Bearing Fault Diagnosis Based on a Novel Adaptive ADSD-gcForest Model
Abstract
:1. Introduction
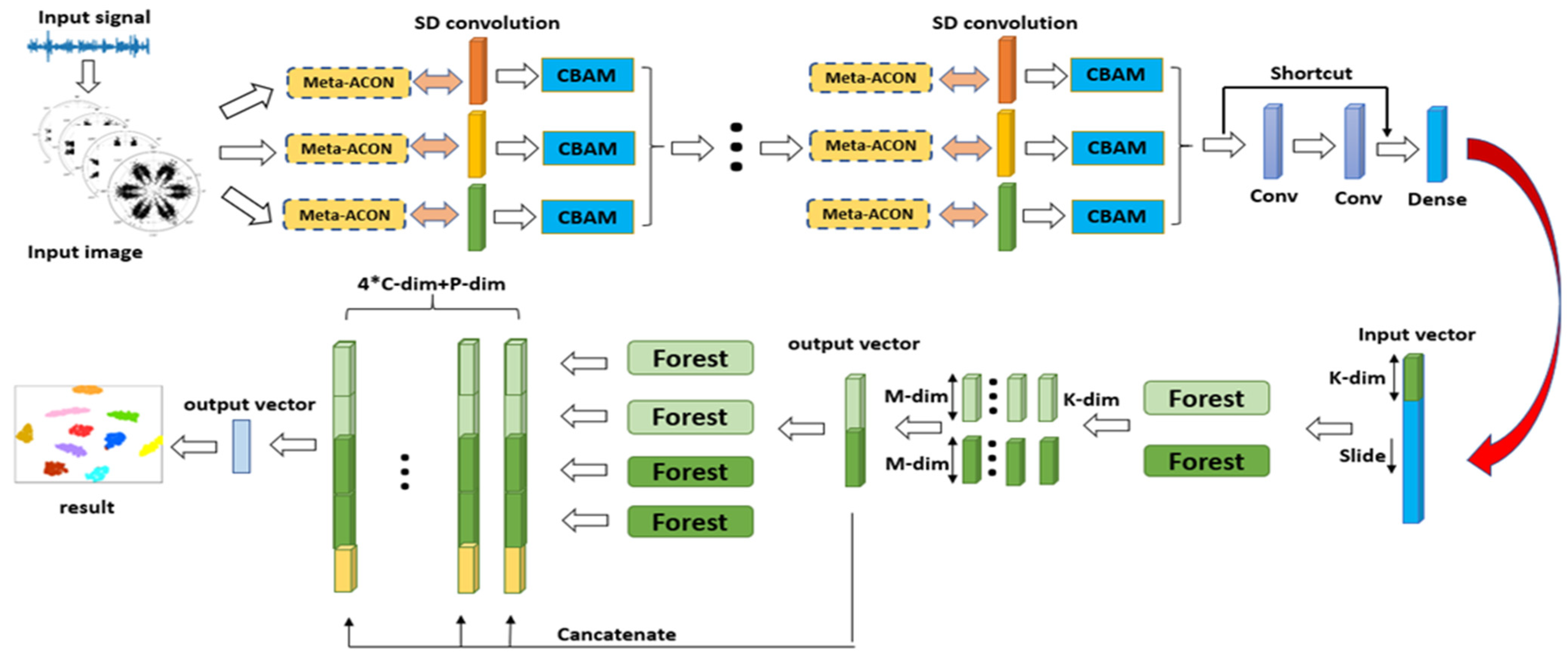
- An adaptive ADSD-gcForest diagnostic model is proposed for the diagnosis of rolling bearing fault diagnosis, allowing the extraction of features under the high-noise and complex working conditions that could be realized. The structure of the diagnosis model achieves adaptive optimization based on the characteristics of the sample data.
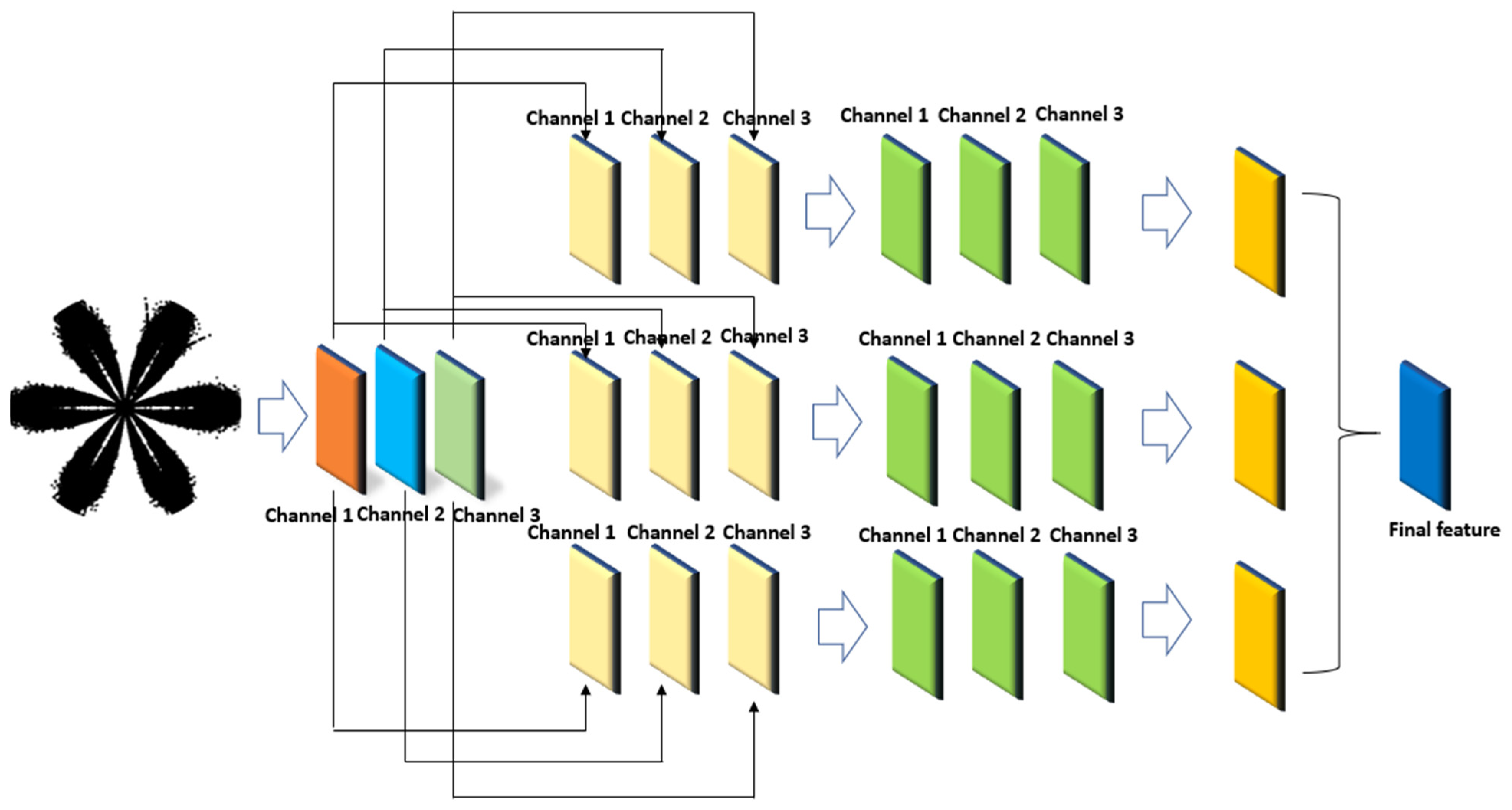
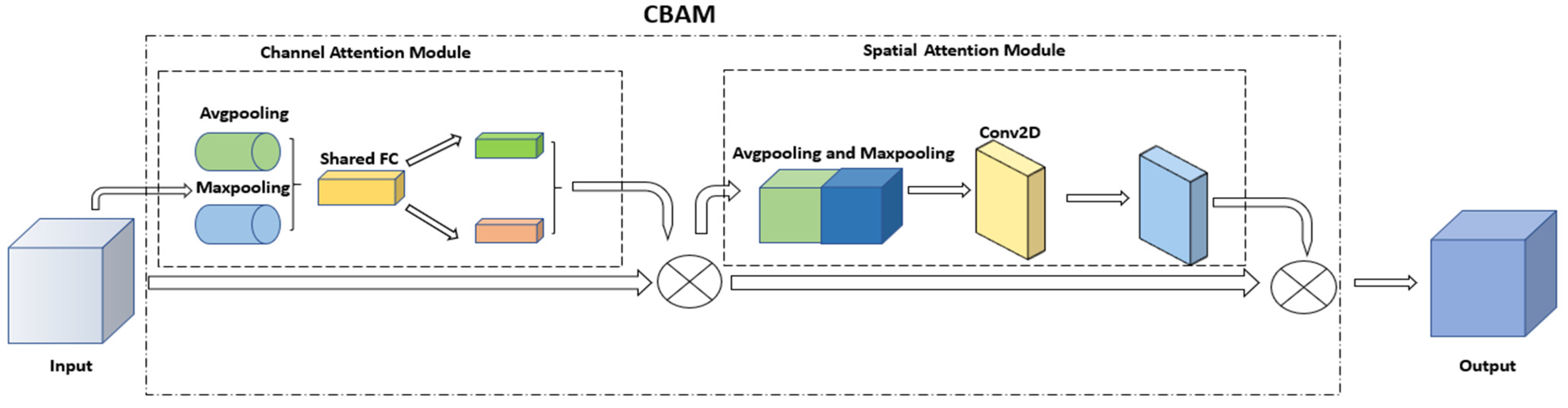
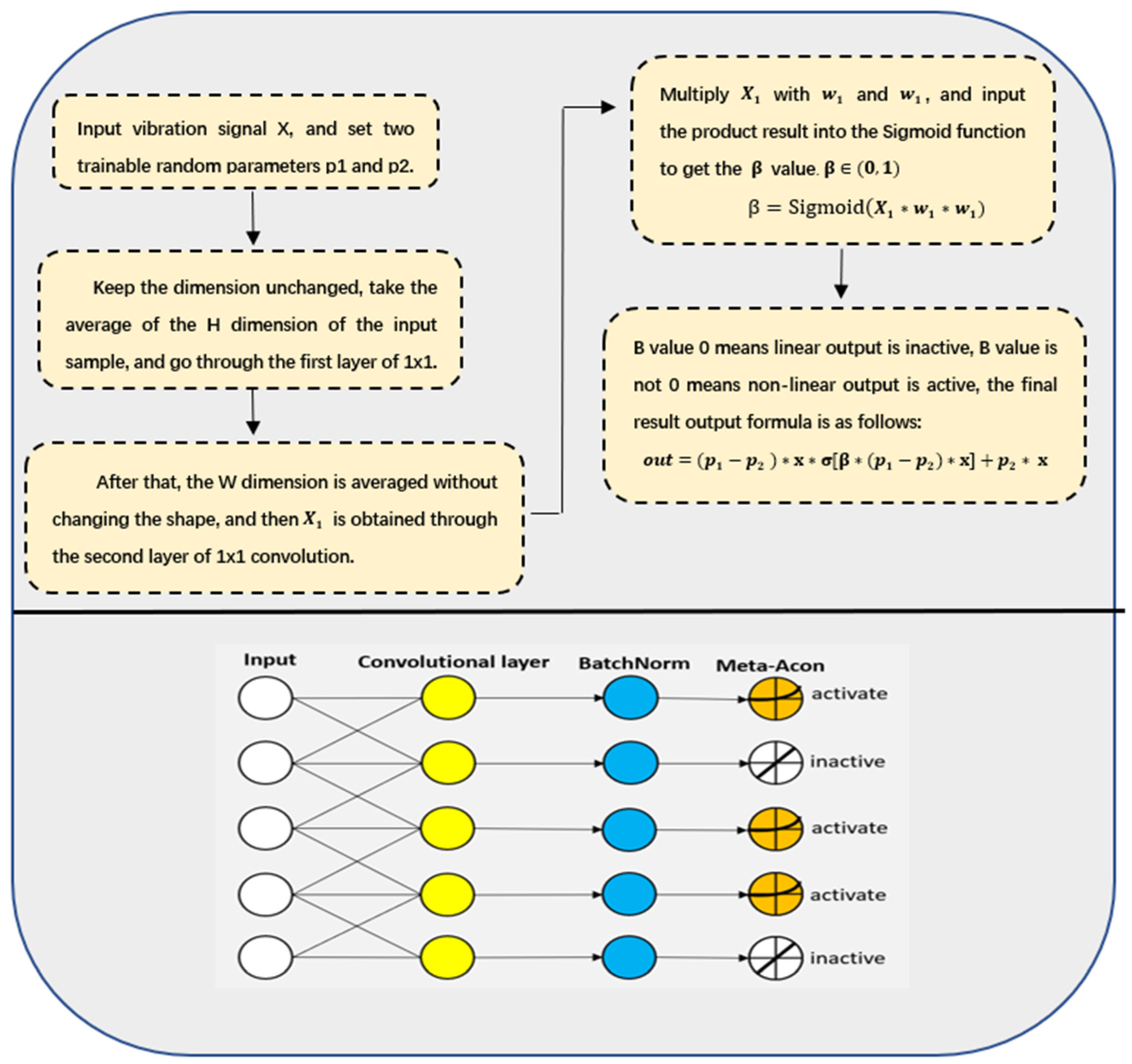
- Combining the multiscale depth-separable dilated convolution with CBAM can effectively extract fault features under strong noise interference. On the basis of the lack of adjust the original structure of the model, the Meta-ACON activation function is introduced into the convolution layer of the model to achieve adaptive optimization of the model structure according to the fault data of different bearings.
