Design of Experiments for Modeling of Fermentation Process Characterization in Biological Drug Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Bacterial Strains, and Vectors
2.2. Protein Expression, Fermentation, and Purification
2.3. Scaling Up Attempts
2.4. Model Building in Design Expert
- Create the new design;
- Use the default regular two-level factorial design to create the table;
- Choose the number of variables (insert additional runs if needed);
- Model building: generate both the factorial design model and the polynomial design models;
- Select candidate models aiming at low AICc and BIC values;
- Validation: compare the models by high R2, high adjusted R2, low predicted residual error sum of squares (PRESS), low p-values in variables, high p-values in lack of fit, low Std. Dev. (also called root mean square error, RMSE) and low (relative) coefficient of variation (C.V.%), monitor summary of fit and ANOVA;
- Perform model diagnostic analysis to study the residuals.
2.5. Model Building in JMP
3. Results
3.1. The Existing Data and the Analyses
3.2. Optimization and Verification
3.3. Final Set of Data and the Characterization of the Model
3.4. Final Set of Data Executed by JMP
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICH. Pharmaceutical Development Q8(R2). Step 4 Version, August 2009. Available online: https://database.ich.org/sites/default/files/Q8_R2_Guideline.pdf (accessed on 26 November 2021).
- U.S. Dood & Drug Administration. What Are the Points to Consider for the Role of Models in Quality by Design (qbd). Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q8-q9-q10-questions-and-answers-appendix-qas-training-sessions-q8-q9-q10-points-consider#V._What_are_the_QBD_ (accessed on 26 November 2021).
- U.S. Dood & Drug Administration. Quality by Design—Fda Lessons Learned and Challenges for International Harmonization. Available online: https://www.fda.gov/media/85369/download (accessed on 25 November 2021).
- Tang, Y.; U.S. Dood & Drug Administration. Quality by Design Approaches to Analytical Methods—Fda Perspective. Available online: https://www.fda.gov/media/84046/download (accessed on 25 November 2021).
- Lebrazi, S.; Niehaus, K.; Bednarz, H.; Fadil, M.; Chraibi, M.; Fikri-Benbrahim, K. Screening and optimization of indole-3-acetic acid production and phosphate solubilization by rhizobacterial strains isolated from acacia cyanophylla root nodules and their effects on its plant growth. J. Genet. Eng. Biotechnol. 2020, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- Lebrazi, S.; Fadil, M.; Chraibi, M.; Fikri-Benbrahim, K. Screening and optimization of indole-3-acetic acid production by rhizobium sp. Strain using response surface methodology. J. Genet. Eng. Biotechnol. 2020, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Giovanni, M. Response surface methodology and product optimization. Food Technol. 1983, 37, 5. [Google Scholar]
- Chidambaram, N. Cder Regulatory Applications—Investigational New Drug and New Drug Applications. Available online: https://www.fda.gov/media/91746/download (accessed on 26 November 2021).
- U.S. Dood & Drug Administration. Pat—A-Framework-for-Innovative-Pharmaceutical-Development-Manufacturing-And-QUALITY-assurance. Available online: https://www.fda.gov/media/71012/download (accessed on 26 November 2021).
- Kelley, B. Industrialization of mab production technology: The bioprocessing industry at a crossroads. Mabs 2009, 1, 443–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appel, R.D.; Bairoch, A.; Hochstrasser, D.F. A new generation of information retrieval tools for biologists: The example of the expasy www server. Trends Biochem. Sci. 1994, 19, 258–260. [Google Scholar] [CrossRef]
- Available online: Http://au.Expasy.Org/tools/protparam-doc.Html, http://au.Expasy.Org/cgi-bin/protparam (accessed on 26 November 2021).
