Abstract
Malaria management remains a challenge, due to the resistance of malaria parasites to current antimalarial agents. This resistance consequently delays the global elimination of malaria throughout the world. Hence, the demand is increasing for new and effective antimalarial drugs. The identification of potential drugs that target Pk-LDH can be obtained through virtual screening analyses, as this has been previously applied to discover Pf-LDH inhibitors. In this study, the selected candidates from our virtual screening analyses were subsequently tested against purified Pk-LDH, and verified through an inhibition of Pk-LDH via enzymatic activity assays. Virtual screening analysis from this study showed that 3,3-Difluoropyrrolidine hydrochloride and 3-hydroxytetrahydrofuran exhibited binding affinity values of −3.25 kcal/mol and −3.74, respectively. These compounds were selected for evaluation towards inhibitory activity against Pk-LDH assays, including two compounds from a previous study which are oxalic acid and glycolamide. The earlier compounds were structurally similar to lactate and pyruvate, and the latter two compounds were structurally similar to a known LDH inhibitor, oxamate. Among all of the compounds tested, oxalic acid showed the highest inhibition activity at 54.12%; interestingly, this correlated well with the virtual screening analyses, which showed that this compound was the best among the oxamate analogues, with a binding affinity value of −2.59 kcal/mol. Hence, further exploration and development of this compound may result in a promising antimalarial drug for malaria treatment, especially for infection involving P. knowlesi.
1. Introduction
Malaria is a neglected tropical disease with a high mortality rate. The latest world malaria report estimates an occurrence of 241 million cases in 2020 [1]. Although mortality caused by malaria has decreased over the past few years, disease management remains a challenge. The causative parasites’ resistance to current antimalarial agents has been identified as one of the factors preventing the eradication of this deadly disease. Thus, the discovery of new, safe, and effective antimalarial drugs is urgently needed.
The causative agents of malaria are from the genus Plasmodium, which spread through infected Anopheles mosquitoes. It is known that malaria in humans is caused by P. falciparum, P. malariae, P. vivax, and P. ovale. Additionally, another species, P. knowlesi, which causes malaria in macaques, has been found to infect humans as well [2]. A large cluster of P. knowlesi infections in humans was initially detected in Sarawak, Malaysia, in 2004. Several cases were later described as being from all over the Southeast Asia region [3], with 91.47% of these being reported from Malaysian Borneo [4]. Recent years have seen an increase in P. knowlesi human infections in Malaysia, and also in China, Indonesia, Myanmar, Philippines, Singapore, Thailand, and Vietnam [4]. Between 2015 and 2017, a total of 3524 cases recorded were P. knowlesi malaria, reported in Sabah state, Malaysia [5]. Recently, eight individuals were found to be infected with P. knowlesi from among 1361 of 14,732 samples of asymptomatic malaria parasite infections that were detected in 23 villages in Pailin and Battambang, western Cambodia [6].
Infection with P. knowlesi can cause a range of potentially fatal diseases, if left untreated [7]. Plasmodium knowlesi has a life cycle of 24 h, which is the shortest among malarial parasites; this results in fatal parasitemia leading to death in infected individuals [4,8]. In contrast, P. malariae multiplies every three days in the blood, and does not cause severe infections [9]. Moreover, most P. knowlesi infections have been misidentified as infections caused by the benign pathogen P. malariae, leading to wrongful diagnoses and consequently delayed parenteral artesunate, with fatal outcomes [10].
Monumental efforts, therefore, are needed to eradicate this disease. Although many of the current antimalarial drugs are used as the treatment for malaria caused by P. knowlesi, their efficacy is compromised by toxicity to mammalian host cells, and the emergence of drug resistance [11]. Unfortunately, highly effective vaccines that are critical to the global eradication of malaria have not yet been developed. These obstacles further emphasize the urgent need to develop novel malaria therapeutics that will avoid drug resistance, and have limited to no toxic effects in mammalian hosts [12].
As an energy provider in the cell, the glycolytic pathway is a likely target for innovative antimalarial drugs, since the Plasmodium spp. depends only on glycolysis for its survival in the hosts, i.e., during its intra-erythrocytic phase. At this point of its complex life cycle, the Plasmodium lacks a functional tricarboxylic acid cycle, and its prolonged pathophysiology is caused by increased glycolysis. Particularly, it has been found that infected erythrocytes use more glucose, and produce more lactate than uninfected cells [13]. The use of glucose analogues, which reduces ATP levels of the parasites, further illustrated the significance of glycolysis [14]. Together, these findings demonstrated the benefits of using the glycolytic pathway as a therapeutic target when developing new antimalarial medications.