- The comparative experiment shows that the ADSD-gcForest model proposed in this paper has strong generalization ability and robustness with certain practical value.
2. Related Works
2.1. SDP Image
2.2. Dilated Convolutional
2.3. Depth-Separable Convolution
2.4. CBAM
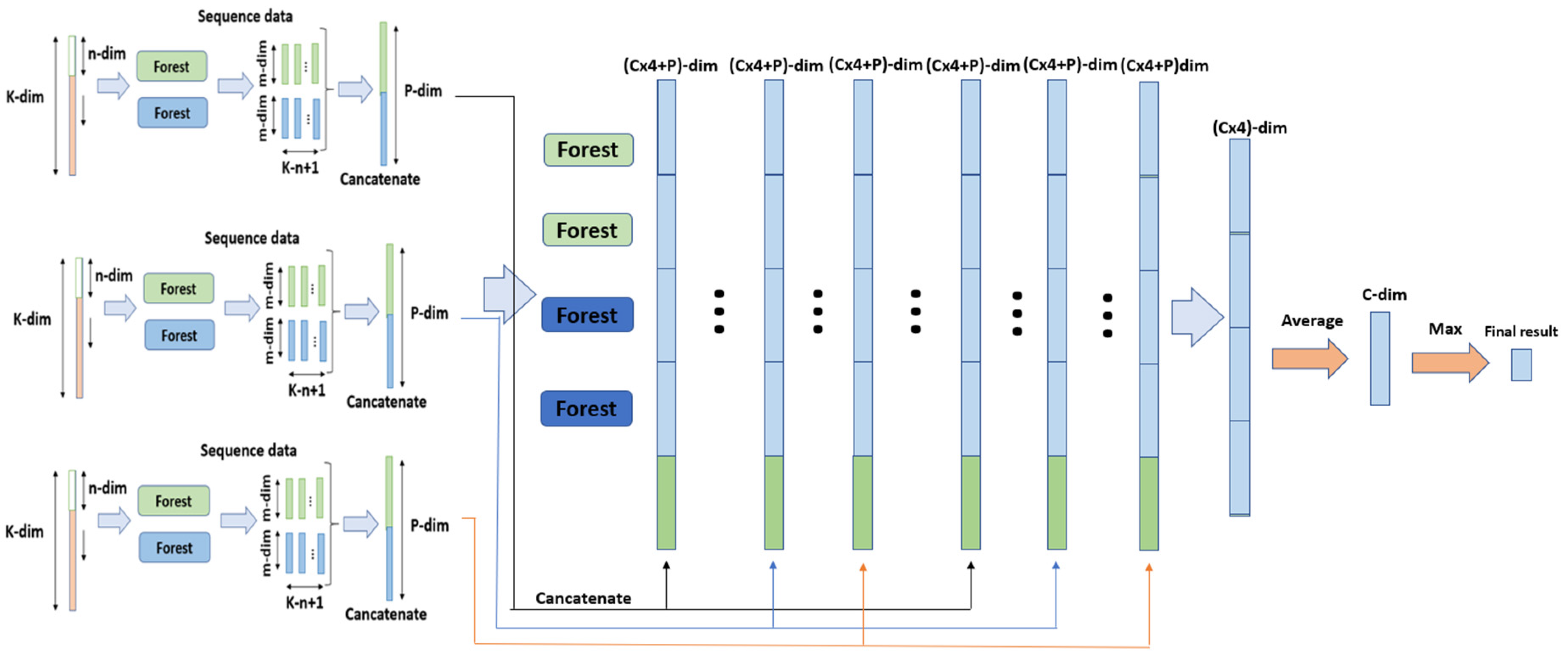
2.5. gcForest
3. Method
3.1. Meta-ACON
3.2. ADSD-gcForest
4. Experiments
4.1. Introduction of Datasets
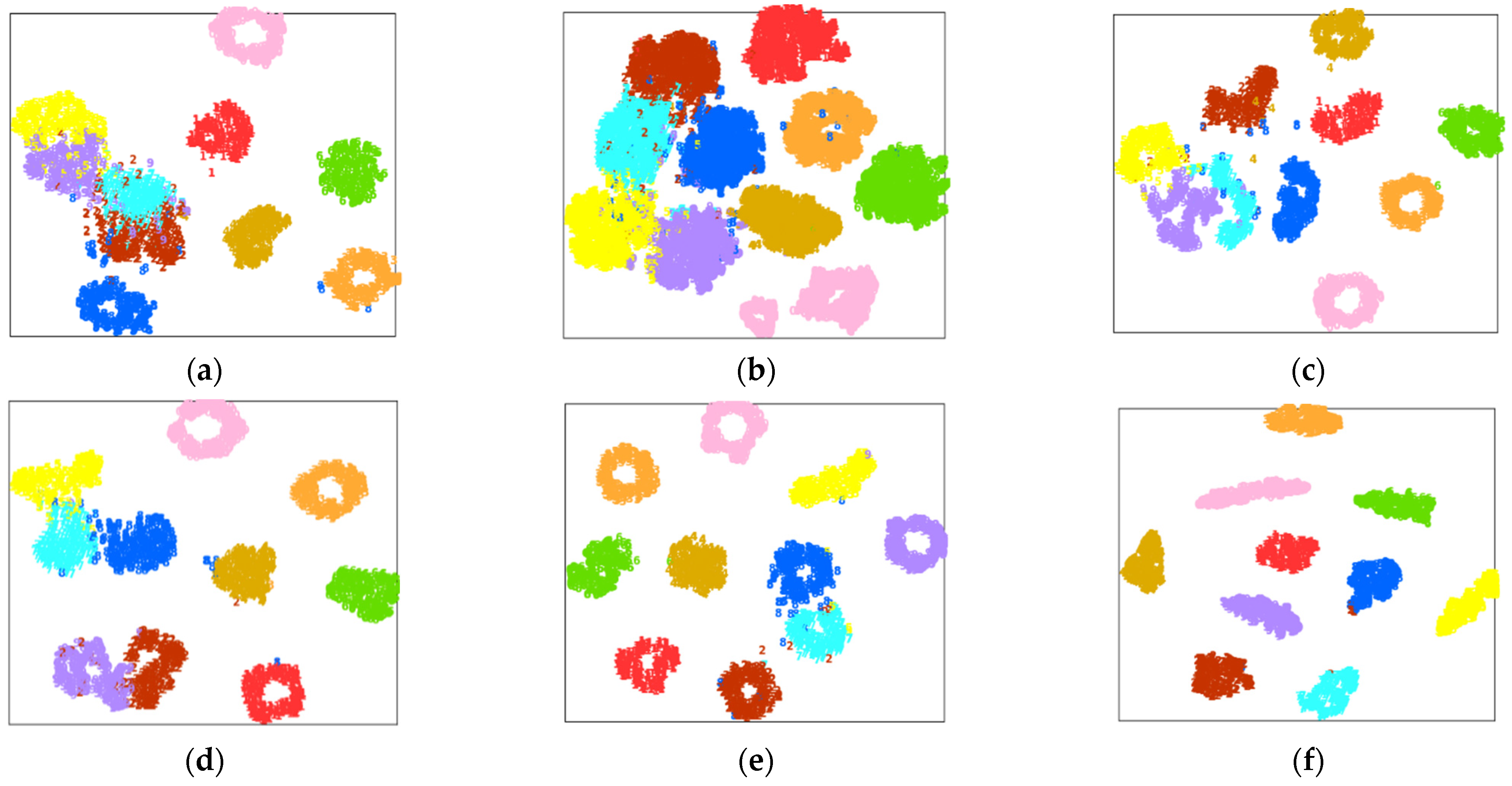
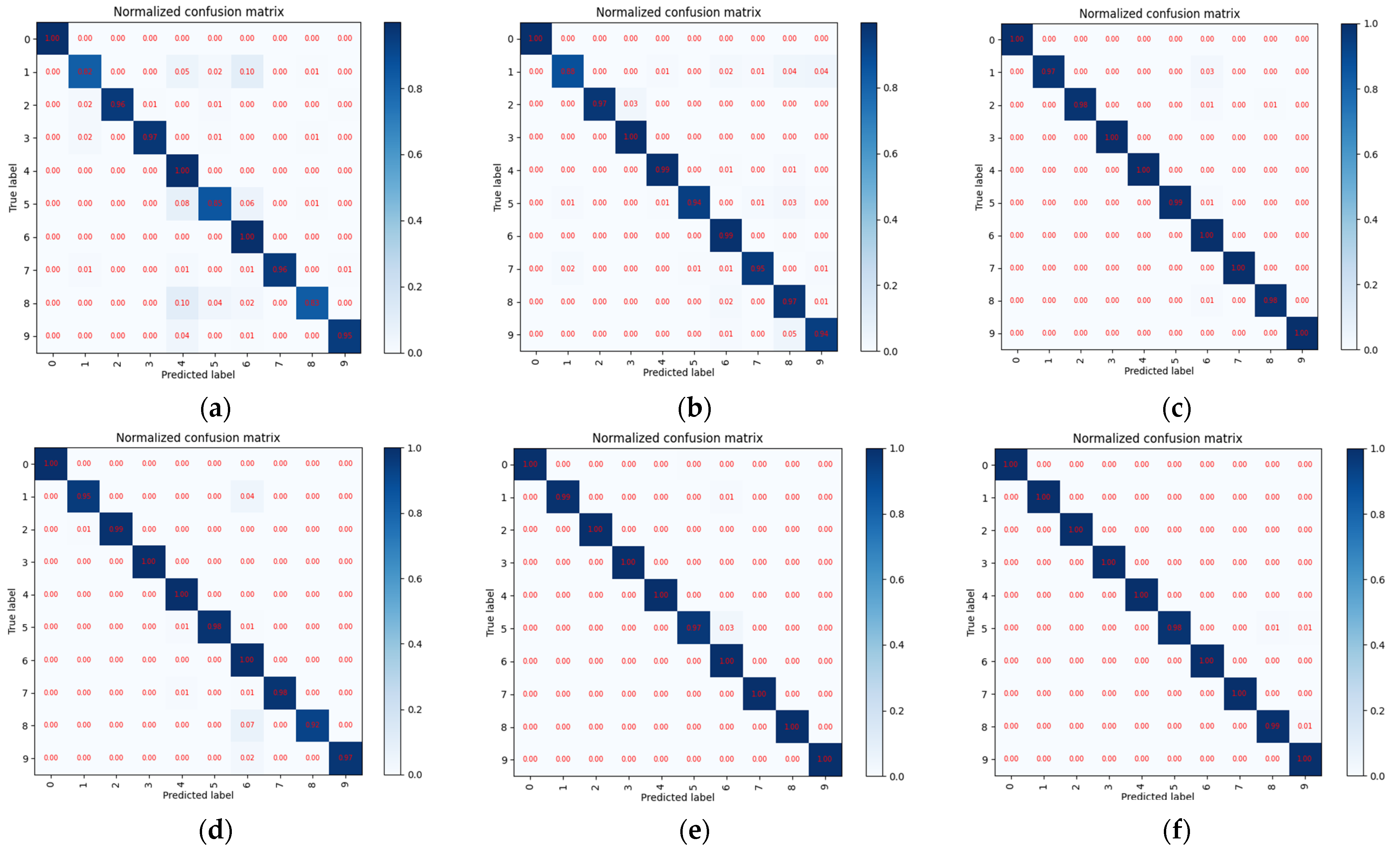
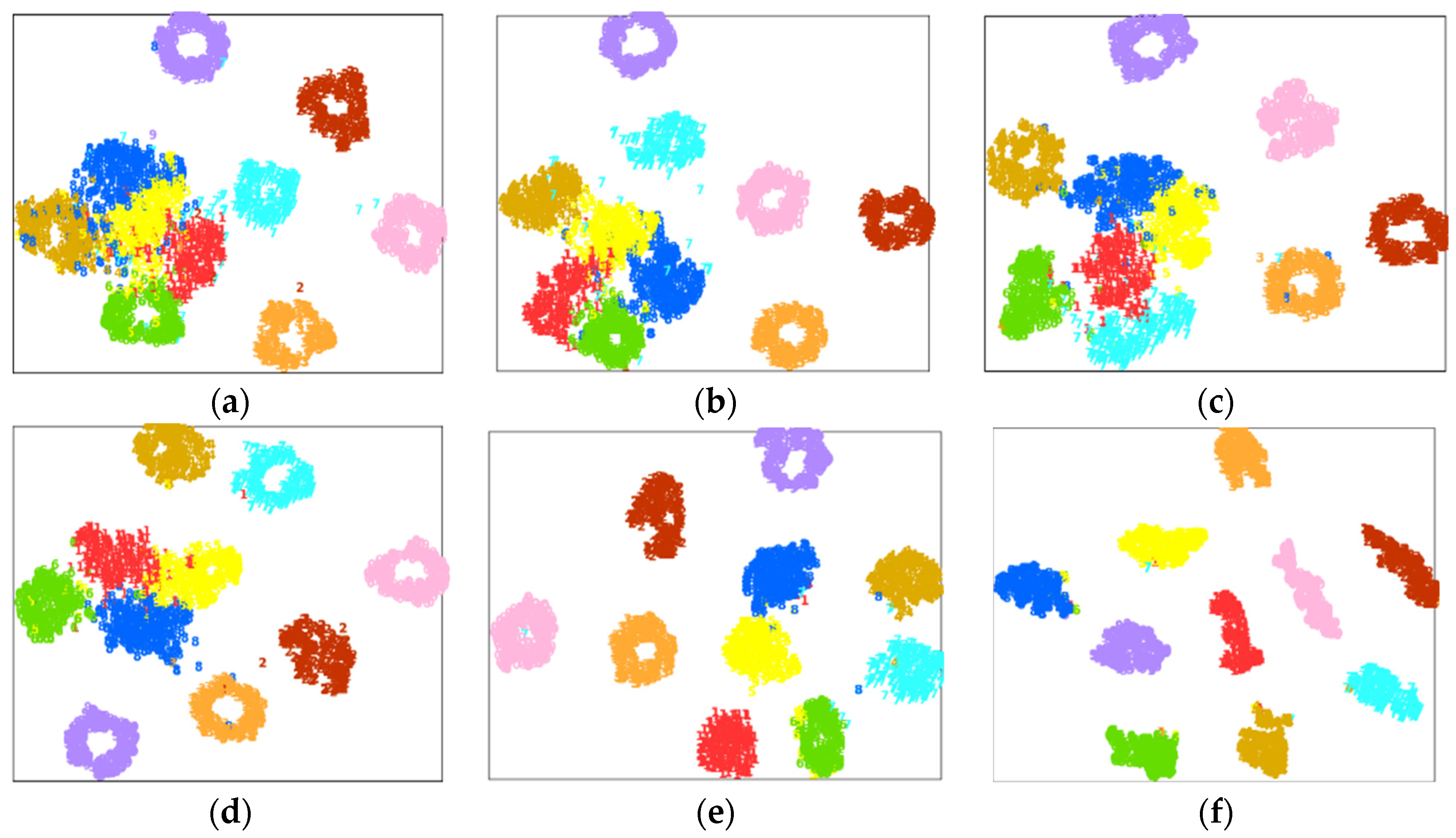
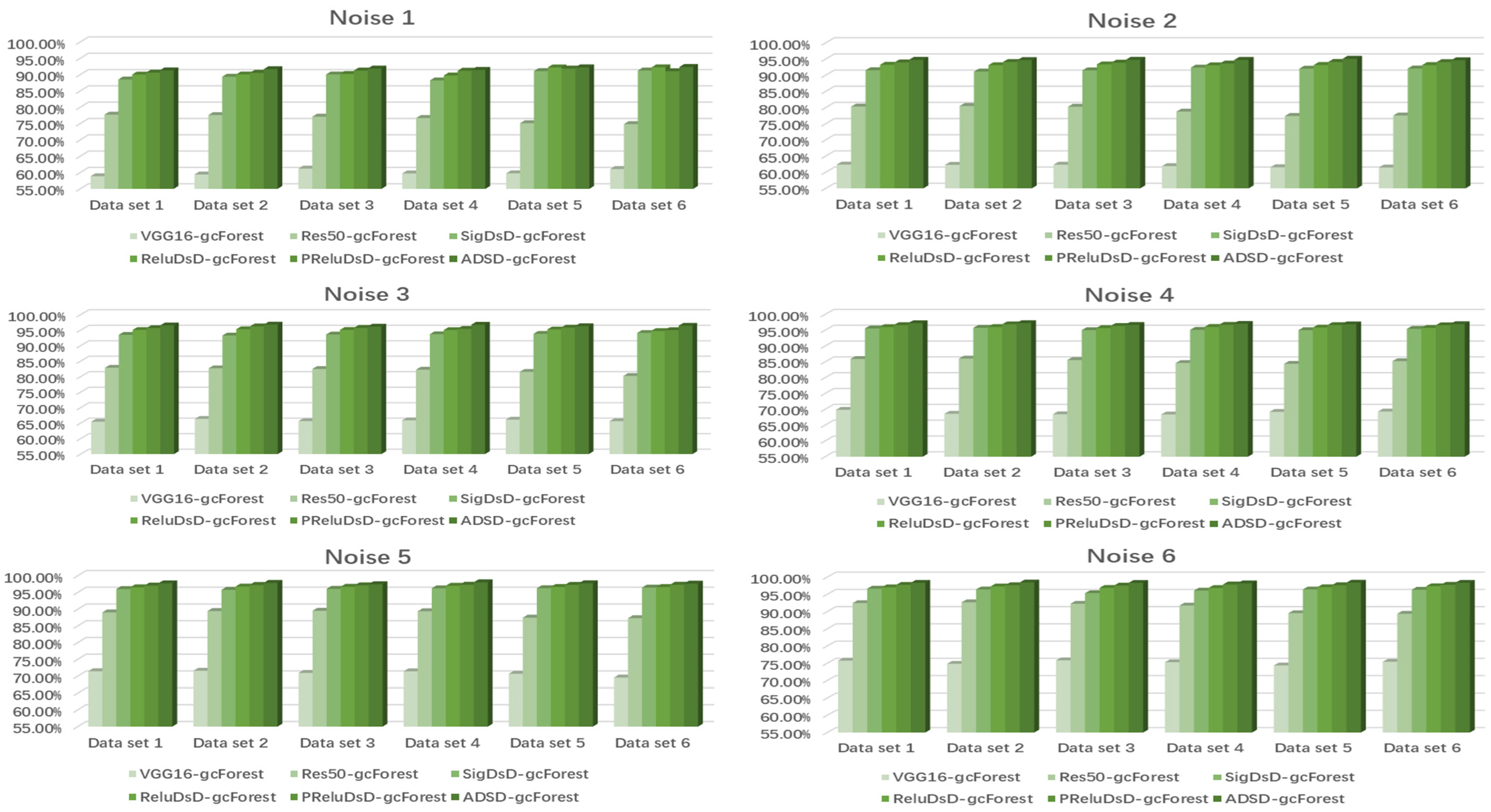
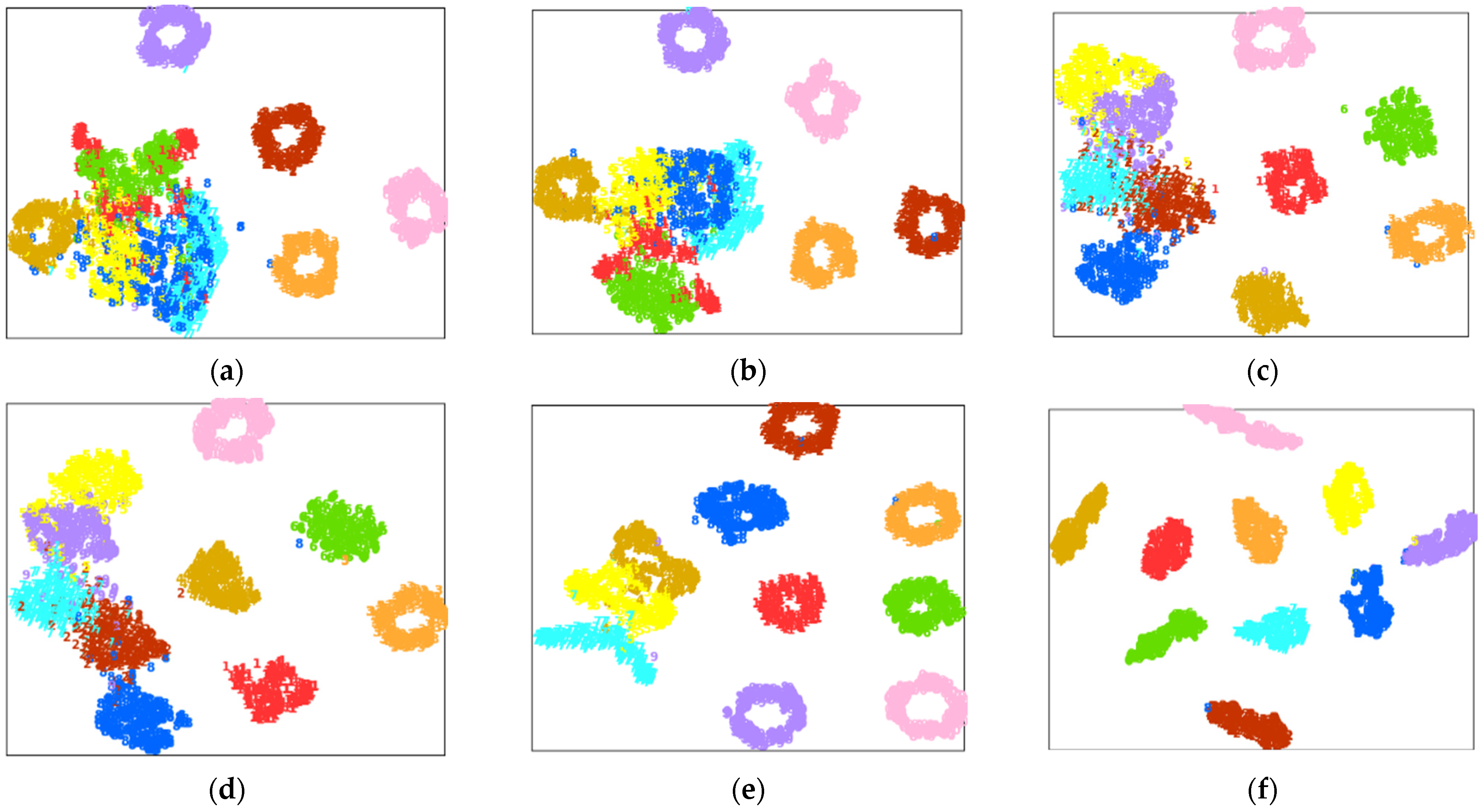
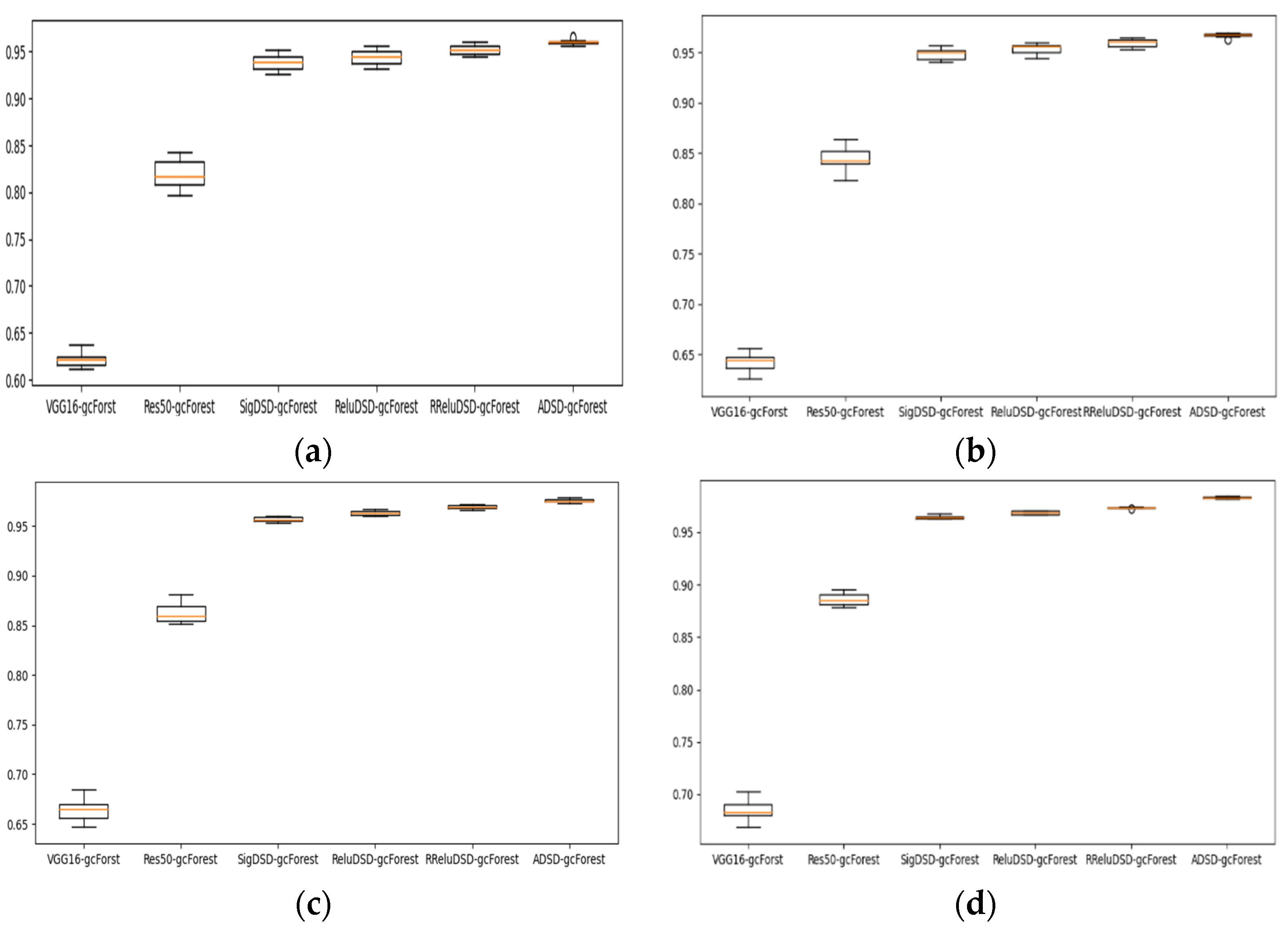
4.2. Case Study 1: Performance of Drive End Bearing Fault Diagnosis
4.3. Case Study 2: Performance of Fan End Bearing Fault Diagnosis
4.4. Case Study 3: Performance of the Ottawa Bearing Dataset
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, J.; Qi, Y.; Liu, L.; Gao, X.; Li, Y. Application of an information fusion scheme for rolling element bearing fault diagnosis. Meas. Sci. Technol. 2021, 32, 075013. [Google Scholar] [CrossRef]
- Niu, G.; Wang, X.; Golda, M.; Mastro, S.; Zhang, B. An optimized adaptive PReLU-DBN for rolling element bearing fault diagnosis. Neurocomputing 2021, 445, 26–34. [Google Scholar] [CrossRef]
- Jie, D.; Zheng, G.; Zhang, Y.; Ding, X.; Wang, L. Spectral kurtosis based on evolutionary digital filter in the application of rolling element bearing fault diagnosis. Int. J. Hydromechatronics 2021, 4, 27–42. [Google Scholar] [CrossRef]
- Zhao, X.; Qin, Y.; Fu, H.; Jia, L.; Zhang, X. Blind source extraction based on EMD and temporal correlation for rolling element bearing fault diagnosis. Smart Resilient Transp. 2021, 3, 52–65. [Google Scholar] [CrossRef]
- Hou, W.; Ye, M.; Li, W. Rolling bearing fault classification based on improved stack noise reduction self-encoding. Chin. J. Mech. Eng. 2018, 54, 87–96. [Google Scholar] [CrossRef]
- Shao, H.; Jiang, H.; Zhang, X.; Niu, M. Rolling bearing fault diagnosis using an optimization deep belief network. Meas. Sci. Technol. 2015, 26, 115002. [Google Scholar] [CrossRef]
- Liang, T.; Wu, S.; Duan, W.; Zhang, R. Bearing fault diagnosis based on improved ensemble learning and deep belief network. J. Phys. Conf. Ser. 2018, 1074, 012154. [Google Scholar] [CrossRef]
- Ma, M.; Chen, X.; Wang, S.; Liu, Y.; Li, W. Bearing degradation assessment based on weibull distribution and deep belief network. In Proceedings of the IEEE International Symposium on Flexible Automation, Cleveland, OH, USA, 1–3 August 2016; pp. 382–385. [Google Scholar]
- Shao, S.; Sun, W.; Wang, P.; Gao, R.X.; Yan, R. Learning features from vibration signals for induction motor fault diagnosis. In Proceedings of the IEEE International Symposium on Flexible Automation, Cleveland, OH, USA, 1–3 August 2016; pp. 71–76. [Google Scholar]
- Han, T.; Yuan, J.H.; Tang, J.; An, L.Z. Intelligent composite fault diagnosis method of rolling bearing based on MWT and CNN. Mech. Transm. 2016, 40, 139–143. [Google Scholar]
- Liang, M.; Cao, P.; Tang, J. Tang. Rolling bearing fault diagnosis based on feature fusion with parallel convolutional neural network. Int. J. Adv. Manuf. Technol. 2020, 112, 819–831. [Google Scholar] [CrossRef]
- Pan, H.; He, X.; Tang, S.; Meng, F. An improved bearing fault diagnosis method using one-dimensional CNN and LSTM. J. Mech. Eng. 2018, 64, 443–452. [Google Scholar]
- Zhang, L.; Jing, L.; Xu, W.; Tan, J. Rolling bearing fault diagnosis based on convolutional noise reduction autoencoder and CNN. Modul. Mach. Tool Autom. Manuf. Technol. 2019, 6, 58–62. [Google Scholar]