- Kenneth, P.; Burnham, D.R.A. Multimodel inference: Understanding aic and bic in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar]
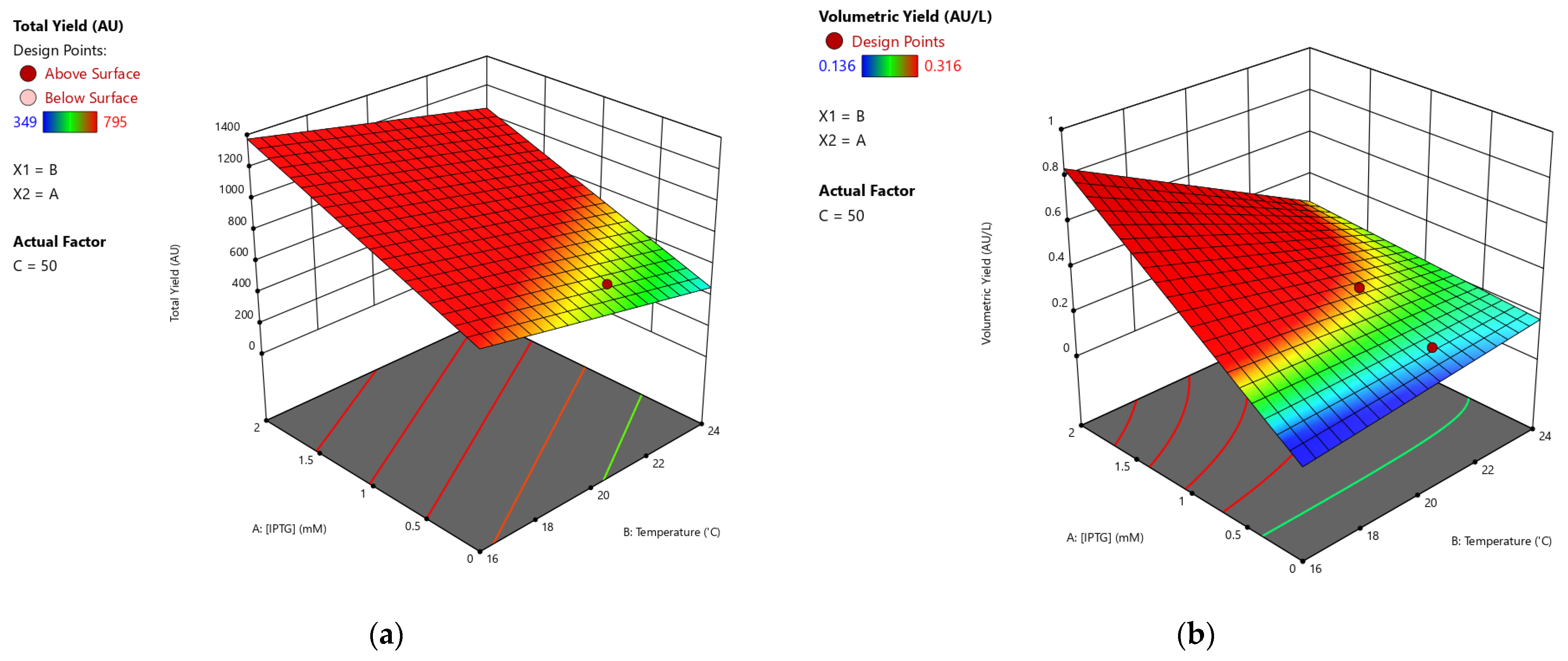

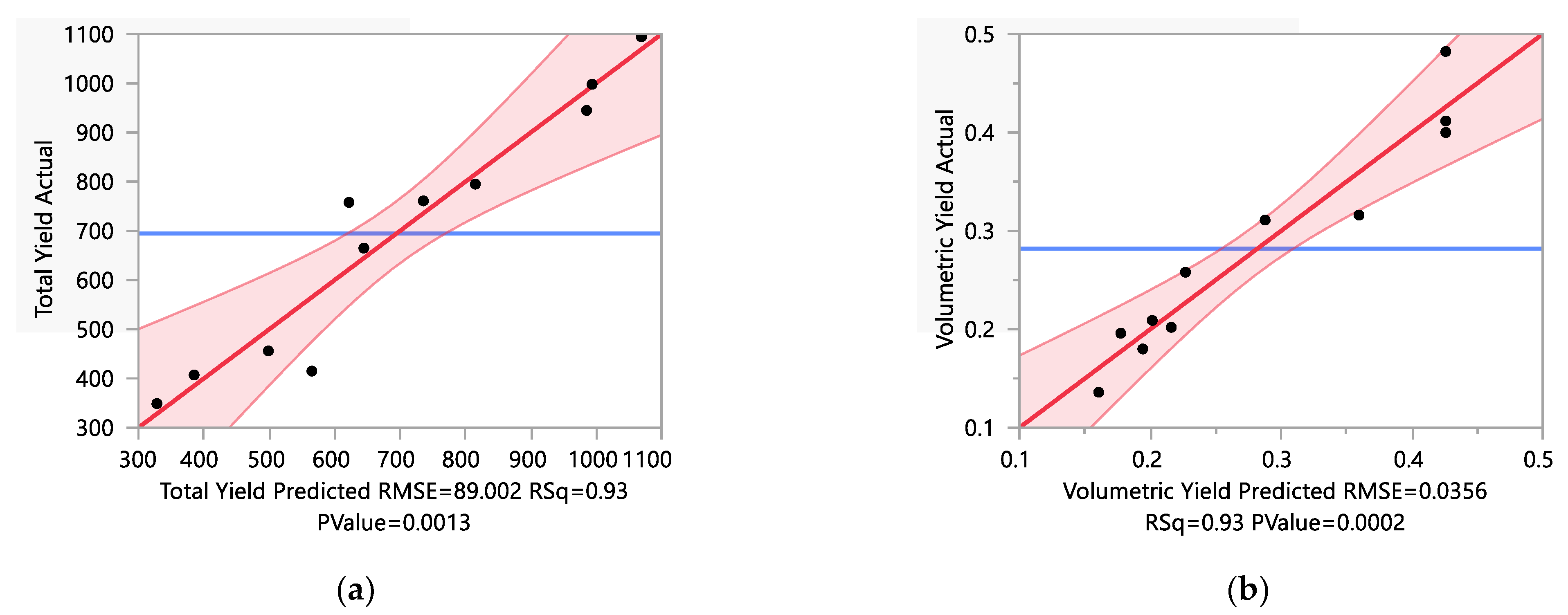
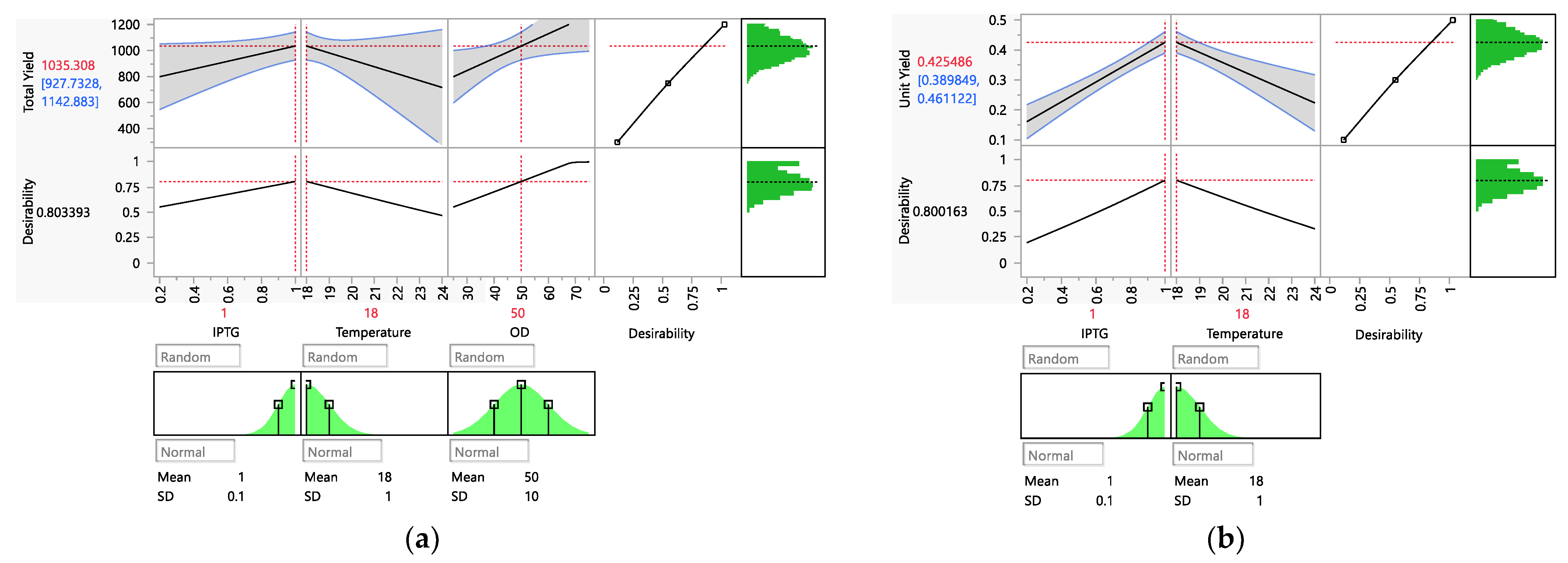
| Run | Variables | Responses | |||
|---|---|---|---|---|---|
| [IPTG] (mM) | Temperature (°C) | OD (AU) | Total Yield (AU) | Volumetric Yield (AU/L) | |
| 1 | 0.2 | 18 | 25 | 415 | 0.136 |
| 2 | 0.4 | 18 | 25 | 758 | 0.258 |
| 3 | 0.8 | 18 | 25 | 761 | 0.316 |
| 4 | 0.2 | 24 | 75 | 349 | 0.180 |
| 5 | 0.8 | 21 | 50 | 795 | 0.311 |
| 6 | 0.2 | 21 | 50 | 665 | 0.196 |
| 7 | 0.8 | 24 | 75 | 456 | 0.202 |
| 8 | 0.4 | 24 | 75 | 407 | 0.209 |
| 9 | 1 | 18 | 48 | 945 | 0.482 |
| 10 | 1 | 18 | 49 | 998 | 0.400 |
| 11 | 1 | 18 | 58 | 1094 | 0.412 |
| Analysis of Variance | ||||
| Source | DF | Sum of Squares | Mean Squares | F Ratio |
| Model | 4 | 623,649.62 | 155,912 | 19.6824 |
| Error | 6 | 47,528.51 | 7921 | |
| C. total | 10 | 671,178.14 | ||
| Parameter Estimates | ||||
