotic organic contaminants in an aerobic aquifer. Appl. Environ. Microbiol. 1992, 58, 3020–3026. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.L.; Ghiorse, W.C. Aquatic Microbiology—An Ecological Approach; Ford, T.E., Ed.; Blackwell Scientific Publication: Hoboken, NJ, USA, 1993; pp. 167–213. [Google Scholar]
- Brad, T.; Van Breukelen, B.M.; Braster, M.; Van Straalen, N.M.; Röling, W.F. Spatial heterogeneity in sediment-associated bacterial and eukaryotic communities in a landfill leachate-contaminated aquifer. FEMS Microbiol. Ecol. 2008, 65, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.R.; Lee, H.Y.; Chen, K.; Chang, C.C.; Tsai, D.S.; Fu, C.C.; Fann, W. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat. Nanotechnol. 2008, 3, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Market Publishers. The Global Market for Nanodiamonds 2020–2030; Future Markets Inc.: London, UK, 2021; p. 114. [Google Scholar]
- Dolmatov, V.Y.; Ozerin, A.N.; Kulakova, I.I.; Bochechka, O.O.; Lapchuk, N.M.; Myllymäki, V.; Vehanen, A. Detonation nanodiamonds: New aspects in the theory and practice of synthesis, properties and applications. Russ. Chem. Rev. 2020, 89, 1428. [Google Scholar] [CrossRef]
- Yotinov, I.; Todorova, Y.; Schneider, I.; Daskalova, E.; Topalova, Y. The effect of nanodiamonds on phenol biodegradation by Pseudomonas sp. strain isolated from polluted sediments. J. Nanosci. Nanotechnol. 2016, 16, 7696–7706. [Google Scholar] [CrossRef]
- Perevedentseva, E.; Cheng, C.Y.; Chung, P.H.; Tu, J.S.; Hsieh, Y.H.; Cheng, C.L. The interaction of the protein lysozyme with bacteria E. coli observed using nanodiamond labelling. Nanotechnology 2007, 18, 315102. [Google Scholar] [CrossRef]
- Huang, H.; Pierstorff, E.; Osawa, E.; Ho, D. Active nanodiamond hydrogels for chemotherapeutic delivery. Nano Lett. 2007, 7, 3305–3314. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jaeeun, K.; Jaeyoung, K.; Hyorang, K. Novel Electrode for Water Purification based on Nanodiamonds. In ECS Meeting Abstracts; IOP Publishing: Bristol, UK, 2011; Volume 38, p. 2346. [Google Scholar]
- Cordova-Rosa, S.M.; Dams, R.I.; Cordova-Rosa, E.V.; Radetski, M.R.; Corrêa, A.X.R.; Radetski, C.M. Remediation of phenol-contaminated soil by a bacterial consortium and Acinetobacter calcoaceticus isolated from an industrial wastewater treatment plant. J. Hazard. Mater. 2009, 164, 61–66. [Google Scholar] [CrossRef] [PubMed]
- ISO. BDS EN ISO 6439-1990 Water Quality—Determiantion of Phenol Index—4-Aminoantipyrine Spectrophotometric Methods after Distillation; ISO: Geneva, Switzerland, 1990. [Google Scholar]
- Kopp, J.F.; McKee, G.D. Methods for Chemical Analysis of Water and Wastes; US Environmental Protection Agency: Washington, DC, USA, 1983; p. 491.