- Yu, L.; Qu, J.; Gao, F.; Tian, Y. A novel hierarchical algorithm for bearing fault diagnosis based on stacked LSTM. Shock. Vib. 2019, 2019, 2756284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Yan, R.; Wang, J.; Mao, K. Learing to monitor machine health with convolutional bi-directional lstm networks. Sensors 2017, 17, 273. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Xu, Y.; Jia, M. Intelligent Fault Diagnosis of Rolling-Element Bearings Using a Self-Adaptive Hierarchical Multiscale Fuzzy Entropy. Entropy 2021, 23, 1128. [Google Scholar] [CrossRef] [PubMed]
- Yong, Z.; Zhang, X.; Da, N. Research on 3D Object Detection Method Based on Convolutional Attention Mechanism. J. Phys. Conf. Ser. 2021, 1848, 012097. [Google Scholar] [CrossRef]
- Cao, Q.; Yu, L.; Wang, Z.; Zhan, S.; Quan, H.; Yu, Y.; Khan, Z.; Koubaa, A. Wild Animal Information Collection Based on Depthwise Separable Convolution in Software Defined IoT Networks. Electronics 2021, 10, 2091. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, X.; Liu, M.; Sun, J. Activate or Not: Learning Customized Activation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Zhang, N.; Cui, F.; Jiang, B.; He, X. Rotating machinery gearbox fault diagnosis method integrating SDP and CNN. Ind. Control. Comput. 2021, 34, 89–91. [Google Scholar]
- Zhao, L.; Xu, L.; Liu, Y.; Liu, J.; Huang, X. Transformer mechanical fault diagnosis method based on point symmetry transformation and image matching. Trans. Chin. Soc. Electr. Eng. 2021, 36, 3614–3626. [Google Scholar]
- Yin, Q.; Yang, W.; Ran, M.; Wang, S. FD-SSD: An improved SSD object detection algorithm based on feature fusion and dilated convolution. Signal Processing Image Commun. 2021, 98, 116402. [Google Scholar] [CrossRef]
- Yuhui, Z.; Mengyao, C.; Yuefen, C.; Zhaoqian, L.; Yao, L.; Kedi, L. An Automatic Recognition Method of Fruits and Vegetables Based on Depthwise Separable Convolution Neural Network. J. Phys. Conf. Ser. 2021, 1871, 012075. [Google Scholar] [CrossRef]
- Teng, Y.; Gao, P. Generative Robotic Grasping Using Depthwise Separable Convolution. Comput. Electr. Eng. 2021, 94, 107318. [Google Scholar] [CrossRef]
- Liu, T.; Pang, B.; Zhang, L.; Yang, W.; Sun, X. Sea Surface Object Detection Algorithm Based on YOLO v4 Fused with Reverse Depthwise Separable Convolution (RDSC) for USV. J. Mar. Sci. Eng. 2021, 9, 753. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, X.; Chen, W.; Li, Y.; Wang, J. Research on Recognition of Fly Species Based on Improved RetinaNet and CBAM. IEEE Access 2020, 8, 102907–102919. [Google Scholar] [CrossRef]
- Canayaz, M. C+EffxNet: A novel hybrid approach for COVID-19 diagnosis on CT images based on CBAM and EfficientNet. Chaos Solitons Fractals 2021, 151, 111310. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Nan, F.; Wang, X. A super resolution frontal face generation model based on 3DDFA and CBAM. Displays 2021, 69, 102043. [Google Scholar] [CrossRef]
- Sun, Z.; Li, M.; Zhang, J.; Hu, B.; Qi, G.; Zhu, Y. Transient Voltage Stability Assessment Method based on gcForest. J. Phys. Conf. Ser. 2021, 1914, 012025. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, N.; Jin, S.; Xu, D.; Gao, W. Small sample color fundus image quality assessment based on gcforest. Multimed. Tools Appl. 2020, 80, 17441–17459. [Google Scholar] [CrossRef]





| Layers | Filters | Kernel_Size/Dilation Rate | Trainable Parameters | Input_Shape | Output_Shape |
|---|---|---|---|---|---|
