LabelFlow Framework for Annotating Workflow Provenance
Abstract
1. Introduction
- Study Subjects; such as the species, the stellar objects or the geographical regions that the data is about.
- Study Factors; these correspond to controlled variations within the research method used. One example could be the space of parameter values for a simulation. Another could be the different diets fed to lab mice prior to data collection.
- Data Origins; if the data is generated first hand from lab/field work, then origin often corresponds to certain attributes of study subjects, e.g., the gender and age of humans from which DNA has been sampled. If data is derived from already existing datasets then origin corresponds to attributes such as, data catalog names, versions, or access end-points.
- Workflow provenance is generic. In order to allow processing of diverse scientific datasets using equally diverse tools; workflows systems are designed to be oblivious to what data and tasks internally represent. This is also known as the “black-box approach”, which generates provenance graphs comprised of opaque nodes [13]. Meanwhile, for reporting, we require domain-specific information on data and tasks. Henceforth workflow provenance requires further annotation in order to be useful in reporting.
- Workflows as automation artefacts proliferate data generation. A workflow is rarely run once, typically ran several times to explore the effects of parameter or input changes on outputs. As a result manual annotation of workflow outputs can be a daunting task for scientists. In a recent survey scientists have stated that they receive ample automation support for performing the analyses, much less so for post-analysis activities such as result management, annotation and sharing [14].
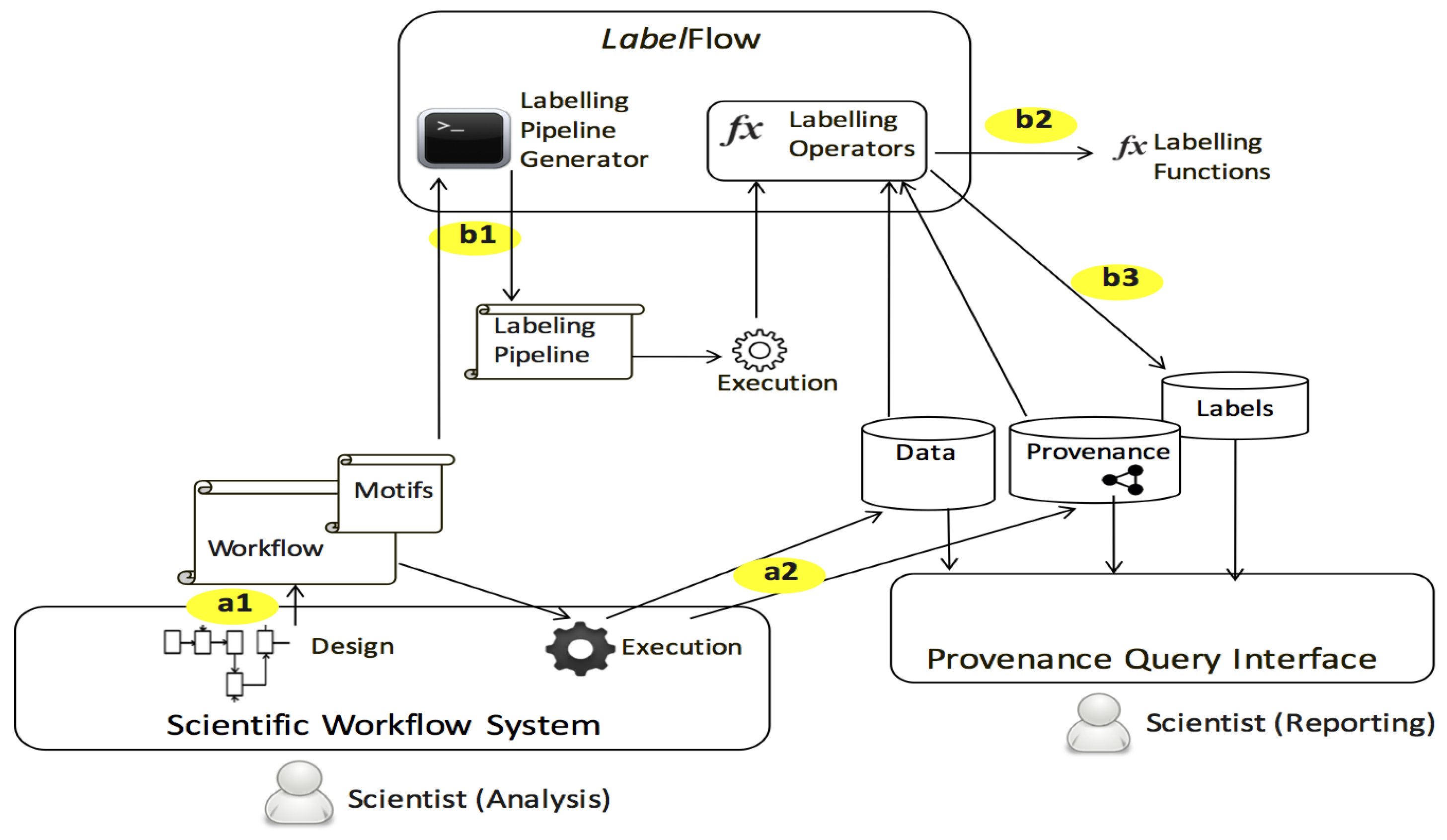
- LabelFlow, a generic framework, that can be plugged with discipline-specific metadata extractors, for annotating datasets. Central to this framework is a set of labelling operators for minting and propagating annotations.
- Labelling Pipelines, which are shadow annotator processes deduced per scientific workflow. The labelling pipeline of a workflow can be used for annotating results for all executions of that workflow. We provide a practical algorithm that consumes a workflow description and its activities’ functional categories and produces a labelling pipeline.
- An implementation of LabelFlow based on technology-independent standard provenance models and its validation using a case study.
2. Workflows, Provenance and Reporting: A Case
- It contains the commonly observed type of activities in workflows and reflects their occurrence percentage. As we discuss in Section 3, a comprehensive survey has categorised activity functionalities to certain Motifs, our workflow’s activities have the data analysis and data retrieval motifs as well as extensive data adaptation.
- Analyses and data retrievals in this workflow are configured by input parameters, which, as we discuss in Section 2.3, become an important hook for querying provenance.
- It illustrates the genericity of workflow provenance, our prime motivation, which limits the use of provenance for reporting. It also illustrates the pattern in provenance, which can be observed in workflows and negatively impacts the utility of both generic and annotated provenance.
2.1. Layers of Provenance
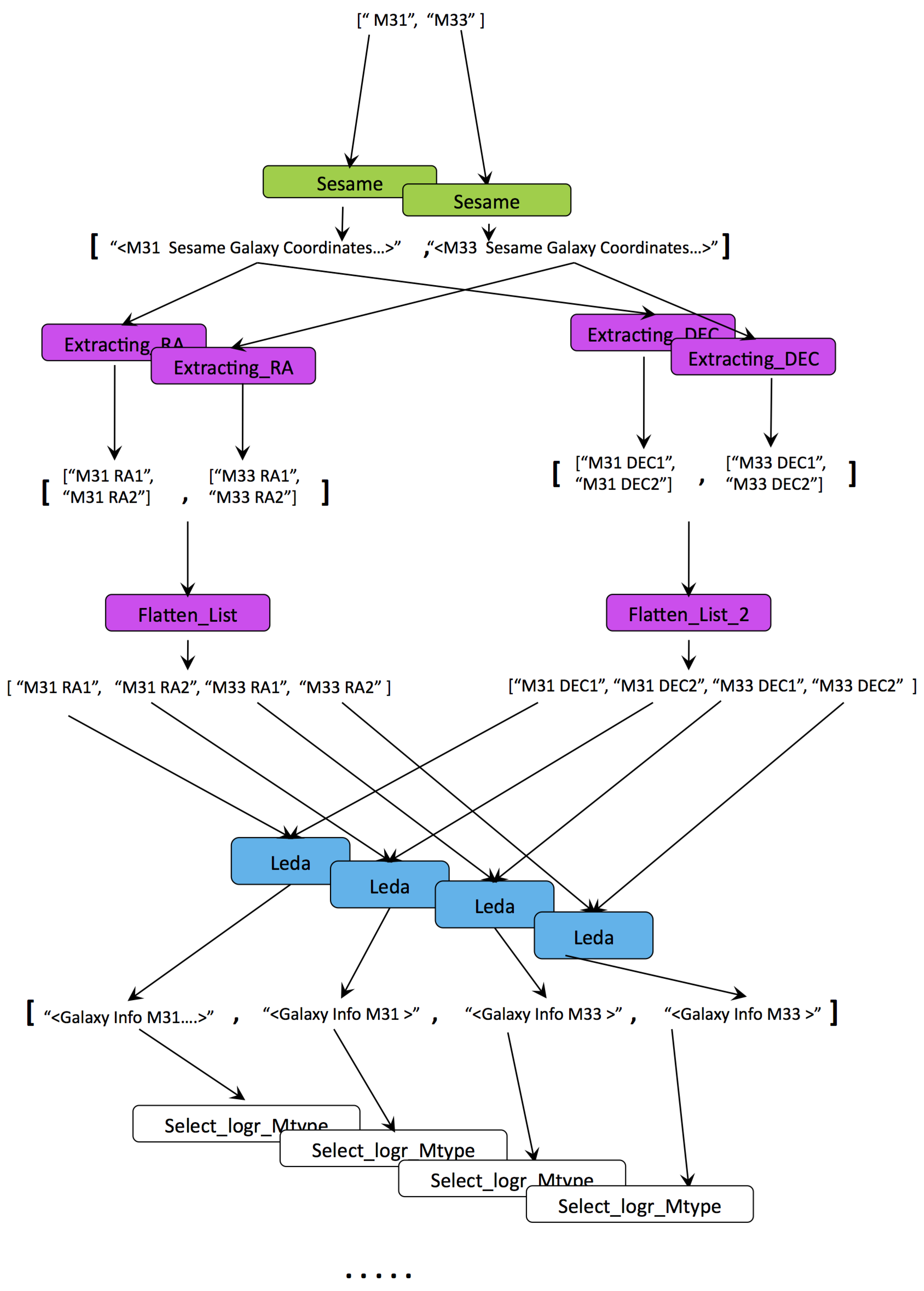
- Iteration proliferates data generation. In our example in Figure 2 the number of outputs increases linearly with inputs (n inputs produce outputs). In cases of complex factorised analyses, where tasks are run on (cartesian) combinations of inputs, the increase in outputs becomes polynomial. As such, automation in annotation is a crucial requirement.