Lactate dehydrogenase (LDH) is essential for the anaerobic lifestyle of Plasmodium, and thus serves as a potential drug target. The enzyme is an oxidoreductase [EC 1.1.1.27], and as the last enzyme of the glycolytic pathway, converts pyruvate to lactate, thereby regenerating NAD+ for continued use in glycolysis. Thus, LDH plays a key role in the energy metabolism of malaria parasites. The enzyme is present abundantly in all malarial parasites, and was reported to be biochemically, immunologically, and structurally different from mammalian and bacterial counterparts [13]. The plasmodial LDH has also been shown to be a potential target for chemotherapy; furthermore, P. falciparum LDH (Pf-LDH) has been used in docking studies for screening for anti-malarial chemical compounds [14].
It was often thought that drug-resistant malarial parasites developed because of mutations in the drug targets’ active sites, or due to adjustments in the drug receptors. Consideration has been given to Pf-LDH as a potential therapeutic target. One of the medications used to treat malaria, chloroquine, has been shown to competitively inhibit Pf-LDH by preferentially engaging in the NADH binding pocket, and taking up a position resembling the adenyl ring [15]. Furthermore, fifty commercially available compounds that are structurally similar to chloroquine were tested against Pf-LDH, and three compounds, itraconazole, atorvastatin, and posaconazole, which showed binding energies that were similar to that of NADH, were selected for testing on chloroquine-resistant P. falciparum. All three compounds proved to be active in the two immunoenzymatic assays performed in parallel using Pf-LDH or histidine-hich protein (HRP2)-specific monoclonal antibodies. The IC50 values for each drug were similar in both tests, and are 40- to 100-fold less active than that of chloroquine. Posaconazole showed the lowest IC50 at 5 mM [15]. These results confirmed that molecular docking is a valuable approach for finding novel antimalarial drugs.
Following these recent discoveries of Pf-LDH inhibitors, we screened for potential drugs that target Pk-LDH through ligand- and structure-based virtual screening analyses. In this study, we report the selected candidates from the virtual screens that were subsequently tested against purified Pk-LDH, and validated through an LDH activity assay.
2. Materials and Methods
2.1. Ligand-Based, Structure-Based Drug Design and Toxicity Tests
Ligand-based drug design (LBDD) was performed on a pool of existing compounds, in order to identify novel compounds that are similar to the query molecule. In this analysis, the substrates of LDH, pyruvate, or lactate, were used as a query molecule. LBDD was carried out according to Nurhainis and Fuad (2017) [16], with some modifications. In this study, USRCAT, an extension to the ultrafast shape recognition (USR) algorithm (available at http://usr.marseille.inserm.fr/ (accessed on 12 November 2020)), was used to search for molecules with similarities to pyruvate or lactate. A webserver for ligand-based virtual screening, USR, using ultrafast shape recognition techniques with a total of 23,129,049 molecules, were collected for the screening library, from 2012-04-26, 2013-01-10, and 2013-12-18 versions of ZINC. ZINC is a free database of over 35 million commercially available compounds in ready-to-dock 3D formats [17]. The predicted similarity of the molecules was calculated, along with the candidates, based on the geometric descriptors of the query molecule. Next, a scoring function was applied, in order to generate a single numerical datum (0 < score ≤ 1). Analyses of the compounds that were selected from LBDD were followed with structure-based drug design (SBDD), where the compounds obtained from USRCAT were docked into the active site of the Pk-LDH model [16]. Selection of candidates for experimental validation in in vitro Pk-LDH inhibition analyses were based on the lowest binding energies, and also with the best dock poses when binding to Pk-LDH. The toxicity of the compounds was predicted using Toxicity Estimation Software Tool (TEST). Furthermore, compounds that were published earlier [16], were also used for the experiments.
2.2. Compounds for Experimental Tests
Four compounds, oxalic acid, glycolamide, 3-hydroxytetrahydrofuran, and 3,3-Difluoropyrrolidine hydrochloride, were selected. Oxalic acid and glycolamide are similar to a known inhibitor of LDH, oxamate, while 3-hydroxytetrahydrofuran (ZINC04716349) and 3,3-Difluoropyrrolidine hydrochloride (ZINC01690282) were selected in our virtual screening study. Stock solutions of the compounds (Sigma-Aldrich, St. Louis, MO, USA) were prepared in 50% dimethyl sulfoxide (DMSO), and were stored at room temperature until use. A final concentration of 1 mM was used for assays against Pk-LDH. Control reactions, that lacked either enzyme (to test for effect of DMSO) or the compound, were set for each test well.