- Kuznetzov, S.I.; Dubinina, G. Methods of Investigation of Water Microorganisms; Acad. Sc. USSR: Moskow, Russia, 1989. [Google Scholar]
- Furukawa, K.; Simon, J.R.; Chakrabarty, A.M. Common induction and regulation of biphenyl, xylene/toluene, and salicylate catabolism in Pseudomonas paucimobilis. J. Bacteriol. 1983, 154, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Belouhova, M.; Topalova, Y. Structural and functional changes in biofilm during adaptation towards amaranth biodegradation. Biotechnol. Biotechnol. Equip. 2021, 35, 1769–1777. [Google Scholar] [CrossRef]
- Belouhova, M.; Dinova, N.; Yotinov, I.; Lincheva, S.; Schneider, I.; Topalova, Y. FISH investigation of the bacterial groups anammox and Azoarcus-Thauera at treatment of landfill leachate. Bulg. Chem. Commun. 2021, 53, 27. [Google Scholar] [CrossRef]
- Loy, A.; Maixner, F.; Wagner, M.; Horn, M. probeBase—an online resource for rRNA-targeted oligonucleotide probes: New features 2007. Nucleic Acids Res. 2007, 35, D800–D804. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.-H.; Amann, R.; Ludwig, W.; Rothemund, C.; Springer, N.; Dorn, S. Nucleic acid probes for the identification and in situ detection of pseudomonads. In Pseudomonas: Molecular Biology and Biotechnology; Galli, E., Silver, S., Witholt, B., Eds.; American Society for Microbiology: Washington, DC, USA, 1992; pp. 127–134. [Google Scholar]
- Stavrev, S.; Karaguiozova, Z.; Karadjov, Y.; Dragieva, I. Formation of Metal layer—Covers on Nanodiamonds Powder. In Proceedings of the 7th Workshop Nanoscience & Nanotechnology, Sofia, Bulgaria, 25–26 November 2005. [Google Scholar]
- Yur’ev, G.S.; Dolmatov, V.Y. X-ray diffraction study of detonation nanodiamonds. J. Superhard Mater. 2010, 32, 311–328. [Google Scholar] [CrossRef]
- Prokhorov, V.; Kulibaba, V.; Korobov, A.; Stavrev, S.; Kolev, S. Preparation and microstructure, elasticity and hardness investigation of alalloy/nanodiamonds composite. In Proceedings of the 8th International Congress-Mashines, Technologies Materials, Varna, Bulgaria, 18–21 September 2011; Volume 2, pp. 86–89. [Google Scholar]
- Yotinov, I.; Todorova, Y.; Schneider, I.; Daskalova, E.; Topalova, Y. Comparison of the Influence of Nanodiamonds and Single-Walled Nanotubes on Phenol Biodetoxification by Pseudomonas sp. J. Nanosci. Nanotechnol. 2017, 17, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Lincheva, S.; Schneider, I.; Daskalova, E.; Topalova, Y. Modelling of risk effect of mercury on nutrient transformation in lake sediments. Biotechnol. Biotechnol. Equip. 2014, 28, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Topalova, Y. Biological Control and Management of Water Treatment; ScieSet-Eco: Sofia, Bulgaria, 2008; p. 352. [Google Scholar]
- Lincheva, S.; Shneider, I.; Topalova, Y. Dynamics of the Functional Structure of the Sediment Microflora during the Model Process of Phenol Biodegradation. Bulg. J. Agric. Sci. 2013, 19, 139–142. [Google Scholar]
- Belouhova, M. Management of Water Purification from Biofilm Communities in Innovative Detoxification Technologies. Ph.D. Dissertation, Sofia University “St. Kliment Ohridski”, Sofia, Bulgaria, 2015; p. 201. [Google Scholar]
- Grekova-Vasileva, M. Bioalgorithms for Water Treatment Management from the Textile Industry. Ph.D. Disertation, Sofia University ‘St. Kliment Ohridski”, Sofia, Bulgaria, 2009; p. 172. [Google Scholar]