- Iteration is an intricate feature of workflows. It requires understanding nested collection structures as well as creating cartesian combinations of items from collections. When utilised correctly, iteration is the primary mechanism making provenance an index linking the subjects and factors of an analysis to corresponding results. However, in certain cases, such as to avoid repeated and costly invocation of remote services or for quicker data adaptation, iteration can be by-passed. This is illustrated with the “Flatten_List” step in Figure 2. All the steps in the workflow are repeated for each input, whereas “Flatten_List” is executed once, processing data of multiple galaxies at a single step. This (anti)pattern, also known as the “ problem”, breaks input-to-output traceability; and, as we shall identify later in this section, it is one of the main factors that reduce the utility of provenance for reporting.
2.2. Modelling Workflows and Provenance
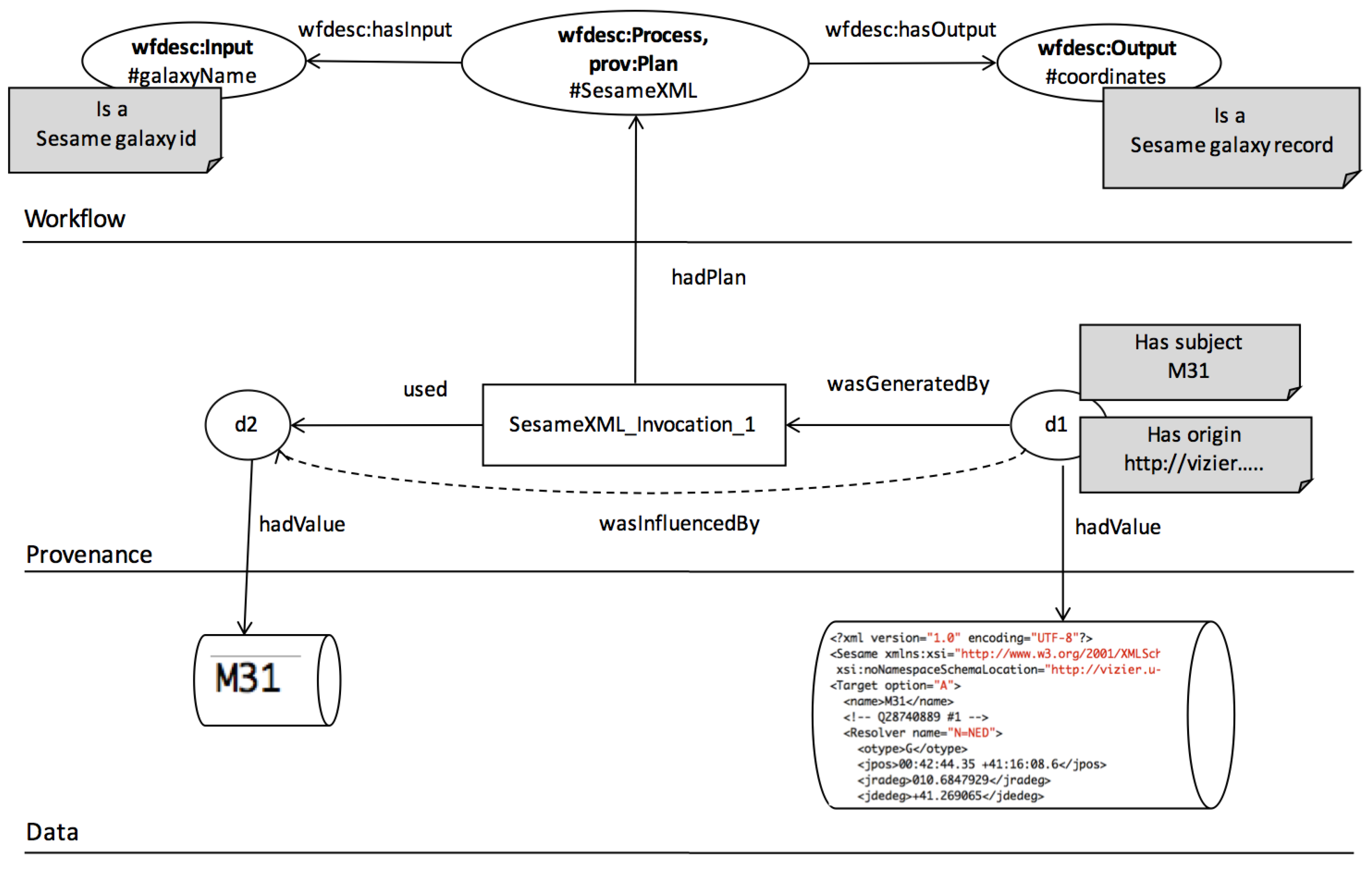
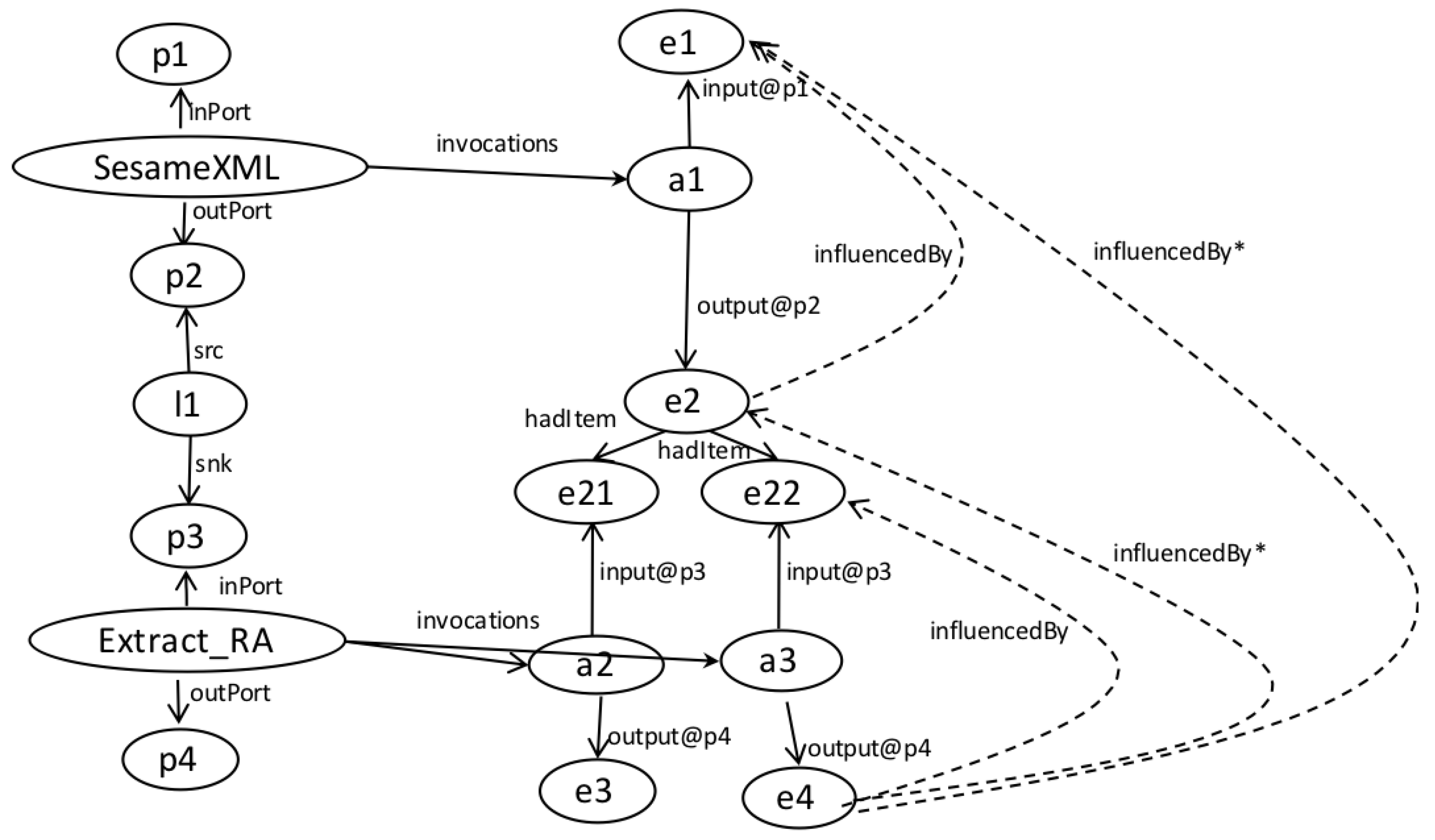
- We assume that a workflow is a directed acyclic graph of analytical tasks and dataflow dependencies among ports of tasks. We assume that provenance is a directed acyclic graph of (data) entities and influence relations among entities (produced/consumed by activities).
- We exclude information on the semantics of task iteration. Workflow systems all provide their own means to repeatedly apply tasks to data. The lack of iteration configuration specifics does not affect LabelFlow’s ability to operate. LabelFlow operates at the level of individual task invocations, and a task’s execution footprint in provenance is the same in all mentioned workflow systems [31]. Meanwhile workflow systems differ in the way they reflect data granularity in provenance [31]. In Taverna an iterated task would be consuming individual items in a collection, whereas in Vistrails there is not collection modelling therefore all iterations would appear to consume/produce a single entity.
2.3. Using Provenance for Reporting
- lineage based on value-copying/data adaptation.