| Term | Estimate | Std Error | t Ratio | Prob > |t| |
| Intercept | 758.4354 | 64.0169 | 11.85 | <0.0001 |
| IPTG (0.2,1) | 113.7417 | 46.8964 | 2.43 | 0.0515 |
| Temperature (18,24) | −129.9934 | 95.2465 | −1.36 | 0.2213 |
| OD (25,75) | 11.5113 | 100.2911 | 0.11 | 0.9124 |
| Temperature*OD | −198.0394 | 72.6126 | −2.73 | 0.0343 * |
| Analysis of Variance | ||||
| Source | DF | Sum of Squares | Mean Squares | F Ratio |
| Model | 3 | 0.11546754 | 0.038489 | 30.3715 & |
| Error | 7 | 0.00887095 | 0.001267 | |
| C. total | 10 | 0.12433849 | ||
| Lack of Fit | ||||
| Lack of fit | 5 | 0.00489903 | 0.00098 | 0.4934 |
| Pure error | 2 | 0.00397192 | 0.001986 | |
| Total error | 7 | 0.00887095 | ||
| Parameter Estimates | ||||
| Term | Estimate | Std Error | t Ratio | Prob > |t| |
| Intercept | 0.2510369 | 0.012421 | 20.21 | <0.0001 |
| IPTG (0.2,1) | 0.0736129 | 0.016402 | 4.49 | 0.0028 * |
| Temperature (18,24) | −0.042237 | 0.013629 | −3.1 | 0.0173 * |