| separable_conv_1 | 64 | 3/1 | 137 | 28 × 28 × 1 | 28 × 28 × 64 |
| CBAM_1 | 677 | 28 × 28 × 64 | 28 × 28 × 64 | ||
| separable_conv_2 | 64 | 3/2 | 137 | 28 × 28 × 1 | 28 × 28 × 64 |
| CBAM_2 | 677 | 28 × 28 × 64 | 28 × 28 × 64 | ||
| separable_conv_3 | 64 | 3/3 | 137 | 28 × 28 × 1 | 28 × 28 × 64 |
| CBAM_3 | 677 | 28 × 28 × 64 | 28 × 28 × 64 | ||
| Add_1 | 0 | 28 × 28 × 64, 28 × 28 × 64, 28 × 28 × 64 | 28 × 28 × 64 | ||
| separable_conv_4 | 128 | 3/1 | 8896 | 28 × 28 × 64 | 28 × 28 × 128 |
| CBAM_4 | 2277 | 28 × 28 × 128 | 28 × 28 × 128 | ||
| separable_conv_5 | 128 | 3/2 | 8896 | 28 × 28 × 64 | 28 × 28 × 128 |
| CBAM_5 | 2277 | 28 × 28 × 128 | 28 × 28 × 128 | ||
| separable_conv_6 | 128 | 3/3 | 8896 | 28 × 28 × 64 | 28 × 28 × 128 |
| CBAM_6 | 2277 | 28 × 28 × 128 | 28 × 28 × 128 | ||
| Add_2 | 0 | 28 × 28 × 128, 28 × 28 × 128, 28 × 28 × 128 | 28 × 28 × 128 | ||
| separable_conv_7 | 256 | 3/1 | 34,176 | 28 × 28 × 128 | 28 × 28 × 256 |
| CBAM_7 | 8949 | 25 × 28 × 256 | 28 × 28 × 256 | ||
| separable_conv_8 | 256 | 3/2 | 34,176 | 28 × 28 × 128 | 28 × 28 × 256 |
| CBAM_8 | 8949 | 25 × 28 × 256 | 28 × 28 × 256 | ||
| separable_conv_9 | 256 | 3/3 | 34,176 | 28 × 28 × 128 | 28 × 28 × 256 |
| CBAM_9 | 8949 | 25 × 28 × 256 | 28 × 28 × 256 | ||
| Add_3 | 0 | 28 × 28 × 256, 28 × 28 × 256, 28 × 28 × 256 | 28 × 28 × 256 | ||
| separable_conv_10 | 256 | 3/1 | 68,096 | 28 × 28 × 256 | 28 × 28 × 256 |
| separable_conv_11 | 256 | 3/1 | 68,096 | 28 × 28 × 256 | 28 × 28 × 256 |
| Add_4 | 0 | 28 × 28 × 256 | 28 × 28 × 256 | ||
| Flatten | 0 | 28 × 28 × 256 | 200,704 × 1 | ||
| dense_1(1000) | 200,705,000 | 200,704 × 1 | 1000 × 1 | ||
| dense_2 (256) | 256,256 | 1000 × 1 | 256 × 1 |
| Bearing Number | Dataset 1 | Dataset 2 | Dataset 3 | Dataset 4 | Dataset 5 | Dataset 6 |
|---|---|---|---|---|---|---|
| SKF6205 | Normal-1 | Normal-2 | Normal-3 | Normal-1 | Normal-2 | Normal-3 |
| B007-1 | B007-3 | B007-3 | B007-2 | B007-1 | B007-2 | |
| B014-1 | B014-3 | B014-3 | B014-2 | B014-3 | B014-3 | |
| B021-1 | B021-3 | B021-2 | B021-3 | B021-2 | B021-1 | |
| IR007-1 | IR007-1 | IR007-2 | IR007-3 | IR007-2 | IR007-1 | |
| IR014-2 | IR014-2 | IR014-2 | IR014-3 | IR014-3 | IR014-2 | |
| IR021-3 | IR021-2 | IR021-2 | IR021-2 | IR021-3 | IR021-3 | |
| OR007-1 | OR007-2 | OR007-1 | OR007-1 | OR007-3 | OR007-1 | |
| OR014-2 | OR014-2 | OR014-1 | OR014-1 | OR014-2 | OR014-1 | |
| OR021-3 | OR021-1 | OR021-2 | 0R021-2 | OR021-1 | OR021-3 | |
| SKF6203 | Normal-2 | Normal-2 | Normal-1 | Normal-3 | Normal-1 | Normal-2 |
| B007-1 | B007-1 | B007-3 | B007-2 | B007-1 | B007-3 | |
| B014-3 | B014-2 | B014-2 | B014-3 | B014-2 | B014-3 | |
| B021-3 | B021-1 | B021-2 | B021-3 | B021-2 | B021-1 | |
| IR007-3 | IR007-2 | IR007-2 | IR007-3 | IR007-2 | IR007-1 | |
| IR014-3 | IR014-1 | IR014-3 | IR014-1 | IR014-1 | IR014-1 | |
| IR021-3 | IR021-3 | IR021-1 | IR021-1 | IR021-3 | IR021-3 | |
| OR007-2 | OR007-2 | OR007-1 | OR007-1 | OR007-1 | OR007-2 | |
| OR014-2 | OR014-2 | OR014-3 | OR014-2 | OR014-3 | OR014-2 | |
| OR021-3 | OR021-3 | OR021-1 | OR021-2 | OR021-3 | OR021-1 |
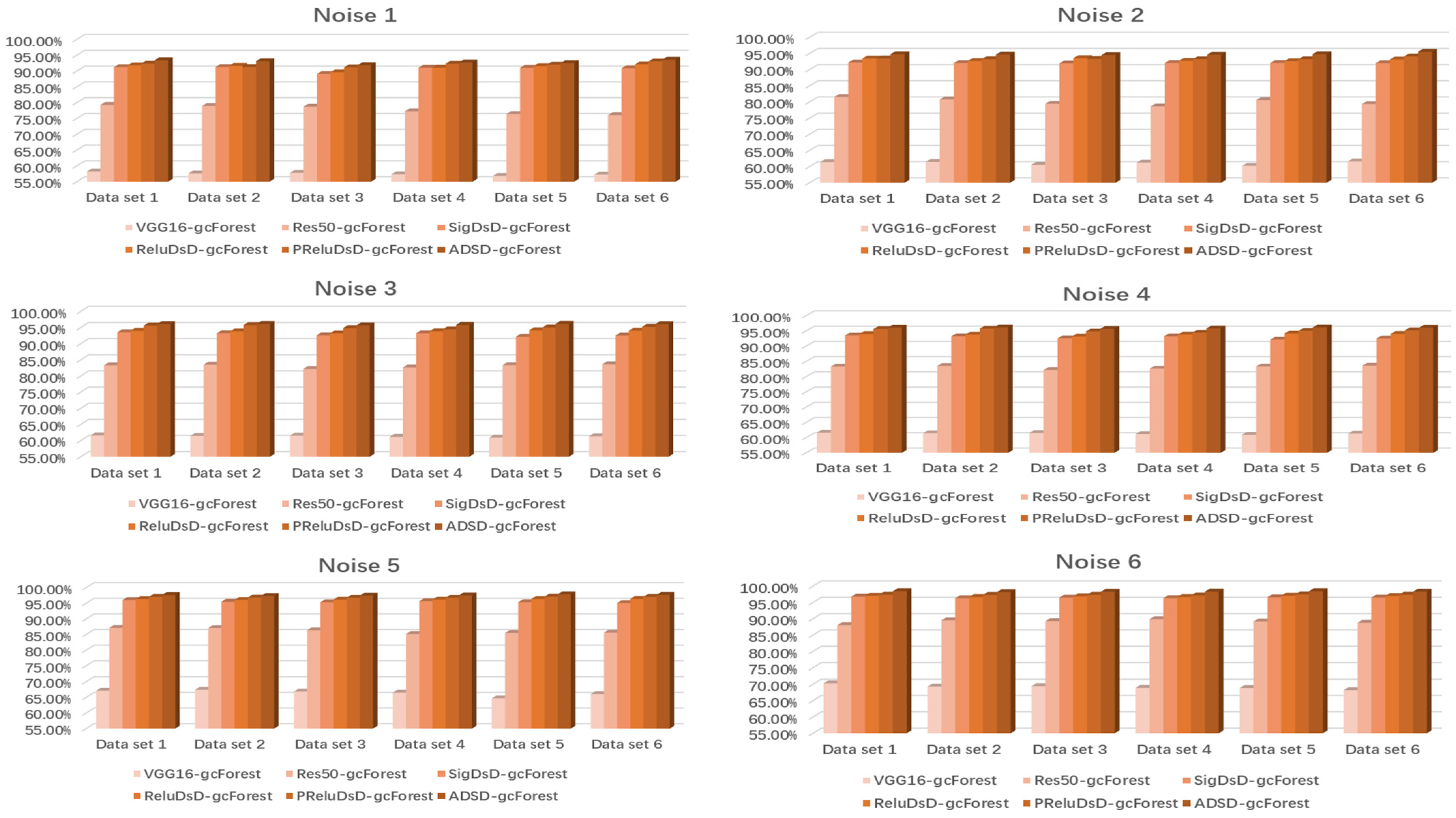
| Dataset | Methods | N1(AC/F1) | N2(AC/F1) | N3(AC/F1) | N4(AC/F1) | N5(AC/F1) | N6(AC/F1) |
|---|---|---|---|---|---|---|---|
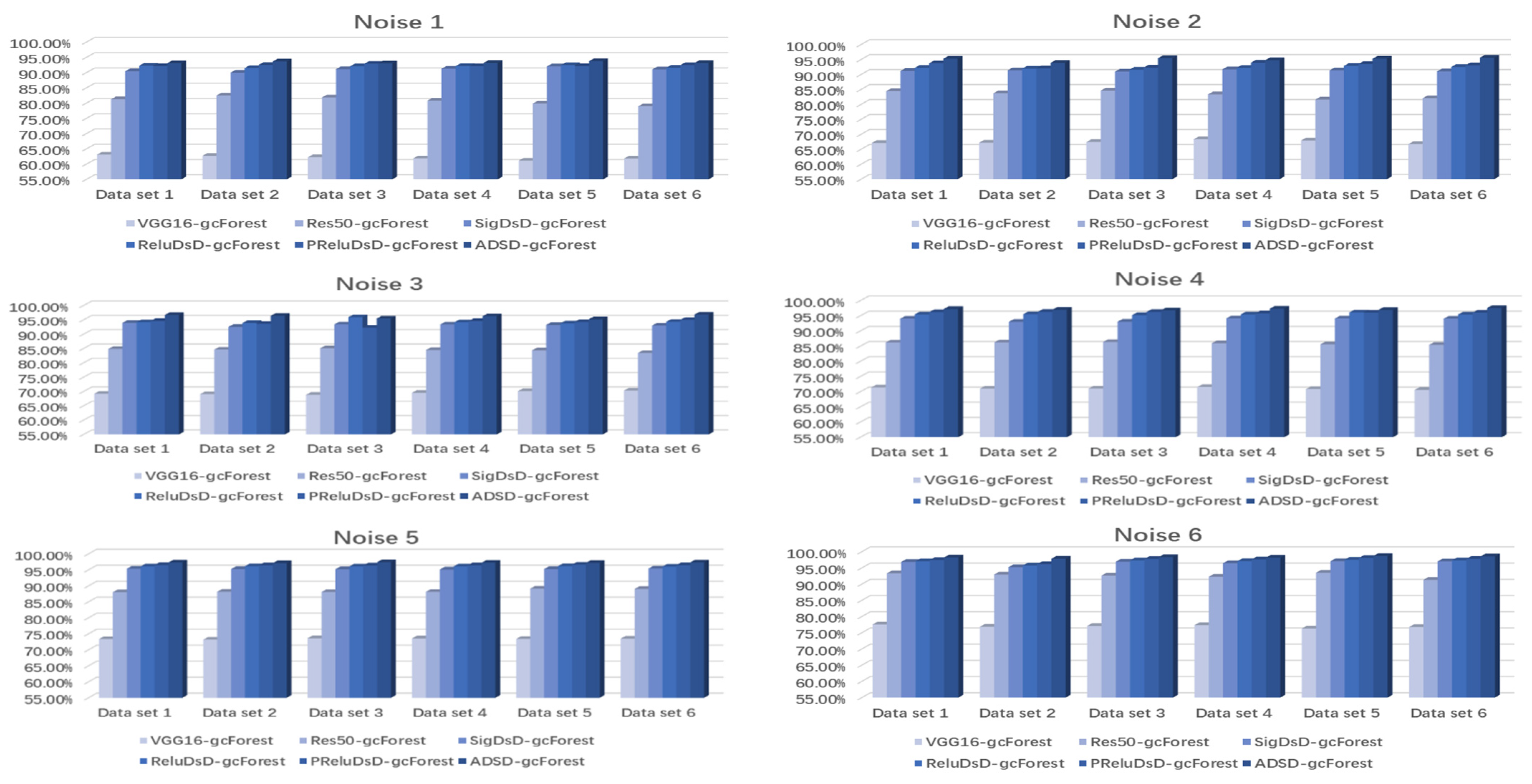
| Dataset 1 | VGG16-gcForest | 65.10%/66.20% | 69.13%/68.78% | 71.20%/72.02% | 72.41%/73.10% | 75.36%/74.48% | 77.54%/76.98% |
| Res50- gcForest | 83.25%/82.86% | 86.42%/85.96% | 87.85%/88.12% | 89.22%/90.03% | 92.02%/92.05% | 93.33%/94.40% | |