- lineage based on any other analytical computation
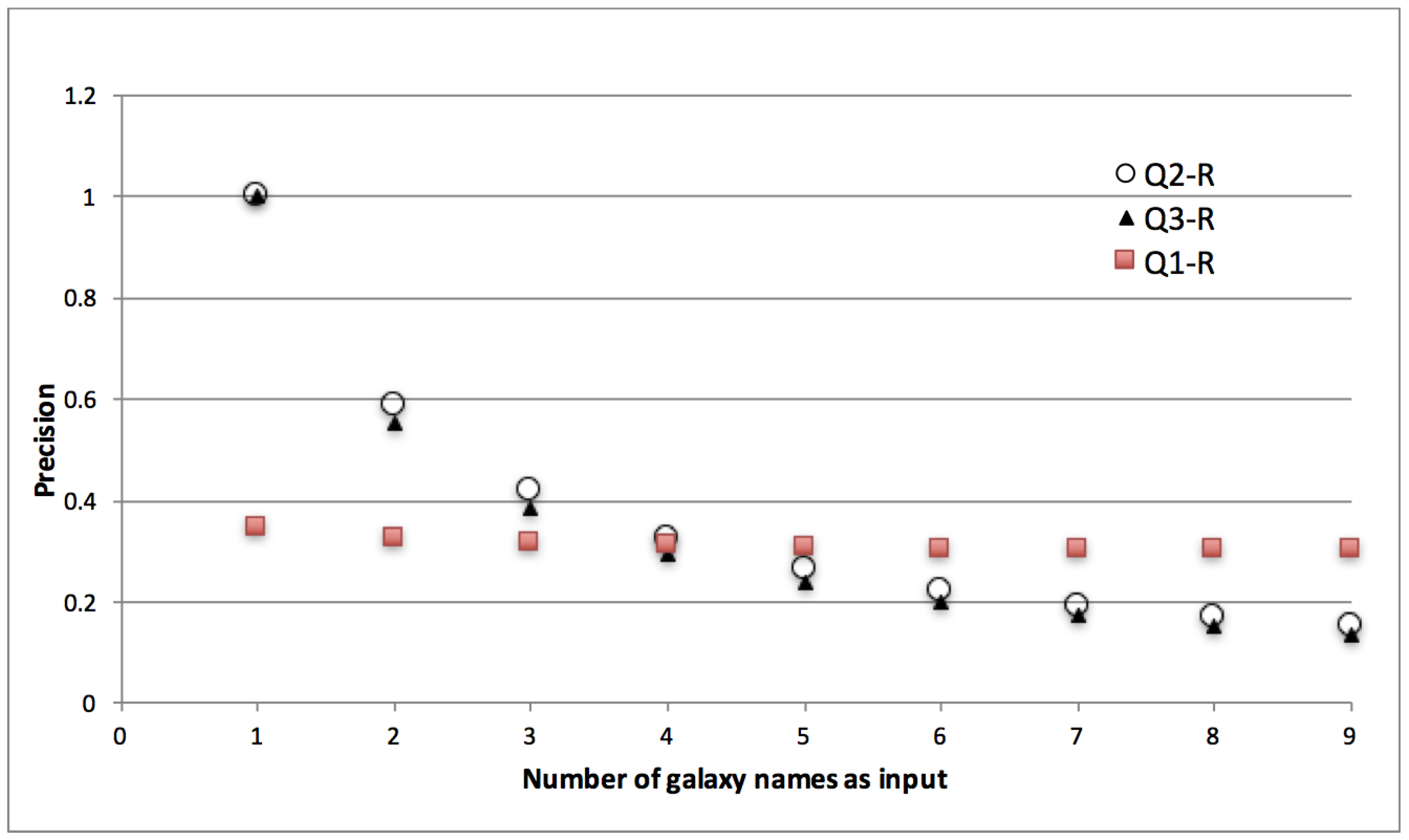
- Correct implementation of iteration is a pre-requisite for provenance being useful in reporting. Queries that seek results belonging to a particular input (subject or factor) require discrete reachability between inputs and respective outputs. When traceability is broken, provenance, either generic or annotated, is of little use. The lack of discrete traceability that causes the sharp loss of precision in Q2-G and Q3-G is not a problem that we’re trying to solve with LabelFlow. In prior work we’ve tackled this problem and shown that workflows can be analyzed to check whether their provenance will have the n-by-m pattern, i.e., lack of discrete traceability [31]. We find it important to highlight this pattern in the context of this paper, because, as we shall see in Section 6 if it exists in provenance it equally reduces provenance utility even after labelling.
- Domain specific information is key for reporting. Q1 seeks , whereas Q2 and Q3 seek attributes, yet we realised our queries over generic provenance. In the absence of metadata, in order to find nodes of interest, we were forced to put selective criteria on data values (Q2-G and Q3-G), or in attributes like name (Q1-G). This approach proved to have the following disadvantages:
- -
- Due to the separated storage of data and provenance seamless implementation of provenance queries was not possible. In fact for queries Q2-G and Q3-G, as a precursor, we identified which node in the provenance graph corresponds to the galaxy by first scanning through the data values stored in the file system.
- -
- As we realise queries over implicit information, and as there is no structure or vocabulary restrictions on this information, our approach is adhoc. e.g., the informativeness of a name for a workflow port or activity is at the disposal of workflow designer, names can be freely given and the same activity (e.g., Sesame database lookup) can have different names across workflows. Similarly in Q3-G we were unable to implement morphology criteria part of query as the data values were not self-describing and structured enough to allow a systematic implementation of this criteria.
- Transparent lineage is needed for reporting. One of our queries (Q1-G) was seeking data based on its origin. In our implementation we represented this with a query where we sought nodes that have some lineage relation to designated origin node. Our answers to this query were partly correct. This is because of we were using opaque lineage relations, a typical result of black-box workflow provenance. Opaque lineage tells us that one data artefact influences the other, but it does not specify the specific nature of this influence. On the other hand Q1-G requires more transparency, it seeks those data artefacts that descend from an origin artefact via a particular influence relation, i.e., value-copying.
- she can ask for outputs of analytical tasks based on input configurations, (the quintessential workflow provenance query)
- she can inquire data origin for data retrieved from external databases (a typical motif in scientific workflows)
3. Architecture and Assumptions
- are available for domains and support an invocation interface that allows them to be plugged intoLabelFlow.
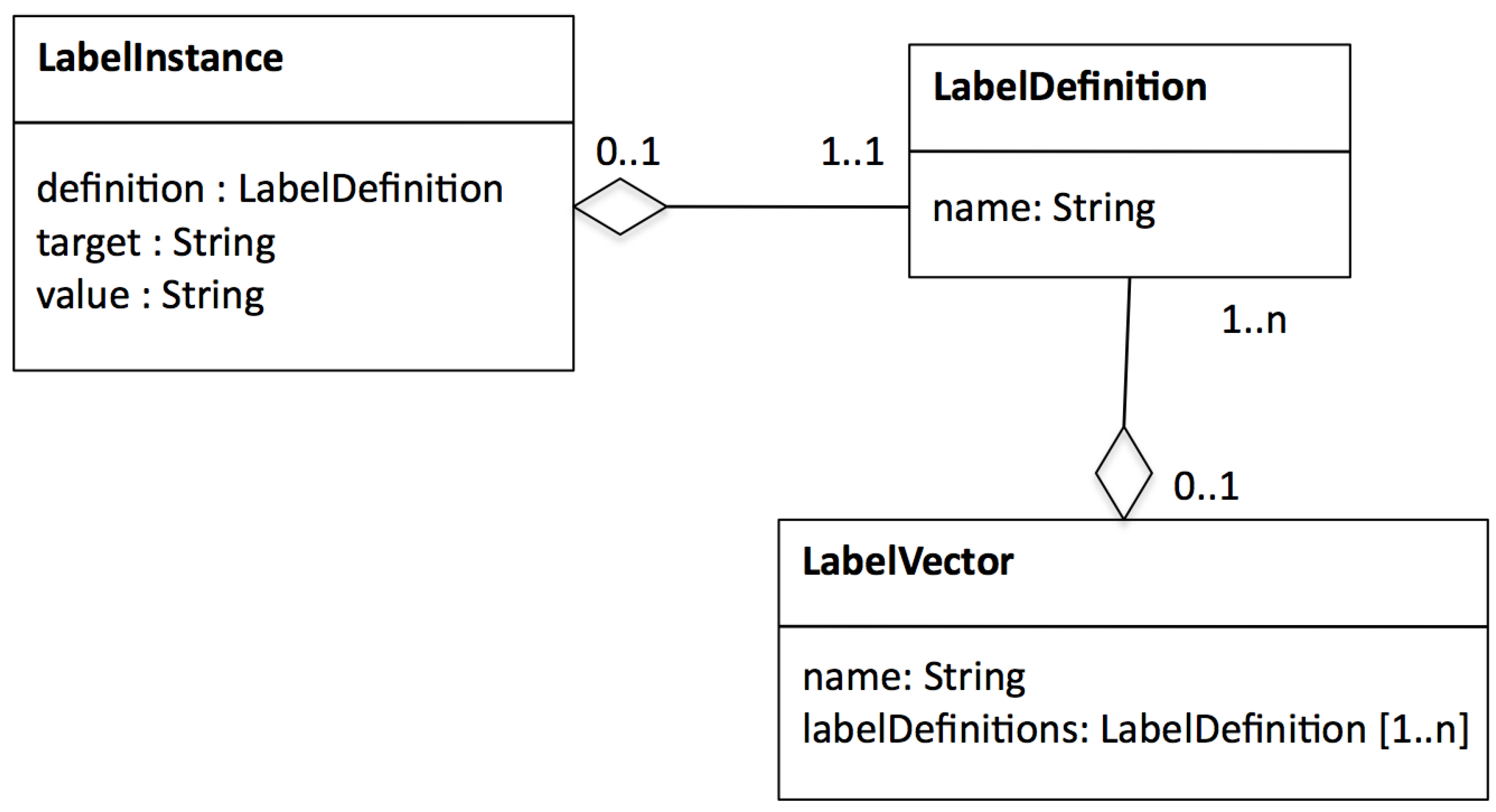
- Workflow tasks are annotated with an extended form of annotation called a . Note that only describe a task’s function. Meanwhile, tasks have input/output ports and we require information on which ports shall receive labels and if/how ports are related. We discuss the information within in the next section. The mechanism to create is left out of scope in this paper.