| IPTG * Temperature | −0.059079 | 0.017984 | −3.29 | 0.0134 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsueh, K.-L.; Lin, T.-Y.; Lee, M.-T.; Hsiao, Y.-Y.; Gu, Y. Design of Experiments for Modeling of Fermentation Process Characterization in Biological Drug Production. Processes 2022, 10, 237. https://doi.org/10.3390/pr10020237
Hsueh K-L, Lin T-Y, Lee M-T, Hsiao Y-Y, Gu Y. Design of Experiments for Modeling of Fermentation Process Characterization in Biological Drug Production. Processes. 2022; 10(2):237. https://doi.org/10.3390/pr10020237
Chicago/Turabian StyleHsueh, Kuang-Lung, Tzung-Yi Lin, Meng-Tse (Gabriel) Lee, Ya-Yun Hsiao, and Yesong Gu. 2022. "Design of Experiments for Modeling of Fermentation Process Characterization in Biological Drug Production" Processes 10, no. 2: 237. https://doi.org/10.3390/pr10020237
APA StyleHsueh, K.-L., Lin, T.-Y., Lee, M.-T., Hsiao, Y.-Y., & Gu, Y. (2022). Design of Experiments for Modeling of Fermentation Process Characterization in Biological Drug Production. Processes, 10(2), 237. https://doi.org/10.3390/pr10020237