2.3. Pk-LDH Enzyme
Recombinant Pk-LDH was cloned, expressed, and purified as described elsewhere [18].
2.4. Steady-State Enzymatic Assays Monitored with Pk-LDH Enzyme Reaction
In order to determine the specific activity of the isolated Pk-LDH, an LDH enzyme test was carried out. At 340 nm, the oxidation of NADH to NAD+ was observed. A reaction mixture of 100 μL was made up of 2.0 mM sodium pyruvate (Sigma-Aldrich), 0.3 mM NADH, and 100 mM sodium phosphate buffer (pH 7.5). Prior to measurement, samples were incubated in a spectrophotometer for 10 min at 27 °C, in order to acclimate to the temperature and establish blank rates. At time equal to zero, 10 μL of diluted Pk-LDH (corresponding to approximately 0.05 mg/mL) was added to the reaction mixture and thoroughly stirred. The EON BIOTEK spectrophotometer was used in this experiment to measure the change at 340 nm. The specific activity for Pk-LDH was estimated in units, where one unit is equal to the quantity of enzyme needed to convert 1 mol of substrate per minute per mg of enzyme, under the conditions of a standard assay condition.
2.5. Inhibition of Pk-LDH Activity by Oxalic Acid, Glycolamide, 3-Hydroxytetrahydrofuran, and 3,3-Difluoropyrrolidine Hydrochloride
The effect of the selected compounds from virtual screening (oxalic acid, glycolamide, 3-hydroxytetrahydrofuran, and 3,3-Difluoropyrrolidine hydrochloride) against a Pk-LDH assay was determined by observing the oxidation of NADH to NAD+ at 340 nm. In this experiment, the compounds were added at a final concentration of 1 mM into the mixtures, and incubated for 1 h. A negative control reaction without inhibitors, and a positive control reaction with a known LDH inhibitor, oxamate, were set. The efficiency of inhibition of LDH activity by the compounds was presented as the percentage of remaining activity of the enzymes after the addition of the compounds. The Pk-LDH inhibition experiments were performed in triplicate for each compound in parallel with oxamate, a known LDH inhibitor. The test was performed in a 96-well microtiter plate (Corning, Santa Clara, CA, USA).
3. Results and Discussion
3.1. Identification of Small Molecule Analogues of Pyruvate and Lactate by USRCAT and Toxicity Test
The USRCAT database was searched for candidates that were similar to the substrates of LDH, lactate, or pyruvate. In USRCAT, a similarity score close to zero indicates less similar compounds, while a score near one proposes closer similarity. The top six compounds with the highest similarities to pyruvate showed scores that ranged from 0.859 to 0.882 (Table 1), while the top six compounds that resembled lactate showed similarity scores that ranged from 0.822 to 0.87 (Table 2).

Table 1.
Similarity scores from ligand-based drug design, and minimum binding energies from structure-based drug design for the compounds that were structurally similar to pyruvate (P). The row in bold indicates the compound that was selected for inhibition analysis.

Table 2.
Similarity scores from ligand-based drug design, and minimum binding energies from structure-based drug design for compounds that were structurally similar to lactate (L). The row in bold indicates the compound that was selected for inhibition analysis.
The toxicities of all of the selected compounds were estimated using TEST, and Table 3 shows results for quantitative structure activity relationships (QSAR). Normally, the predicted values, which were obtained from the seven different types of test endpoint analyses, showed that the toxicity of the compound was high when the value was smaller (Ruiz, et al., 2012). Table 3 shows the largest values (according to the test endpoints) among all of the compounds that were tested for toxicity prediction. Only compounds P5 and L5 were predicted to be nontoxic on the basis of the developmental toxicity assay. Notably, compounds P2, P4, P5, L1–L3, L5, and L6 were negative for mutagenicity. Prior to the docking of the selected compounds, validation of docking was performed according to Nurhainis and Fuad (2017) [17].

Table 3.
Estimated toxicity values for compounds that were selected in QSAR structure-based drug design. Highest toxicity values are presented for all the compounds. The compounds that were selected for inhibition analyses are highlighted in bold.