| SigDSD- gcForest | 92.43%/91.73% | 94.17%/93.92% | 95.95%/94.65% | 96.04%/96.11% | 96.35%/96.30% | 96.85%/96.80% | |
| ReluDSD-gcforest | 92.85%/93.05% | 94.22%/93.88% | 96.20%/96.22% | 96.47%/96.50% | 96.70%/96.73% | 97.03%/96.98% | |
| PReluDSD-gcForest | 93.09%/92.86% | 94.72%/94.70% | 96.62%/96.65% | 96.73%/96.80% | 96.83%/96.96% | 97.45%/97.52% | |
| ADSD-gcForest | 94.32%/94.30% | 95.85%/95.78% | 97.70%/97.73% | 97.83%/98.03% | 97.92%/98.15% | 98.23%/98.28% | |
| Dataset 2 | VGG16-gcForest | 64.72%/63.56% | 69.20%/68.95% | 69.96%/70.14% | 72.02%/71.92% | 73.46%/73.45% | 76.85%/76.88% |
| Res50- gcForest | 83.50%/83.47% | 86.72%/86.75% | 87.60%/87.59% | 89.27%/89.30% | 91.26%/91.33% | 92.96%/93.05% | |
| SigDSD- gcForest | 89.98%/90.13% | 91.40%/91.48% | 92.53%/92..60% | 93.06%/93.10% | 94.12%/94.08% | 95.23%/95.26% | |
| ReluDSD-gcforest | 90.48%/90.43% | 91.92%/92.03% | 92.90%/92.86% | 93.55%/93.57% | 94.67%/94.65% | 95.73%/95.70% | |
| PReluDSD-gcForest | 91.52%/91.55% | 92.03%/92.05% | 93.63%/93.71% | 94.52%/94.50% | 95.03%/95.11% | 96.15%/96.18% | |
| ADSD-gcForest | 92.65%/92.66% | 93.79%/93.82% | 94.42%/94.45% | 95.91%/96.02% | 96.51%/96.47% | 97.83%/97.85% | |
| Dataset 3 | VGG16-gcForest | 66.25%/66.31% | 67.43%/67.40% | 69.84%/69.86% | 72.45%/72.50% | 74.62%/75.06% | 77.11%/78.23% |
| Res50- gcForest | 83.85%/83.80% | 85.62%/86.15% | 86.87%/86.93% | 88.38%/88.43% | 91.67%/91.72% | 92.63%/92.73% | |
| SigDSD- gcForest | 91.61%/91.78% | 92.96%/93.32% | 95.41%/95.60% | 96.08%/96.05% | 96.21%/96.34% | 96.91%/97.01% | |
| ReluDSD-gcforest | 91.95%/91.86% | 93.60%/93.74% | 95.92%/96.02% | 96.52%/96.46% | 96.87%/96.94% | 97.32%/97.28% | |
| PReluDSD-gcForest | 92.07%/91.96% | 94.30%/94.52% | 95.42%/95.40% | 96.90%/96.86% | 97.24%/97.35% | 97.75%/97.92% | |
| ADSD-gcForest | 93.22%/93.25% | 95.43%/95.48% | 96.48%/96.56% | 97.11%/97.16% | 97.84%/98.02% | 98.33%/98.30% | |
| Dataset 4 | VGG16-gcForest | 65.89%/65.92% | 68.34%/69.16% | 70.54%/71.17% | 73.52%/73.58% | 75.66%/76.16% | 77.35%/77.49% |
| Res50- gcForest | 77.82%/77.80% | 80.35%/81.42% | 84.48%/85.53% | 86.76%/87.36% | 90.81%/91.28% | 92.28%/93.16% | |
| SigDSD- gcForest | 92.25%/91.89% | 92.72%/92.70% | 93.78%/93.89% | 94.75%/94.82% | 95.52%/95.64% | 96.44%/97.13% | |
| ReluDSD-gcforest | 92.71%/93.04% | 93.21%/93.19% | 94.36%/94.28% | 95.46%/95.51% | 96.01%/96.33% | 97.08%/97.26% | |
| PReluDSD-gcForest | 93.07%/93.26% | 94.96%/95.14% | 95.58%/96.27% | 96.82%/97.14% | 97.42%/97.59% | 97.64%/98.01% | |
| ADSD-gcForest | 94.18%/95.64% | 95.78%/95.74% | 96.28%/96.32% | 97.34%/97.64% | 97.92%/98.12% | 98.17%/98.34% | |
| Dataset 5 | VGG16-gcForest | 66.14%/66.10% | 67.94%/68.23% | 71.13%/71.06% | 72.86%/72.93% | 75.43%/76.15% | 76.29%/77.35% |
| Res50- gcForest | 79.85%/80.15% | 81.62%/81.64% | 84.96%/84.86% | 88.62%/88.75% | 91.52%/91.67% | 93.53%/93.78% | |
| SigDSD- gcForest | 92.02%/92.35% | 94.40%/94.67% | 95.22%/96.37% | 95.72%/96.89% | 96.51%/97.02% | 97.05%/97.28% | |
| ReluDSD-gcforest | 92.47%/92.43% | 94.86%/94.82% | 95.76%/95.84% | 96.12%/96.15% | 97.02%/97.46% | 97.53%/97.88% | |
| PReluDSD-gcForest | 93.08%/93.47% | 95.48%/94.86% | 96.27%/96.20% | 96.76%/97.16% | 97.67%/98.16% | 98.08%/98.24% | |
| ADSD-gcForest | 94.27%/94.35% | 96.27%/96.37% | 97.19%/97.10% | 97.46%/97.65% | 98.15%/98.10% | 98.67%/98.76% | |
| Dataset 6 | VGG16-gcForest | 65.85%/65.78% | 66.72%/67.05% | 70.32%/71.14% | 72.61%/72.76% | 74.50%/75.65% | 76.76%/77.04% |
| Res50- gcForest | 78.95%/79.13% | 84.06%/84.23% | 85.46%/85.63% | 87.42%/87.25% | 89.73%/89.53% | 91.32%/92.03% | |