4. LabelFlow Framework
5. LabeFlow Model
5.1. Labelling Operators
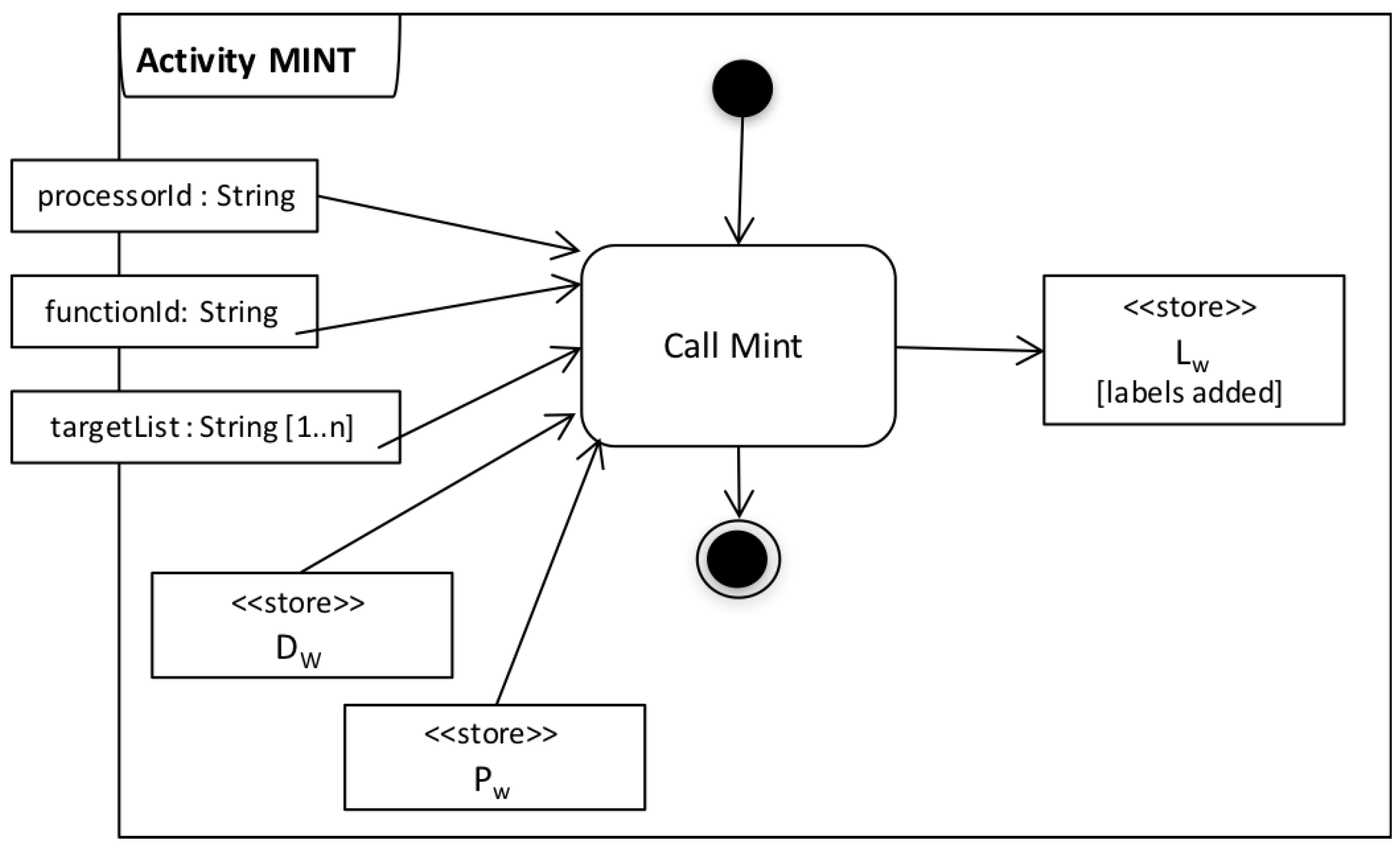
| Algorithm 1: Mint |
 |
| Algorithm 2: Propagate |
 |
| Algorithm 3: Distribute |
 |
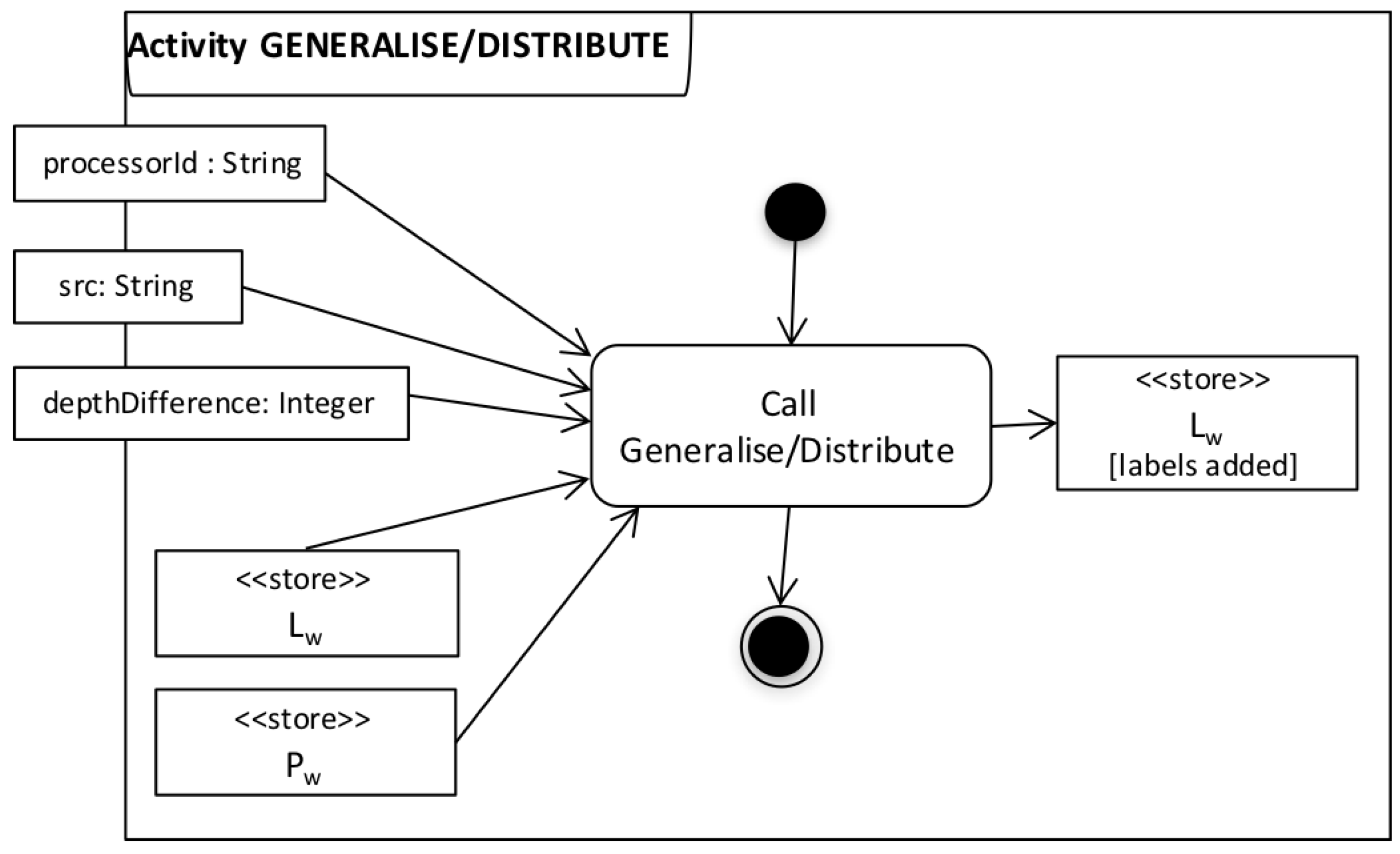
| Algorithm 4: Generalise |
 |
5.2. Labelling Pipelines
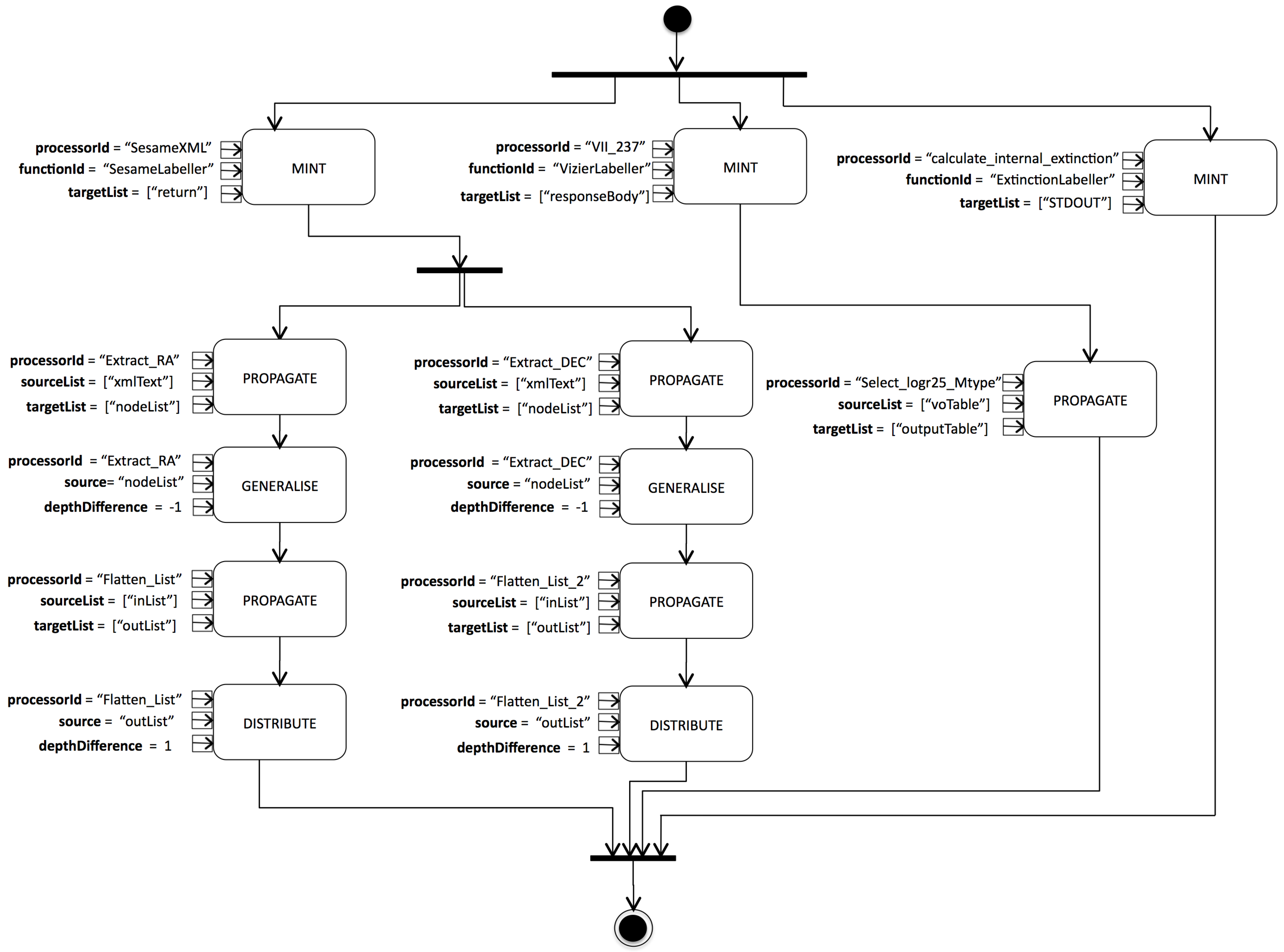
5.2.1. Example Labelling Pipeline
- String value of “SesameXML” for the input parameter.