3.2. Molecular Docking for Structure-Based Drug Design
The binding energies of the highest six compounds (obtained from USRCAT), when docked into the Pk-LDH structural model, are shown in Table 1 for compounds that were structurally similar to pyruvate, and in Table 2 for compounds that were structurally similar to lactate. Pyruvate and lactate were used as positive controls in the docking. Among the compounds that were structurally similar to pyruvate, compound P3 showed the lowest binding energy, at −5.25 kcal/mol, although compound P5 exhibited the highest binding energy, at −1.99 kcal/mol. Similarly, in the compounds that were structurally similar to lactate, compound L1 showed a binding energy of −3.9 kcal/mol, which was closest to that of lactate. However, compounds P4 and L6 were selected to be tested with Pk-LDH inhibition analyses, based on their binding affinity values, −3.25 kcal/mol and −3.74, respectively. Furthermore, these compounds showed minimum binding energies that were similar to those of the positive controls; negative values for mutagenicity are required in high concentrations for most end-point toxicity tests. Since all of the ligands’ binding affinities were far lower than those of NADH, it is likely that they would have been ineffective as competitive inhibitors. The discovery of a different binding site, that had a similar affinity and was unaffected by the cofactor, may suggest that there was a possibility of allosteric non-competitive inhibition occurring [19]. A better understanding of this phenomenon would be intriguing for drug discovery purposes.
3.3. Inhibition of Pk-LDH Activity by Oxalic Acid, Glycolamide, 3-Hydroxytetrahydrofuran, and 3,3-Difluoropyrrolidine Hydrochloride
In order to evaluate inhibitory activities of the compounds that were selected by virtual screening, enzyme activity inhibition tests were performed. Four compounds were selected; two were selected from a previous virtual screening study with similarity to a known inhibitor of LDH, oxamate; meanwhile, the other two compounds were selected from virtual screening that was performed in this study. The two selected compounds in this study were each from the compound that is similar to lactate and pyruvate. The compound that was similar to lactate was compound L3, namely 3-hydroxytetrahydrofuran, and the compound that was similar to pyruvate was compound P4, namely 3,3-difluoropyrrolidine hydrochloride. Meanwhile, compound O4 was oxalic acid, and compound O6 was glycolamide, both of which were similar to oxamate. The enzyme assays were performed, as mentioned in the Materials and Methods section, and the potential inhibition activities were assessed in the presence of the compounds in the enzyme assay. The enzyme activity inhibition tests against Pk-LDH showed that compounds L3, P4, O4, and O6 were active, and effectively inhibited the activity of Pk-LDH (Figure 1). However, from four of the tested compounds, O4 showed the highest inhibition activity at 54.12%, followed by compounds O6 (41.83%), P4 (41.50%), and L3 (22.05%), as compared to an enzyme assay in the absence of inhibitor. Thus, amongst all, compound O4 was the most active. Notably, these results were in accord with an earlier study by Nurhainis and Fuad (2017) [16], which reported that among all of the compounds tested by the docking method, compound O4 showed the highest binding affinity to Pk-LDH.
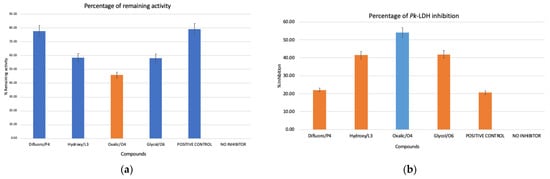
Figure 1.
(a) Specific activities of Pk-LDH in the presence of the indicated compounds: P4: 3,3-difluoropyrrolidine hydrochloride, L3: 3-hydroxytetrahydrofuran, O4: oxalic acid, O6: glycolamide, positive control: OXM and no inhibitor: negative control. (b) Percentage of Pk-LDH inhibition with different compounds labelled the same as in (a). Compound O4 showed the highest inhibition at 54.12%.