| SigDSD- gcForest | 91.06%/91.32% | 92.92%/93.16% | 93.72%/94.07% | 95.03%/95.06% | 96.40%/96.32% | 97.02%/97.14% | |
| ReluDSD-gcforest | 91.62%/91.76% | 93.52%/94.31% | 94.34%/94.54% | 95.43%/95.32% | 97.02%/96.53% | 97.33%/97.27% | |
| PReluDSD-gcForest | 92.07%/92.15% | 94.08%/93.64% | 95.17%/95.67% | 96.06%/96.05% | 97.47%/97.36% | 97.78%/98.05% | |
| ADSD-gcForest | 93.76%/93.48% | 95.61%/95.53% | 96.86%/96.57% | 97.59%/97.42% | 98.34%/98.28% | 98.55%/98.62% |
| Dataset | Methods | N1(AC/F1) | N2(AC/F1) | N3(AC/F1) | N4(AC/F1) | N5(AC/F1) | N6(AC/F1) |
|---|---|---|---|---|---|---|---|
| Dataset 1 | VGG16-gcForest | 61.87%/61.80% | 64.84%/64.75% | 67.45%/68.12% | 69.85%/70.23% | 73.46%/73.32% | 75.75%/75.82% |
| Res50- gcForest | 79.75%/80.04% | 81.28%/81.42% | 84.86%/84.90% | 87.91%/88.34% | 91.03%/91.06% | 92.46%/92.50% | |
| SigDSD- gcForest | 91.52%/91.68% | 92.53%/93.46% | 94.48%/95.62% | 95.56%/95.63% | 96.01%/96.20% | 96.65%/96.60% | |
| ReluDSD-gcforest | 92.03%/92.12% | 93.20%/93.18% | 95.06%/94.92% | 96.01%/95.68% | 96.53%/96.48% | 97.04%/97.06% | |
| PReluDSD-gcForest | 92.62%/92.65% | 93.95%/93.86% | 95.65%/95.60% | 96.62%/96.63% | 97.03%/97.18% | 97.75%/97.70% | |
| ADSD-gcForest | 93.32%/93.34% | 94.74%/94.70% | 96.54%96.46% | 97.23%/97.20% | 97.72%/97.67% | 98.34%/98.25% | |
| Dataset 2 | VGG16-gcForest | 62.41%/63.26% | 64.27%/64.68% | 66.75%/67.35% | 68.58%/69.14% | 71.59%/71.48% | 73.84%/73.76% |
| Res50- gcForest | 79.61%/79.53% | 81.45%/81.39% | 83.68%/83.61% | 86.84%/86.72% | 89.83%/89.76% | 92.69%/92.53% | |
| SigDSD- gcForest | 90.37%/90.28% | 93.12%/93.36% | 94.27%/95.26% | 95.76%/95.63% | 95.67%/95.72% | 96.43%/96.84% | |
| ReluDSD-gcforest | 91.06%/91.26% | 94.08%/94.34% | 95.59%/95.62% | 96.22%/96.32% | 96.75%/97.14% | 97.29%/97.38% | |
| PReluDSD-gcForest | 91.56%/92.36% | 94.67%/94.37% | 96.24%/95.37% | 96.87%/97.08% | 97.32%/97.46% | 97.64%/97.85% | |
| ADSD-gcForest | 92.68%/93.06% | 95.26%/95.37% | 96.81%/96.68% | 97.26%/97.39% | 97.83%/97.80% | 98.44%/98.40% | |
| Dataset 3 | VGG16-gcForest | 63.21%/63.46% | 65.32%/64.89% | 67.63%/67.90% | 68.42%/68.36% | 70.94%/71.26% | 72.84%/73.58% |
| Res50- gcForest | 77.12%/77.42% | 80.23%/80.68% | 82.47%/82.86% | 86.58%/86.69% | 89.52%/90.15% | 92.27%/92.38% | |
| SigDSD- gcForest | 90.46%/91.68% | 93.48%/93.56% | 94.59%/94.75%. | 95.03%/96.15% | 95.67%/96.82% | 95.33%/95.59% | |
| ReluDSD-gcforest | 90.72%/91.08% | 93.73%/94.28% | 95.07%/95.36% | 95.68%/96.06% | 96.28%/96.33% | 96.87%/97.11% | |
| PReluDSD-gcForest | 91.25%/91.20% | 94.36%/94.38% | 95.53%/95.49% | 96.36%/96.42% | 96.82%/96.74% | 97.35%/97.19% | |
| ADSD-gcForest | 91.85%/92.09% | 94.75%/95.13% | 96.15%/96.18% | 96.98%/97.26% | 97.23%/97.46% | 98.31%/98.48% | |
| Dataset 4 | VGG16-gcForest | 61.74%/62.31% | 63.87%/64.26% | 66.88%/67.18% | 68.36%/69.45% | 70.42%/71.26% | 71.29%/72.21% |
| Res50- gcForest | 76.42%/76.48% | 78.73%/78.62% | 82.23%/82.26% | 84.64%/85.04% | 88.37%/88.49% | 91.44%/91.57% | |
| SigDSD- gcForest | 91.23%/91.34% | 92.34%/92.86% | 94.68%/95.71% | 95.82%/96.02% | 96.42%/96.74% | 97.08%/97.06% | |
| ReluDSD-gcforest | 91.75%/91.60% | 93.03%/93.09% | 95.02%/95.16% | 96.35%/96.42% | 96.97%/97.05% | 97.34%/97.48% | |
| PReluDSD-gcForest | 92.20%/92.34% | 93.58%/93.64% | 95.42%/95.56% | 96.72%/96.83% | 97.41%/97.56% | 97.86%/98.01% | |
| ADSD-gcForest | 93.45%/93.40% | 94.71%/94.65% | 96.75%/96.63% | 97.32%/97.46% | 98.03%/98.14% | 98.29%/98.24% | |
| Dataset 5 | VGG16-gcForest | 62.76%/62.64% | 64.52%/65.38% | 67.12%/67.48% | 69.29%/69.70% | 70.68%/71.17% | 71.36%/72.47% |