- String value of “SesameLabeller” for input parameter,
- a Set containing the String value “return” for input parameter.
5.2.2. Pipeline Generation Procedure
5.3. Implementation
| Algorithm 5: Pipeline Generation |
 |
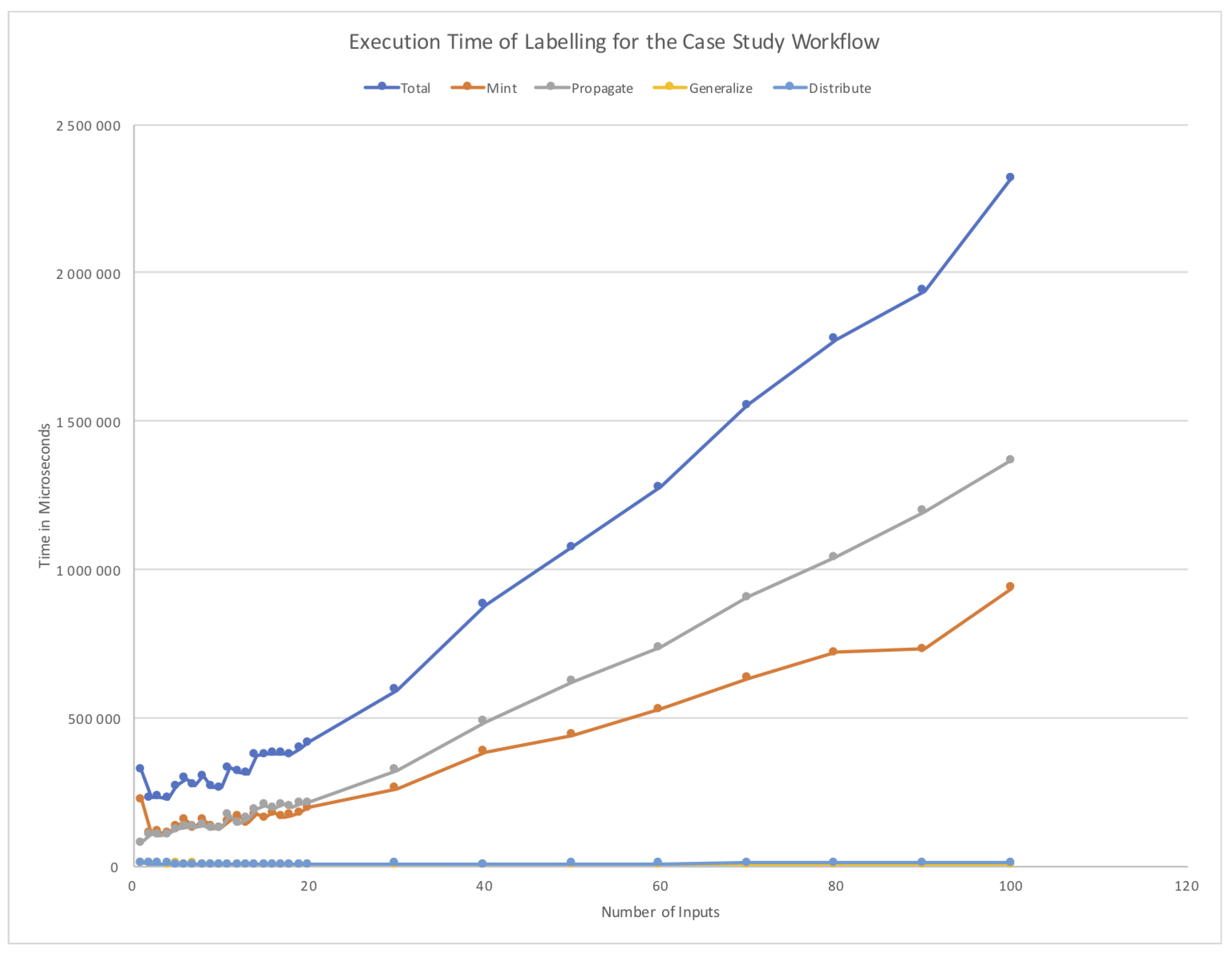
5.4. Performance of LabelFlow
6. Revisiting Case-Study
- we implemented three simple domain-specific labelling functions, one for each scientifically significant step in the workflow (as discussed in Section 5.2.1). These functions can parse the data consumed/generated by these activities and create labels that correspond to either input configurations (context) or data origin.
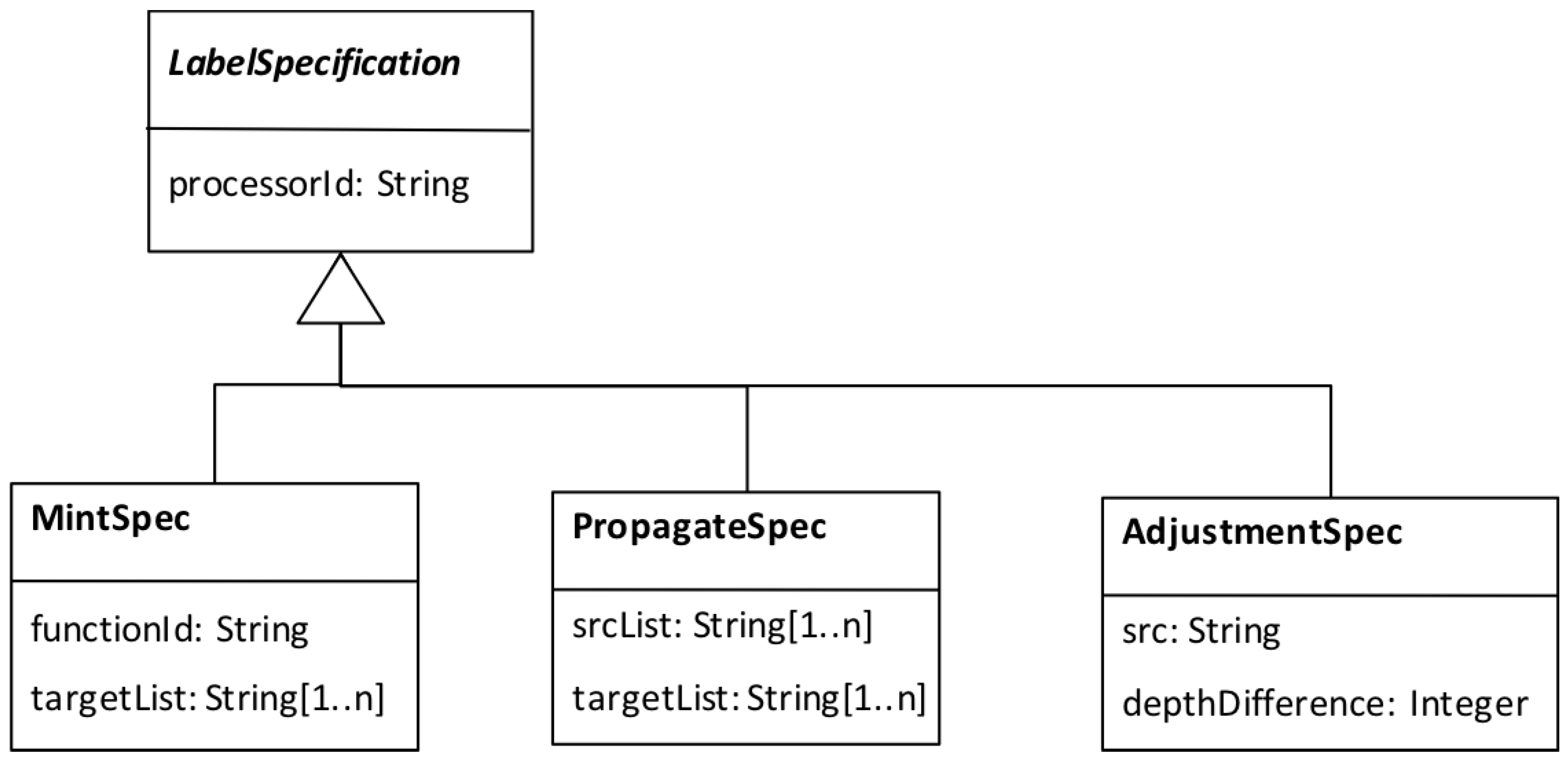
- we associated Labelling Specs with workflow activities according to the information model given in Figure 12. For the data adapter activities, for which a corresponding labelling behaviour is given (Table 3), we created s and for the scientifically significant activities, we created s, pointing to the labelling functions.
7. Related Work
7.1. Obtaining Annotations
- it is non-intrusive to the execution of workflow. As the metadata is sourced from the data, as long as the data values are kept, annotation can take place as an offline process any later time. As it is offline, however, LabelFlow may miss out on metadata that is available only at runtime and does not get serialised into task outputs.
- is not tied to a particular raw data form or indexing technology. In our survey of workflow Motifs [24] we observed that metadata is not always presented as additional columns in a tabular layout. It is often found in dedicated headers as in a Blast Report, a Variant Call File or a NIFTI file header for fMRI data.