- Dimkov, R.; Topalova, Y. Dependence of microbiological degradation of the aryl-containing xenobioics on their concentrations. Acta Microbiol. Bulg. 1993, 29, 9–16. [Google Scholar]
- Topalova, Y.; Dimkov, R. Role of the factors–controlling aryl-biodegradation in the process management, biostatistics and applicability. Biotechnol. Biotechnol. Equip. 2003, 17, 104–111. [Google Scholar]
- Grekova-Vasileva, M.; Topalova, Y. Biological algorithms for textile wastewater management. Biotechnol. Biotechnol. Equip. 2009, 23, 442–447. [Google Scholar] [CrossRef]



| Microbiological Indicator | Nutrient Media | Producer | Incubation Conditions |
|---|---|---|---|
| Aerobic heterotrophic bacteria (AeH) | Nutrient agar in aerobic conditions | HiMedia | 24 h, 37 °C |
| Genus Pseudomonas (Ps.) | Glutamate starch Pseudomonas agar | HiMedia | 24 h, 37 °C |
| Genus Acinetobacter (Ac.) | Sellers differential agar | HiMedia | 24 h, 37 °C |
| Phenol-degrading bacteria (Ph.degr.) | Synthetic salt medium with added phenol | according to Furukawa, 1983 | 2–7 days, 28 °C |
| Parameters | Variants | 48th hour | 214th hour |
|---|---|---|---|
| Rate of biodegradation of phenol /RBP/mg/kgxh | Ph | 1.598 ± 0.166 | 4.578 ± 0.295 |
| PhND | 0.891 ± 0.117 | 5.446 ± 0 | |
| Specific rate of biodegradation of phenol/SRBP/mg/kgxh | Ph | 0.211 ± 0.022 | 0.348 ± 0.027 |
| PhND | 0.100 ± 0.016 | 0.436 ± 0 | |
| Rate of accumulation of biomass /RAB/mg/lxh | Ph | −0.082 ± 0.053 | 0.034 ± 0.008 |
| PhND | −0.054 ± 0.028 | 0.021 ± 0.008 | |
| Effectiveness of elimination of phenol/EFF./% | Ph | 26.15 ± 1.976 | 29.64 ± 1.638 |
| PhND | 16.81 ± 2.260 | 33.42 ± 0.486 |
| Groups of M.O. | Change (%) versus Variant Ph at 48 h | Change (%) versus Variant Ph at 214 h |
|---|---|---|
| AeH | +33.87% | −19.45% |
| Ps. | −76.30% | +71.83% |
| Ac. | +74.65% | −37.58% |
| Ph.degr. | −60.61% | −75.35% |
| 2nd hour | 48th hour | 144th hour | 214th hour |
 Ps. /FISH/- 2nd hour | 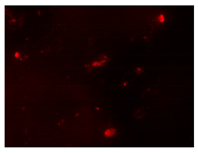 Ps. /FISH/-48th hour | 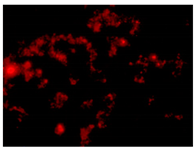 Ps. /FISH/-144th hour | 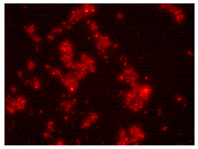 Ps. /FISH/-214th hour |
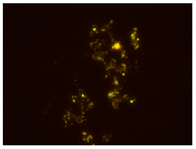 Ac. /FISH/-2nd hour |  Ac. /FISH/-48th hour | 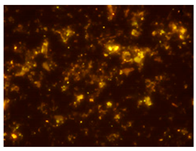 Ac. /FISH/-144th hour | 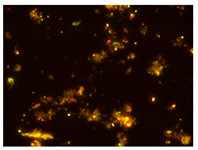 Ac. /FISH/-214th hour |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yotinov, I.; Belouhova, M.; Foteva, A.; Dinova, N.; Todorova, Y.; Schneider, I.; Daskalova, E.; Topalova, Y. Application of Nanodiamonds in Modelled Bioremediation of Phenol Pollution in River Sediments. Processes 2022, 10, 602. https://doi.org/10.3390/pr10030602
Yotinov I, Belouhova M, Foteva A, Dinova N, Todorova Y, Schneider I, Daskalova E, Topalova Y. Application of Nanodiamonds in Modelled Bioremediation of Phenol Pollution in River Sediments. Processes. 2022; 10(3):602. https://doi.org/10.3390/pr10030602
Chicago/Turabian StyleYotinov, Ivaylo, Mihaela Belouhova, Anna Foteva, Nora Dinova, Yovana Todorova, Irina Schneider, Elmira Daskalova, and Yana Topalova. 2022. "Application of Nanodiamonds in Modelled Bioremediation of Phenol Pollution in River Sediments" Processes 10, no. 3: 602. https://doi.org/10.3390/pr10030602
APA StyleYotinov, I., Belouhova, M., Foteva, A., Dinova, N., Todorova, Y., Schneider, I., Daskalova, E., & Topalova, Y. (2022). Application of Nanodiamonds in Modelled Bioremediation of Phenol Pollution in River Sediments. Processes, 10(3), 602. https://doi.org/10.3390/pr10030602