| Res50- gcForest | 75.12%/75.49% | 77.37%/77.25% | 79.52%/79.26% | 82.67%/82.54% | 84.46%/84.69% | 87.53%/88.14% | |
| SigDSD- gcForest | 91.40%/91.42% | 93.01%/92.89% | 94.81%/95.17% | 95.02%/95.43% | 96.24%/96.39% | 96.82%/97.16% | |
| ReluDSD-gcforest | 92.22%/92.47% | 93.50%/93.49% | 95.22%/95.24% | 95.75%/96.78% | 96.64%/96.51% | 97.27%/97.38% | |
| PReluDSD-gcForest | 92.86%/92.92% | 94.08%/94.06% | 95.82%/95.86% | 96.22%/96.36% | 97.25%/97.36% | 97.76%/97.82% | |
| ADSD-gcForest | 93.24%/93.28% | 95.78%/95.83% | 96.28%/97.36% | 96.87%/97.12% | 97.73%/97.68% | 98.38%/98.06% | |
| Dataset 6 | VGG16-gcForest | 61.98%/61.86% | 63.45%/64.58% | 65.62%/65.52% | 68.29%/68.34% | 69.56%/69.96% | 71.42%/71.86% |
| Res50- gcForest | 74.86%/74.92% | 77.53%/77.50% | 80.22%/80.36% | 82.23%/82.18% | 84.29%/84.27% | 86.36%/86.34% | |
| SigDSD- gcForest | 92.25%/92.37% | 93.45%/93.57% | 94.09%/94.16% | 95.42%/95.26% | 96.24%/96.31% | 96.83%/96.80% | |
| ReluDSD-gcforest | 92.86%/92.79% | 93.73%/93.76% | 94.42%/94.39% | 95.73%/95.70% | 96.89%/96.92% | 97.36%/97.34% | |
| PReluDSD-gcForest | 93.03%/93.17% | 94.24%/94.28% | 95.02%/94.88% | 96.36%/97.32% | 97.25%/97.05% | 97.76%/97.65% | |
| ADSD-gcForest | 93.55%/93.64% | 94.61%/94.78% | 95.42%/95.57% | 96.92%/97.18% | 97.83%/98.07% | 98.34%/98.64% |
| The Name of Dataset | Dataset 1 | Dataset 2 | Dataset 3 | Dataset 4 | Dataset 5 | Dataset 6 |
|---|---|---|---|---|---|---|
| University of Ottawa bearing data | HD-1 | HA-1 | HB-1 | HA-1 | HC-1 | HD-1 |
| HA-1 | HB-1 | HC-1 | HB-1 | HA-1 | HB-1 | |
| HB-1 | HD-1 | HA-1 | HD-1 | HB-1 | HC-1 | |
| IC-1 | IA-1 | IC-1 | IB-1 | IB-1 | ID-1 | |
| ID-1 | IB-1 | IA-1 | IA-1 | IC-1 | IA-1 | |
| IB-1 | ID-1 | IB-1 | ID-1 | IA-1 | IB-1 | |
| OB-1 | OD-1 | OB-1 | OB-1 | OA-1 | OD-1 | |
| OD-1 | OC-1 | OA-1 | OD-1 | OC-1 | OB-1 | |
| OA-1 | OB-1 | OD-1 | OC-1 | OD-1 | OC-1 | |
| HC-1 | IA-1 | HA-1 | IB-1 | OD-1 | OB-1 |
| Dataset | Methods | N1(AC/F1) | N2(AC/F1) | N3(AC/F1) | N4(AC/F1) | N5(AC/F1) | N6(AC/F1) |
|---|---|---|---|---|---|---|---|
| Dataset 1 | VGG16-gcForest | 56.23%/56.20% | 60.39%/60.34% | 63.76%/63.72% | 65.63%/65.59% | 68.42%/68.45% | 70.28%/70.36% |
| Res50- gcForest | 78.29%/78.26% | 82.53%/82.57% | 84.31%/84.59% | 85.42%/85.31% | 87.12%/87.18% | 88.03%/87.93% | |
| SigDSD- gcForest | 91.23%/91.36% | 92.39%/92.58% | 94.52%/95.67% | 95.26%/95.68% | 96.01%/96.19% | 96.75%/97.70% | |
| ReluDSD-gcforest | 91.79%/91.66% | 93.42%/93.35% | 95.01%/95.16% | 95.76%/95.72% | 96.42%/96.48% | 97.03%/97.18% | |
| PReluDSD-gcForest | 92.34%/92.46% | 93.44%/93.67% | 95.47%/95.55% | 96.12%/96.39% | 97.03%/96.98% | 97.35%/97.36% | |
| ADSD-gcForest | 93.42%/93.39 | 94.76%/94.58% | 96.08%/96.06% | 96.89%/96.92% | 97.62%/97.58% | 98.42%/98.48% | |
| Dataset 2 | VGG16-gcForest | 57.62%/57.55% | 59.45%/59.52% | 62.45%/62.38% | 64.81%/64.80% | 67.02%/67.28% | 69.31%/69.59% |
| Res50- gcForest | 77.94%/77.80% | 80.66%/81.56% | 83.52%/84.28% | 86.44%/86.50% | 88.18%/88.26% | 89.50%/89.53% | |
| SigDSD- gcForest | 90.46%/90.63% | 92.02%/91.26% | 93.25%/93.18% | 94.11%/94.07% | 95.47%/95.48% | 96.26%/96.31% | |
| ReluDSD-gcforest | 91.04%/91.16% | 92.68%/92.76% | 93.82%/93.96% | 94.45%/94.48% | 96.06./95.89% | 96.63%/96.54% | |
| PReluDSD-gcForest | 91.65%/91.77% | 93.24%/93.20% | 94.75%/94.79% | 95.36%/95.42% | 96.62%/96.59% | 97.30%/97.28% | |
| ADSD-gcForest | 92.42%/92.38% | 94.66%/94.76% | 96.14%/97.28% | 96.76%/97.02% | 97.32%/97.48% | 98.15%/98.24% | |