- it focuses on capturing the context that surrounds a particular analytical activity. All prior approaches focus on extracting metadata that they assume exists within raw data. As illustrated in our case study workflow, context may not be consistently available within the data, therefore we focus on the cases where the context is spread out among input parameters and data copies. As we use labelling functions that consume all data (input/output) associated with an activity, we provide a mechanism to weave back this context and propagate it to data copies.
7.2. Propagating Annotations
8. Discussion
- We act on partial information (grey-box transparency denoting some value-copying occurring between inputs and outputs of an activity). When an activity invocation consumes a collection of items with distinct labels, and produces another collection of items, grey-box transparency does not allow us to accurate propagate labels item-wise. So instead we first labels to the top level input collection and them to the top level output collection.
- To further expand the reach of labels we labels at the top level collection to each item.
- they do not support a fine-grained provenance capability where annotations from distinct fine grained sources need to be managed as in the Galaxy workflow system’s metadata propagation feature [7].
- or they require the user to not only supply the initial annotations but also the rules of propagation per workflow activity as in the Wings workflow system [8].
- Start by identifying a metadata profile that is applicable to your domain. Profiles are lightweight metadata schemas, typically comprised of a set of attributes. Profiles find increasing use in the context of data publishing. In our case-study this was the Astronomy Visualisation Metadata scheme available at the UK Digital Curation Center portal [3].
- Check whether there are existing tools/extractors that can produce metadata conforming to this profile. In our case a tool did not exist. However there were several VOTable parsers, or plain XML parsers we could utilise, we the latter.
- Develop a label minting function that either wraps an existing tool or is built afresh, which returns labels, whose definition correspond to attributes from the applicable metadata profile.
- For a workflow or group of workflows identify provenance querying hooks, these are input parameters that can be permuted at run time, or processor configurations determined during workflow design. Create a labelling vector that is comprised of label definitions that would carry this information. In our case this was simply a subset of the AVM metadata profile.
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A.
Appendix A.1. Auxiliary Methods
| getInvocations(processorId:String, provStore:String):String[0..n] |
| Obtains identifiers of all the nodes in the trace that are documented to have occurred using as a . |
| getAllGeneratedOutputs(processorId:String, port:String, provStore:String):String[0..n] |
| Obtains identifiers of all the (data) nodes in the trace that have been in a qualified relationship with some activity, where the activity has occurred according to a of identifier and the generated data had role () . |
| getAllActivityData(activityId:String, provStore:String) :String[0..n] |
| Obtains identifiers all the (data) nodes in the trace that have been in a or a relationship with the designated . |
| getActivityOutData(activityId:String, port:String, provStore:String):String[0..n] |
| Obtains identifiers of the (data) nodes in the trace that are in a relation with the designated , where the generation is qualified stating that the data node played the role () identified with . |
| bindLabelsToData(dataId:String, labels:LabelInstance[0..n]) :LabelInstance[0..n] |
| Returns a copy of the labels, where the target of each copy is set to the designated data record. |
| clone(labels:LabelInstance[0..n]):LabelInstance[0..n] |
| Creates a copy of all the labels in the input set . |
| submitLabels(labels:LabelInstance[0..n], labelStore:String) |
| Stores all the label instances in the designated label space. |
| getItems(coll:String, depthDifference:Integer, provStore:String):String[0..n] |
| Obtains identifiers of (data) nodes in the trace that are contained () by the designated at level deep. |
| getLabels(item, labelDefinitions, labelStore:String) :LabelInstance[0..n] |
| Obtains all the labels, whose target is the designated item. |
| getEnclosingCollections(items:String[0..n], depthDifference:Integer, provStore:String):String[0..n] |
| Obtains the identifiers of nodes in the trace, which at level deep contain () the designated items. |
Appendix A.2. UML Activity Diagram Syntax
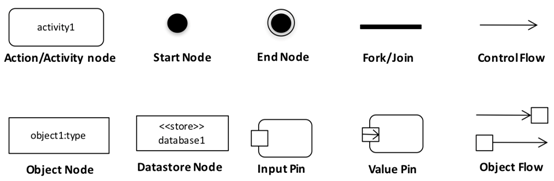 |
|---|
| An activity diagram is a graph of nodes denoting a process comprised of steps of computation and flows of (primarily) control (and optionally) data among steps. |
| An action/activity node (rounded rectangle) denotes a computational step. An action is an atomic step which is not further broken into sub-steps, whereas an activity is a group of actions or sub-activities. |
| Start node (solid circle) is a control node at which flow starts when an activity is invoked. |
| End node (hollow circle with solid circle inside) is a control node that stops all flows in an activity. |
| Fork node (thick line segment) is a control node that has one incoming edge and multiple outgoing edges and is used to split incoming flow into multiple concurrent flows. Join node is a control node that has multiple incoming edges and one outgoing edge and is used to synchronise incoming concurrent flows. |
| Control flow edge (arrow) is an edge denoting flow of control from one activity to another. |
| Object node (rectangle) is an edge denoting flow of data from one activity to another. |
| A data store nodes (rectangle) are stereotyped object nodes, which denote non-transient data that is persisted during the computational process. |
| Object flow edge (arrow) is an edge denoting flow of data during a computational process. An object flow edge is one that connects two nodes, where at least one is an object node. A value pin is special kind of input pin defined to provide constant values as input. |
| Pins (small rectangle at edge of rounded rectangle) are object nodes used to denote inputs/outputs to activities. A value pin is a special kind of input pin, which denotes constant-valued inputs to an activity. |
References
- Hey, T.; Tansley, S.; Tolle, K.M. (Eds.) The Fourth Paradigm: Data-Intensive Scientific Discovery; Microsoft Research: New York, NY, USA, 2009. [Google Scholar]
- Scientific Data, Open-Access Journal; Nature Publishing Group: London, UK, 2015. Available online: http://www.nature.com/sdata/ (accessed on 22 February 2018).
- Davenhall, C. Curation Reference Manual, Chapter on Scientific Metadata; The Digital Curation Centre (DCC): Edinburgh, UK, 2011; Available online: http://www.dcc.ac.uk/resources/curation-reference-manual (accessed on 22 February 2018).
- Taylor, C.F.; Field, D.; Sansone, S.A.; Aerts, J.; Apweiler, R.; Ashburner, M.; Ball, C.A.; Binz, P.; Bogue, M.; Booth, T.; et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: The MIBBI project. Nat. Biotechnol. 2008, 26, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Sansone, S.A.; Rocca-Serra, P.; Field, D.; Maguire, E.; Taylor, C.; Hofmann, O.; Fang, H.; Neumann, S.; Tong, W.; Amaral-Zettler, L.; et al. Toward interoperable bioscience data. Nat. Genet. 2012, 44, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Ludaescher, B.; Altintas, I.; Berkley, C.; Higgins, D.; Altintas, I.; Berkley, C.; Higgins, D.; Jaeger, E.; Jones, M.; Lee, E.A.; et al. Scientific workflow management and the Kepler system. Concurr. Comput. Pract. Exp. 2006, 18, 1039–1065. [Google Scholar] [CrossRef]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Shah, P.; Elnitski, L.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Gil, Y.; Ratnakar, V.; Kim, J.; González-Calero, P.A.; Groth, P.; Moody, J.; Deelman, E. Wings: Intelligent Workflow-Based Design of Computational Experiments. IEEE Intell. Syst. 2011, 26, 62–72. [Google Scholar] [CrossRef]
- Callahan, S.P.; Freire, J.; Santos, E.; Scheidegger, C.E.; Silva, C.T.; Vo, H.T. Vistrails: Visualization meets data management. In ACM SIGMOD; ACM Press: New York, NY, USA, 2006; pp. 745–747. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: https://www.r-project.org (accessed on 22 February 2018).
- Rossum, G. Python Reference Manual; Technical Report; CWI (Centre for Mathematics and Computer Science): Amsterdam, The Netherlands, 1995. [Google Scholar]
- Missier, P.; Paton, N.W.; Belhajjame, K. Fine-grained and Efficient Lineage Querying of Collection-based Workflow Provenance. In Proceedings of the 13th International Conference on Extending Database Technology, Lausanne, Switzerland, 22–26 March 2010; ACM: New York, NY, USA, 2010; pp. 299–310. [Google Scholar]
- Chapman, A.; Jagadish, H.V. Understanding provenance black boxes. Distrib. Parallel Databases 2010, 27, 139–167. [Google Scholar] [CrossRef]
- Tenopir, C.; Allard, S.; Douglass, K.; Aydinoglu, A.U.; Wu, L.; Read, E.; Manoff, M.; Frame, M. Data Sharing by Scientists: Practices and Perceptions. PLoS ONE 2011, 6, e21101. [Google Scholar] [CrossRef] [PubMed]
- Missier, P.; Sahoo, S.S.; Zhao, J.; Goble, C.; Sheth, A. Janus: From Workflows to Semantic Provenance and Linked Open Data. In Proceedings of the 3rd International Provenance and Annotation Workshop (IPAW 2010), Troy, NY, USA, 15–16 June 2010; pp. 129–141. [Google Scholar]
- Cao, B.; Plale, B.; Subramanian, G.; Missier, P.; Goble, C.A.; Simmhan, Y. Semantically Annotated Provenance in the Life Science Grid. In Proceedings of the 1st International Workshop on the role of Semantic Web in Provenance Management (SWPM 2009), Washington DC, USA, 25 October 2009. [Google Scholar]
- Ailamaki, A.; Kantere, V.; Dash, D. Managing Scientific Data. Commun. ACM 2010, 53, 68–78. [Google Scholar] [CrossRef]
- Belhajjame, K.; Zhao, J.; Garijo, D.; Garrido, A.; Soiland-Reyes, S.; Alper, P.; Corcho, O. A Workflow PROV-corpus Based on Taverna and Wings. In Proceedings of the Joint EDBT/ICDT 2013 Workshops, Genoa, Italy, 18–22 March 2013; ACM: New York, NY, USA, 2013; pp. 331–332. [Google Scholar]
- Hull, D.; Stevens, R.; Lord, P.; Wroe, C.; Goble, C. Treating shimantic web syndrome with ontologies. In Proceedings of the 1st Advanced Knowledge Technologies Workshop on Semantic Web Services (AKT-SWS04) KMi, Milton Keynes, UK, 8 December 2004. [Google Scholar]
- Alagiannis, I.; Borovica, R.; Branco, M.; Idreos, S.; Ailamaki, A. NoDB: Efficient Query Execution on Raw Data Files. In Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data, Scottsdale, AZ, USA, 20–24 May 2012; ACM: New York, NY, USA, 2012; pp. 241–252. [Google Scholar]
- Wu, K. FastBit: An efficient indexing technology for accelerating data-intensive science. J. Phys. Conf. Ser. 2005, 16, 556. [Google Scholar] [CrossRef]
- Alawini, A.; Maier, D.; Tufte, K.; Howe, B.; Nandikur, R. Towards Automated Prediction of Relationships Among Scientific Datasets. In Proceedings of the 27th International Conference on Scientific and Statistical Database Management, La Jolla, CA, USA, 29 June–1 July 2015; ACM: New York, NY, USA, 2015. [Google Scholar]
- Sousa, V.S.; de Oliveira, D.; Mattoso, M. Exploratory Analysis of Raw Data Files through Dataflows. In Proceedings of the 2014 International Symposium on Computer Architecture and High Performance Computing Workshop (SBAC-PADW), Paris, France, 22–24 October 2014; pp. 114–119. [Google Scholar]
- Garijo, D.; Alper, P.; Belhajjame, K.; Corcho, O.; Gil, Y.; Goble, C. Common motifs in scientific workflows: An empirical analysis. Future Gener. Comput. Syst. 2014, 36, 338–351. [Google Scholar] [CrossRef]
- Zhao, J.; Sahoo, S.S.; Missier, P.; Sheth, A.P.; Goble, C.A. Extending Semantic Provenance into the Web of Data. IEEE Internet Comput. 2011, 15, 40–48. [Google Scholar] [CrossRef]
- Alper, P.; Goble, C.A.; Belhajjame, K. On assisting scientific data curation in collection-based dataflows using labels. In Proceedings of the 8th Workshop On Workflows in Support of Large-Scale Science, (WORKS), Denver, CO, USA, 17 November 2013; pp. 7–16. [Google Scholar]
- Alper, P.; Belhajjame, K.; Goble, C.A.; Karagoz, P. LabelFlow: Exploiting Workflow Provenance to Surface Scientific Data Provenance. In Proceedings of the 5th International Provenance and Annotation Workshop (IPAW), Cologne, Germany, 9–13 June 2014; pp. 84–96. [Google Scholar]
- Exposito, S.S. Workflow: Calculating the Internal Extinction with Data from Leda. myExperiment Repository. 2012. Available online: http://www.myexperiment.org/workflows/2920/versions/2.html (accessed on 22 February 2018).