Oxalic acid was reported to act on Pk-LDH by competitively inhibiting the reduction of pyruvate to lactate [19]. An earlier study reported that LDH from other sources was inhibited by oxalic acid and oxamic acid [20]. Selective inhibition of the final step of glycolysis in P. knowlesi or other malarial parasites could possibly offer harmless and effective antimalarial compounds [19]. In addition, many novel glycolamide esters were synthesized and tested for their in vitro inhibition of cyclooxygenase (COX-1 and COX-2), and were proven to be selective for COX-2 inhibitors [21]. On the other hand, inhibition of LDH by 3-hydroxytetrahydrofuran and 3,3-difluoropyrrolidine hydrochloride has not been previously reported. In the glycolytic pathway, Pk-LDH is the most active enzyme for energy production, producing ample lactate during the erythrocytic phase of the parasite life cycle. The enzyme uses NAD+ or NADH as a cofactor to convert pyruvate to lactate, and vice versa. Its catalytic mechanism has been extensively investigated, and consists of three stages: the binding of the cofactor in the first stage, the binding of the substrate in the second stage, and the closure of the active site in the third stage [22]. The LDH enzyme of Plasmodium has unique amino acid residues, and kinetic properties relative to other LDHs, especially human isoforms; this makes it a potential target for the development of antimalarial drugs [23].
4. Conclusions
In summary, this study determined the likelihood of developing specific inhibitors against enzymes that regulate the activity of P. knowlesi, which is the lactate dehydrogenase enzyme. The structural variances between Pk-LDH and human LDH allow better targeted inhibition of the parasite enzyme. Among all of the compounds tested, oxalic acid was the most active compound compared to others, and showed the best results when using the docking method. Further exploration and development of the compound may lead to a promising antimalarial drug treatment in the future, especially with P. knowlesi infections.
Author Contributions
Conceptualization, F.A.A.F. and N.O.S.; methodology, F.A.A.F. and N.O.S.; formal analysis, N.O.S.; writing—original draft preparation, N.O.S.; writing—review and editing, F.A.A.F.; supervision, F.A.A.F.; project administration, F.A.A.F.; funding acquisition, F.A.A.F. These authors contributed equally. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by International Islamic University Malaysia Research Initiative Grants, grant number RIGS15-141-0141.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank the Director-General of Health Malaysia, the Ministry of Health, Malaysia, and the Director, Institute for Medical Research (IMR), for permission to publish this article. We also would like to thank the Parasitology Unit, Institute for Medical Research, Kuala Lumpur, Malaysia, for giving us an opportunity to perform our experiments in their facilities. We are also indebted to the colleagues at the Malaysian Genome Institute, Selangor, Malaysia, for their assistance in protein preparation. This study was supported by the International Islamic University Malaysia Research Initiative Grants (RIGS15-141-0141).
Conflicts of Interest
The authors declare that there is no conflicts of interest regarding the publication of this manuscript.
References
- WHO. The E-2020 Initiative of 21 Malaria-Eliminating Countries: 2019 Progress Report; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Cox, F.E. History of the Discovery of the Malaria Parasites and Their Vectors. Parasit Vectors 2010, 3, 5. [Google Scholar] [CrossRef]
- Sabbatani, S.; Fiorino, S.; Manfredi, R. The Emerging of the Fifth Malaria Parasite (Plasmodium Knowlesi). A Public Health Concern? Braz. J. Infect. Dis. 2010, 14, 299–309. [Google Scholar] [CrossRef][Green Version]
- Amir, A.; Cheong, F.W.; de Silva, J.R.; Liew, J.W.K.; Lau, Y.L. Plasmodium Knowlesi Malaria: Current Research Perspectives. Infect. Drug Resist. 2018, 11, 1145–1155. [Google Scholar] [CrossRef]