| Dataset 3 | VGG16-gcForest | 56.80%/56.68% | 58.55%/59.04% | 62.53%/62.68% | 64.33%/65.22% | 66.86%/66.78% | 68.42%/68.40% |
| Res50- gcForest | 76.73%/76.55% | 78.42%/78.61% | 81.18%/81.26% | 83.92%/83.90% | 86.35%/86.42% | 88.26%/88.37% | |
| SigDSD- gcForest | 89.08%/88.89% | 90.91%/91.26% | 92.58%/92.87% | 94.27%/94.38% | 95.31%/95.36% | 96.26%/96.28% | |
| ReluDSD-gcforest | 89.62%/89.52% | 91.54%/91.69% | 93.18%/92.89% | 94.82%/94.80% | 96.05%/96.18% | 96.86%/96.76% | |
| PReluDSD-gcForest | 90.16%/91.02% | 92.32%/92.21% | 94.84%/94.56% | 95.47%/95.32% | 96.76%/96.79% | 97.32%/97.42% | |
| ADSD-gcForest | 91.85%/91.29% | 93.49%/93.74% | 95.68%/95.88% | 96.31%/96.28% | 97.45%/97.52% | 98.27%/98.38% | |
| Dataset 4 | VGG16-gcForest | 57.34%/57.49% | 59.56%/59.63% | 61.16%/61.19% | 63.47%/63.30% | 65.52%/65.71% | 67.92%/67.94% |
| Res50- gcForest | 75.26%/75.36% | 77.56%/77.49% | 80.64%/80.79% | 83.94%/84.06% | 85.16%/85.67% | 87.85%/88.06% | |
| SigDSD- gcForest | 91.53%/91.68% | 92.20%/92.34% | 93.23%/94.36% | 94.78%/94.82% | 95.63%/95.56% | 96.28%/96.34% | |
| ReluDSD-gcforest | 92.06%/92.64% | 92.76%/92.84% | 93.74%/93.70% | 95.58%/95.88% | 96.17%/96.24% | 96.65%/96.69% | |
| PReluDSD-gcForest | 92.60%/91.65% | 93.26%/93.36% | 94.43%/94.58% | 96.03%/96.15% | 96.76%/96.72% | 97.22%/97.36% | |
| ADSD-gcForest | 93.42%/93.62% | 94.60%/94.26% | 95.81%/95.68% | 96.63%/96.58% | 97.45%/97.26% | 98.32%/98.30% | |
| Dataset 5 | VGG16-gcForest | 56.85%/56.79% | 58.25%/58.16% | 61.83%/62.05% | 62.55%/63.18% | 64.67%/65.25% | 66.86%/66.79% |
| Res50- gcForest | 76.40%/76.59% | 78.62%/78.59% | 82.35%/83.59% | 84.54%/85.69% | 85.46%/85.96% | 89.14%/89.19% | |
| SigDSD- gcForest | 91.01%/91.08% | 93.06%/93.28% | 95.15%/95.28% | 95.70%/95.76% | 96.02%/96.34% | 96.55%/96.64% | |
| ReluDSD-gcforest | 91.57%/91.68% | 93.62%/94.68% | 95.65%/95.78% | 96.04%/96.29% | 96.66%/96.64% | 97.05%/97.14% | |
| PReluDSD-gcForest | 92.02%/91.89% | 94.22%/94.26% | 96.03%/96.38% | 96.48%/96.58% | 97.21%/97.36% | 97.43%/97.58% | |
| ADSD-gcForest | 92.52%/93.76% | 94.76%/94.88% | 96.56%/96.49% | 97.03%/96.69% | 97.85%/97.68% | 98.42%/98.64% | |
| Dataset 6 | VGG16-gcForest | 57.25%/57.64% | 59.60%/60.12% | 61.33%/61.24% | 64.58%/64.88% | 66.02%/65.89% | 68.20%/67.96% |
| Res50- gcForest | 75.05%/76.18% | 77.33%/78.38% | 79.62%/80.19% | 82.32%/83.49% | 85.58%/84.99% | 88.75%/89.06% | |
| SigDSD- gcForest | 92.08%/93.16% | 94.01%/93.86% | 94.52%/95.67% | 95.27%/95.86% | 96.06%/96.18% | 96.47%/96.34% | |
| ReluDSD-gcforest | 92.52%/92.36% | 94.63%/94.59% | 95.02%/94.98% | 95.72%/95.64% | 96.58%/96.59% | 96.95%/96.85% | |
| PReluDSD-gcForest | 93.05%/92.96% | 95.06%/94.89% | 95.62%/95.57% | 96.34%/96.78% | 97.10%/96.68% | 97.36%/97.29% | |
| ADSD-gcForest | 93.60%/93.84% | 95.52%/95.67% | 96.06%/95.98% | 96.85%/97.93% | 97.67%/97.58% | 98.29%/98.36% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, S.; Wang, Z.; Gao, D. Bearing Fault Diagnosis Based on a Novel Adaptive ADSD-gcForest Model. Processes 2022, 10, 209. https://doi.org/10.3390/pr10020209
Zhai S, Wang Z, Gao D. Bearing Fault Diagnosis Based on a Novel Adaptive ADSD-gcForest Model. Processes. 2022; 10(2):209. https://doi.org/10.3390/pr10020209
Chicago/Turabian StyleZhai, Shuo, Zhenghua Wang, and Dong Gao. 2022. "Bearing Fault Diagnosis Based on a Novel Adaptive ADSD-gcForest Model" Processes 10, no. 2: 209. https://doi.org/10.3390/pr10020209
APA StyleZhai, S., Wang, Z., & Gao, D. (2022). Bearing Fault Diagnosis Based on a Novel Adaptive ADSD-gcForest Model. Processes, 10(2), 209. https://doi.org/10.3390/pr10020209