- Missier, P.; Soiland-Reyes, S.; Owen, S.; Tan, W.; Nenadic, A.; Dunlop, I.; Williams, A.; Oinn, T.; Goble, C. Taverna, Reloaded. In Proceedings of Scientific and Statistical Database Management Conference (SSDBM), Lecture Notes in Computer Science, Heidelberg, Germany, 30 June–2 July 2010; Gertz, M., Ludäscher, B., Eds.; Springer: Berlin, Germany, 2010; Volume 6187, pp. 471–481. [Google Scholar]
- Moreau, L.; Ludäscher, B.; Altintas, I.; Barga, R.S.; Bowers, S.; Callahan, S.; Chin, G., Jr.; Clifford, B.; Cohen, S.; Cohen-Boulakia, S.; et al. The First Provenance Challenge. CCPE 2008, 20, 409–418. [Google Scholar]
- Alper, P.; Belhajjame, K.; Goble, C.A. Static analysis of Taverna workflows to predict provenance patterns. Future Gener. Comput. Syst. 2017, 75, 310–329. [Google Scholar] [CrossRef]
- Belhajjame, K.; Zhao, J.; Garijo, D.; Gamble, M.; Hettne, K.; Palma, R.; Mina, E.; Corcho, O.; Gómez-Pérez, J.M.; Bechhofer, S.; et al. Using a suite of ontologies for preserving workflow-centric research objects. Web Semant. Sci. Serv. Agents World Wide Web 2015, 32, 16–42. [Google Scholar] [CrossRef]
- Wood, D.; Lanthaler, M.; Cyganiak, R. RDF 1.1 Concepts and Abstract Syntax. W3C Recommendation. 2014. Available online: https://www.w3.org/TR/rdf11-concepts/ (accessed on 22 February 2018).
- Groth, P.; Editors, L.M. PROV-Overview: An Overview of the PROV Family of Documents; W3C, 2013. Available online: http://www.w3.org/TR/2013/NOTE-prov-overview-20130430/ (accessed on 22 February 2018).
- Missier, P.; Dey, S.; Belhajjame, K.; Cuevas-Vicenttın, V.; Ludäscher, B. D-PROV: Extending the PROV provenance model with workflow structure. In Proceedings of the 5th USENIX Workshop on the Theory and Practice of Provenance, Lombard, IL, USA, 2–3 April 2013. [Google Scholar]
- Brandizi, M.; Melnichuk, O.; Bild, R.; Kohlmayer, F.; Rodriguez-Castro, B.; Spengler, H.; Kuhn, K.A.; Kuchinke, W.; Ohmann, C.; Mustonen, T.; et al. Orchestrating differential data access for translational research: A pilot implementation. BMC Med. Inf. Decis. Mak. 2017, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.; Arenas, M.; Benedikt, M. SPARQLByE: Querying RDF Data by Example. Proc. VLDB Endow. 2016, 9, 1533–1536. [Google Scholar] [CrossRef]
- Garijo, D.; Alper, P.; Belhajjame, K. The Workflow Motif Ontology. UPM Ontology Engineering Group, Revision 1.02. 2013. Available online: http://vocab.linkeddata.es/motifs/ (accessed on 22 February 2018).
- Booch, G.; Rumbaugh, J.; Jacobson, I. Unified Modeling Language User Guide, 2nd ed.; Addison-Wesley Object Technology Series; Addison-Wesley Professional: Boston, MA, USA, 2005. [Google Scholar]
- Alper, P. LabelFlow Evaluation Datasets. 2015. Available online: https://github.com/pinarpink/phd-sources/tree/master/labeling-workflow-generator (accessed on 22 February 2018).
- Belhajjame, K.; Cheney, J.; Corsar, D.; Garijo, D.; Soiland-Reyes, S.; Zednik, S.; Zhao, J. PROV-O: The PROV Ontology; W3C, 2012. Available online: http://www.w3.org/TR/prov-o/ (accessed on 22 February 2018).
- Group, P.W. PROV Implementation Report. 2013. Available online: https://www.w3.org/TR/prov-implementations/ (accessed on 22 February 2018).
- Carroll, J.J.; Dickinson, I.; Dollin, C.; Reynolds, D.; Seaborne, A.; Wilkinson, K. Jena: Implementing the Semantic Web Recommendations. In Proceedings of the 13th International World Wide Web Conference on Alternate Track Papers & Amp, New York, NY, USA, 17–20 May 2004; Posters; ACM: New York, NY, USA, 2004; pp. 74–83. [Google Scholar]
- Moreau, L.; Huynh, T.D.; Michaelides, D. An Online Validator for Provenance: Algorithmic Design, Testing, and API. In Fundamental Approaches to Software Engineering; Gnesi, S., Rensink, A., Eds.; Springer: Berlin, Germany, 2014; pp. 291–305. [Google Scholar]
- Missier, P.; Sahoo, S.S.; Zhao, J.; Goble, C.; Sheth, A. Janus: From Workflows to Semantic Provenance and Linked Open Data. In Provenance and Annotation of Data and Processes; Springer: Berlin, Germany, 2010; Volume 6378, pp. 129–141. [Google Scholar]
- Zhao, J.; Wroe, C.; Goble, C.; Stevens, R.; Quan, D.; Greenwood, M. Using Semantic Web Technologies for Representing e-Science Provenance. In Proceedings of the ISWC 2004, Hiroshima, Japan, 7–11 November 2004; Springer: Berlin, Germany, 2004; Volume 3298, pp. 92–106. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sahoo, S.S.; Sheth, A.; Henson, C. Semantic provenance for escience: Managing the deluge of scientific data. IEEE Internet Comput. 2008, 12, 46–54. [Google Scholar] [CrossRef]
- De Oliveira, D.; Silva, V.; Mattoso, M. How Much Domain Data Should Be in Provenance Databases? In Proceedings of the 7th USENIX Workshop on the Theory and Practice of Provenance (TaPP 15), Edinburgh, UK, 8–9 July 2015; USENIX Association: Edinburgh, UK, 2015.
- Halper, M.; Geller, J.; Perl, Y. Value Propagation in Object-oriented Database Part Hierarchies. In Proceedings of the Second International Conference on Information and Knowledge Management, ACM, CIKM’93, Washington, DC, USA, 1–5 November 1993; pp. 606–614. [Google Scholar]
- Artale, A.; Franconi, E.; Guarino, N.; Pazzi, L. Part-whole Relations in Object-centered Systems: An Overview. Data Knowl. Eng. 1996, 20, 347–383. [Google Scholar] [CrossRef]
- Greenberg, J. Theoretical Considerations of Lifecycle Modelling: An Analysis of the Dryad Repository Demonstrating Automatic Metadata Propagation, Inheritance, and Value System Adoption. Cat. Classif. Q. 2009, 47, 380–402. [Google Scholar]
- Bhagwat, D.; Chiticariu, L.; Tan, W.-C.; Vijayvargiya, G. An Annotation Management System for Relational Databases. In Proceedings of the Thirtieth International Conference on Very Large Data Bases, Toronto, ON, Canada, 31 August–3 September 2004; Nascimento, M.A., Ozsu, M.T., Nascimento, M.A., Özsu, M.T., Kossmann, D., Miller, R.J., Blakeley, J.A., Schiefe, B., Eds.; VLDB Endowment Inc.: San Fransisco, CA, USA, 2004; pp. 900–911. [Google Scholar]
- Bowers, S.; Ludäscher, B. A Calculus for Propagating Semantic Annotations Through Scientific Workflow Queries. In Proceedings of the 2006 International Conference on Current Trends in Database Technology, Munich, Germany, 26–31 March 2006; Springer: Berlin, Germany, 2006; pp. 712–723. [Google Scholar]
- Bhagat, J.; Tanoh, F.; Nzuobontane, E.; Laurent, T.; Orlowski, J.; Roos, M.; Wolstencroft, K.; Aleksejevs, S.; Stevens, R.; Pettifer, S.; et al. BioCatalogue: A universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010, 38, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Hitzler, P.; Krötzsch, M.; Parsia, B.; Rudolph, S. (Eds.) OWL 2 Web Ontology Language: Primer. W3C Recommendation. 27 October 2009. Available online: http://www.w3.org/TR/owl2-primer/ (accessed on 22 February 2018).