- Cooper, D.J.; Rajahram, G.S.; William, T.; Jelip, J.; Mohammad, R.; Benedict, J.; Alaza, D.A.; Malacova, E.; Yeo, T.W.; Grigg, M.J.; et al. Plasmodium Knowlesi Malaria in Sabah, Malaysia, 2015–2017: Ongoing Increase in Incidence Despite Near-Elimination of the Human-Only Plasmodium Species. Clin. Infect. Dis. 2019, 70, 361–367. [Google Scholar] [CrossRef]
- Imwong, M.; Madmanee, W.; Suwannasin, K.; Kunasol, C.; Peto, T.J.; Tripura, R.; von Seidlein, L.; Nguon, C.; Davoeung, C.; Day, N.P.J.; et al. Asymptomatic Natural Human Infections with the Simian Malaria Parasites Plasmodium Cynomolgi and Plasmodium Knowlesi. J. Infect. Dis. 2019, 219, 695–702. [Google Scholar] [CrossRef]
- Singh, B.; Daneshvar, C.; Siner, A.; Ahmed, M.A.; Woon, L.C.; Pasini, E.M.; Kocken, C.H.; Singh, B.; Cox-Singh, J.; Krishna, S.; et al. Human Infections and Detection of Plasmodium Knowlesi. Clin. Microbiol. Rev. 2013, 26, 165–184. [Google Scholar] [CrossRef]
- Parija, S.; Janagond, A.; Jeremiah, S. Challenges in Diagnosis of Plasmodium Knowlesi Infections. Trop. Parasitol. 2014, 4, 25. [Google Scholar] [CrossRef]
- Antinori, S.; Galimberti, L.; Milazzo, L.; Corbellino, M. Plasmodium Knowlesi: The Emerging Zoonotic Malaria Parasite. Acta Trop. 2013, 125, 191–201. [Google Scholar] [CrossRef]
- Mahittikorn, A.; Masangkay, F.R.; Kotepui, K.U.; Milanez, G.D.J.; Kotepui, M. Quantification of the Misidentification of Plasmodium Knowlesi as Plasmodium Malariae by Microscopy: An Analysis of 1569 P. Knowlesi Cases. Malar. J. 2021, 20, 1–11. [Google Scholar] [CrossRef]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of Artemisinin Resistance in Plasmodium Falciparum Malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef]
- Andrews, K.T.; Fisher, G.; Skinner-Adams, T.S. Drug Repurposing and Human Parasitic Protozoan Diseases. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Kaushal, D.C.; Rathaur, S.; Kumar, N.; Kaushal, N.A. Cloning, Overexpression, Purification and Characterization of Plasmodium Knowlesi Lactate Dehydrogenase. Protein. Expr. Purif. 2012, 84, 195–203. [Google Scholar] [CrossRef] [PubMed]
- de Souza, N.B.; Carmo, A.M.L.; da Silva, A.D.; França, T.C.C.; Krettli, A.U. Antiplasmodial Activity of Chloroquine Analogs against Chloroquine-Resistant Parasites, Docking Studies and Mechanisms of Drug Action. Malar. J. 2014, 13, 469. [Google Scholar] [CrossRef][Green Version]
- Penna-Coutinho, J.; Cortopassi, W.A.; Oliveira, A.A.; França, T.C.C.; Krettli, A.U. Antimalarial Activity of Potential Inhibitors of Plasmodium Falciparum Lactate Dehydrogenase Enzyme Selected by Docking Studies. PLoS ONE 2011, 6, e21237. [Google Scholar] [CrossRef] [PubMed]
- Nurhainis, O.S.; Fuad, F.A.A. Screening Potential Inhibitors of Lactate Dehydrogenase from Plasmodium Knowlesi via Virtual Screening Approaches. Trop. Biomed. 2017, 34, 841–854. [Google Scholar] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Salim, N.O.; Fuad, F.A.A.; Khairuddin, F.; Seman, W.M.K.W.; Jonet, M.A. Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium Knowlesi for the Development of Anti-Malarial Drugs. Molecules 2021, 26, 6625. [Google Scholar] [CrossRef]
- Saxena, N.; Pandey, V.; Dutta, G.; Ghatak, S. Characterization of Lactate Dehydrogenase of Plasmodium Knowlesi. Mol. Biochem. Parasitol. 1986, 21, 199–202. [Google Scholar] [CrossRef]
- Goldberg, E. Lactate Dehydrogenases in Spermatozoa: Subunit Interactions in Vitro. Arch. Biochem. Biophys. 1965, 109, 134–141. [Google Scholar] [CrossRef]
- Khanna, S.; Madan, M.; Vangoori, A.; Banerjee, R.; Thaimattam, R.; Jafar Sadik Basha, S.K.; Ramesh, M.; Casturi, S.R.; Pal, M. Evaluation of Glycolamide Esters of Indomethacin as Potential Cyclooxygenase-2 (COX-2) Inhibitors. Bioorg. Med. Chem. 2006, 14, 4820–4833. [Google Scholar] [CrossRef]
- Quaytman, S.L.; Schwartz, S.D. Comparison Studies of the Human Heart and Bacillus Stearothermophilus Lactate Dehydrogreanse by Transition Path Sampling. J. Phys. Chem. A 2009, 113, 1892–1897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, S.; Pradhan, A.; Hammond, N.L.; Chittiboyina, A.G.; Tekwani, B.L.; Avery, M.A. Design, Synthesis, and Biological Evaluation of Plasmodium Falciparum Lactate Dehydrogenase Inhibitors. J. Med. Chem. 2007, 50, 3841–3850. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).