- Bechhofer, S.; Buchan, I.; De Roure, D.; Missier, P.; Ainsworth, J.; Bhagat, J.; Couch, P.; Cruickshank, D.; Delderfield, M.; Dunlop, I.; et al. Why linked data is not enough for scientists. Special section: Recent advances in e-Science. Future Gener. Comput. Syst. 2013, 29, 599–611. [Google Scholar] [CrossRef]
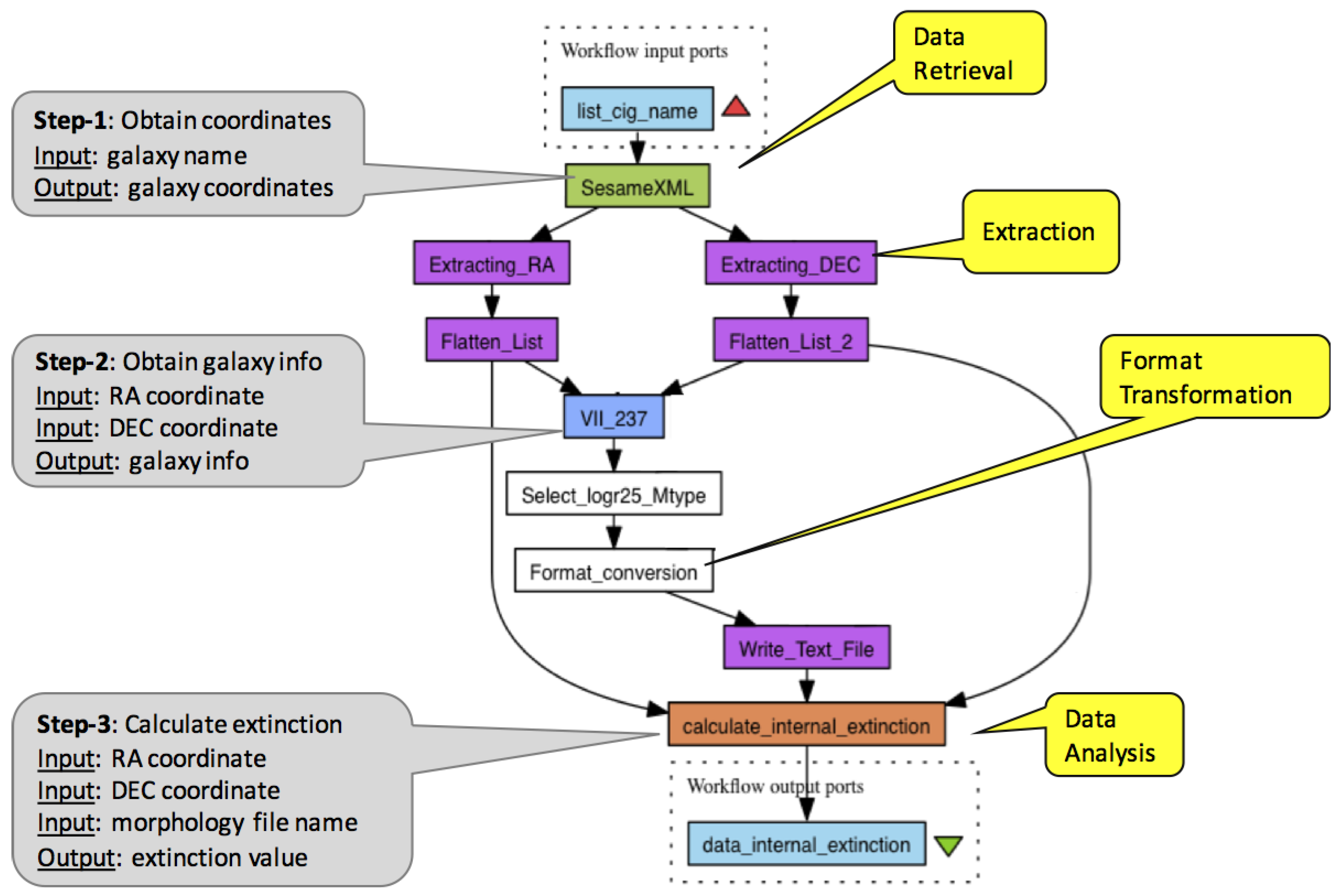


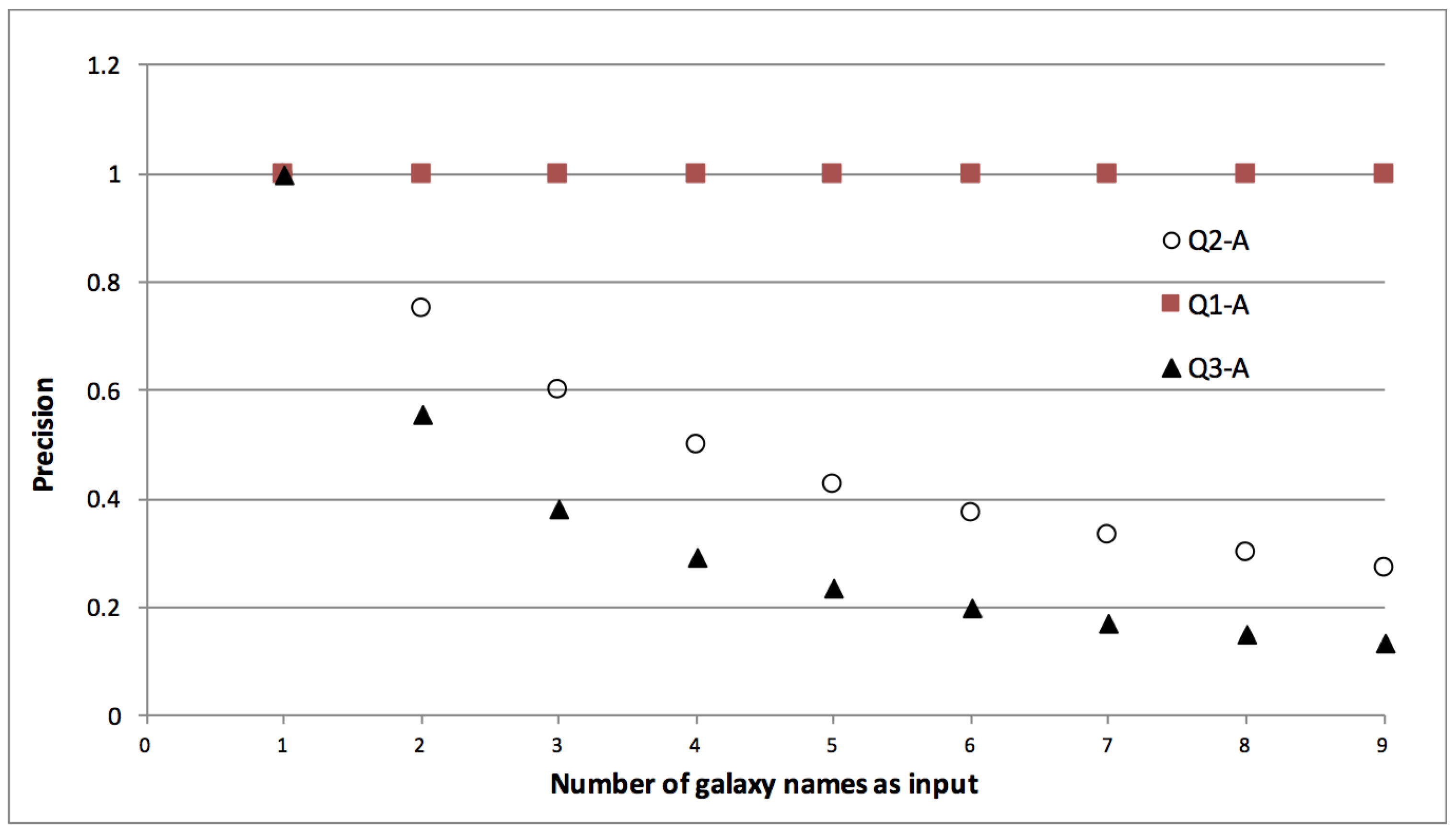
| Q.1 Which results are coordinates obtained from the Sesame database, from which database catalogs are they obtained. |
| Q.2 Select all results belonging to the Andromeda Galaxy. |
| Q.3 Select extinction calculation results for the Andromeda Galaxy, where the morphology parameter setting was . |
| Motif | Value-Copying | Example in Case | Labelling Behaviour |
|---|---|---|---|
| Data Analysis Data Retrieval Data Visualization | N/A | “SesameXML”, “VII_237”, “calculate_int_extinction” | mint |
| Augmentation | I O | Not present in case. | propagate |
| Extraction | I O | “Extract_DEC”, “Extract_RA” | propagate |
| Split | I O | Not present in case. | propagate |
| Merge | I O | “Flatten_List” | propagate |
| Filter | I O | “Select_logr25_Mtype” | propagate |
| Combine | I O | Not present in case. | propagate |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alper, P.; Belhajjame, K.; Curcin, V.; Goble, C.A. LabelFlow Framework for Annotating Workflow Provenance. Informatics 2018, 5, 11. https://doi.org/10.3390/informatics5010011
Alper P, Belhajjame K, Curcin V, Goble CA. LabelFlow Framework for Annotating Workflow Provenance. Informatics. 2018; 5(1):11. https://doi.org/10.3390/informatics5010011
Chicago/Turabian StyleAlper, Pinar, Khalid Belhajjame, Vasa Curcin, and Carole A. Goble. 2018. "LabelFlow Framework for Annotating Workflow Provenance" Informatics 5, no. 1: 11. https://doi.org/10.3390/informatics5010011
APA StyleAlper, P., Belhajjame, K., Curcin, V., & Goble, C. A. (2018). LabelFlow Framework for Annotating Workflow Provenance. Informatics, 5(1), 11. https://doi.org/10.3390/informatics5010011





