Insights into Microbiome and Metabolic Signatures of Children Undergoing Peanut Oral Immunotherapy
Abstract
:1. Introduction
2. Materials and Methods
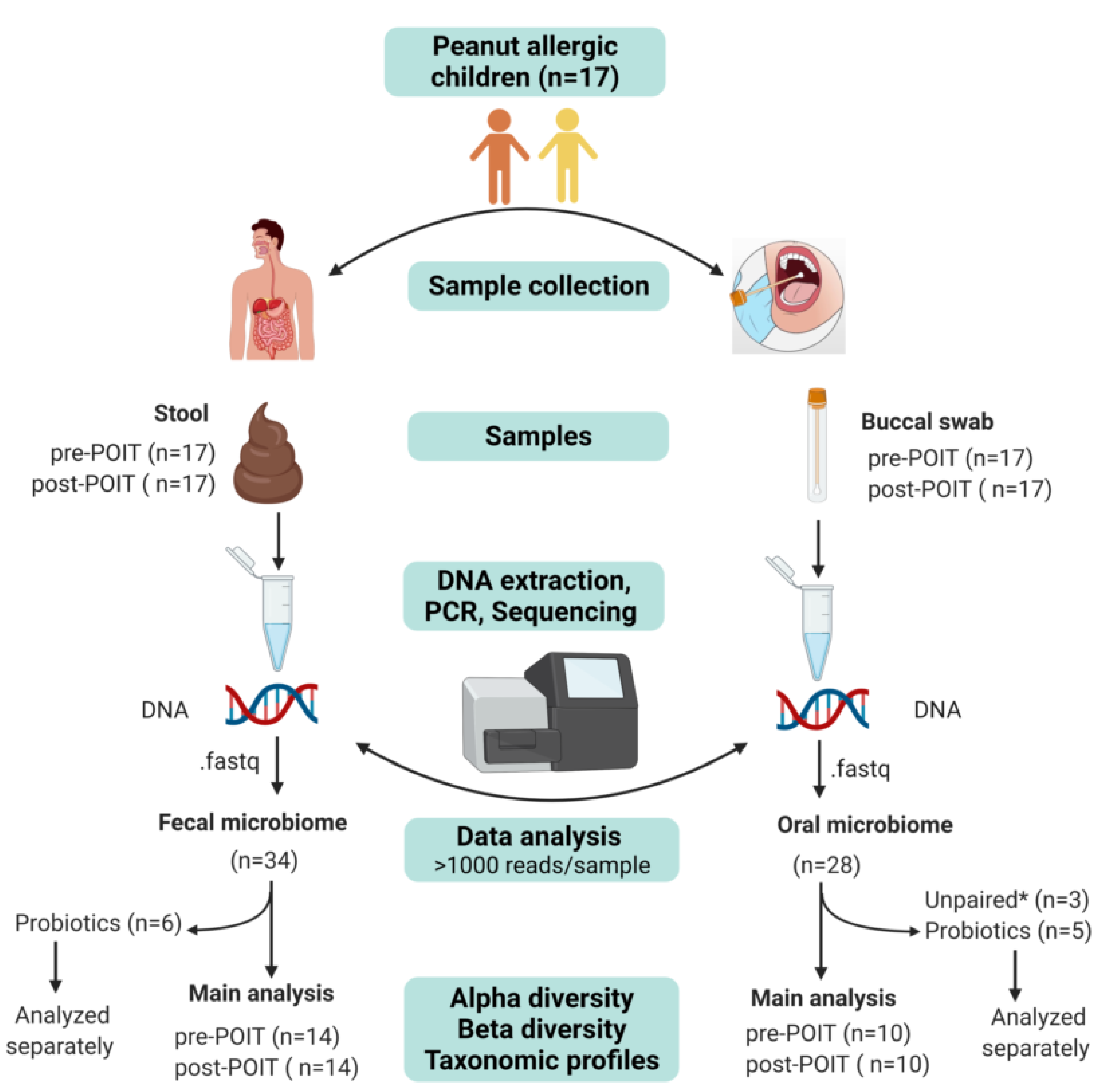
2.1. Patient Enrollment and Ethical Approval
2.2. Sample Collection
2.2.1. Buccal Samples
2.2.2. Stool Samples
2.3. DNA Extraction
2.4. 16S rRNA Sequencing
2.5. Bioinformatic Analysis of 16S Data
2.6. Fecal SCFAs Measurement
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Study Cohort
3.2. Microbiome Sequencing Results
3.3. Effects of Demographics and Allergic Co-Morbidities on the Microbiome
3.4. Oral Microbiome before and after POIT
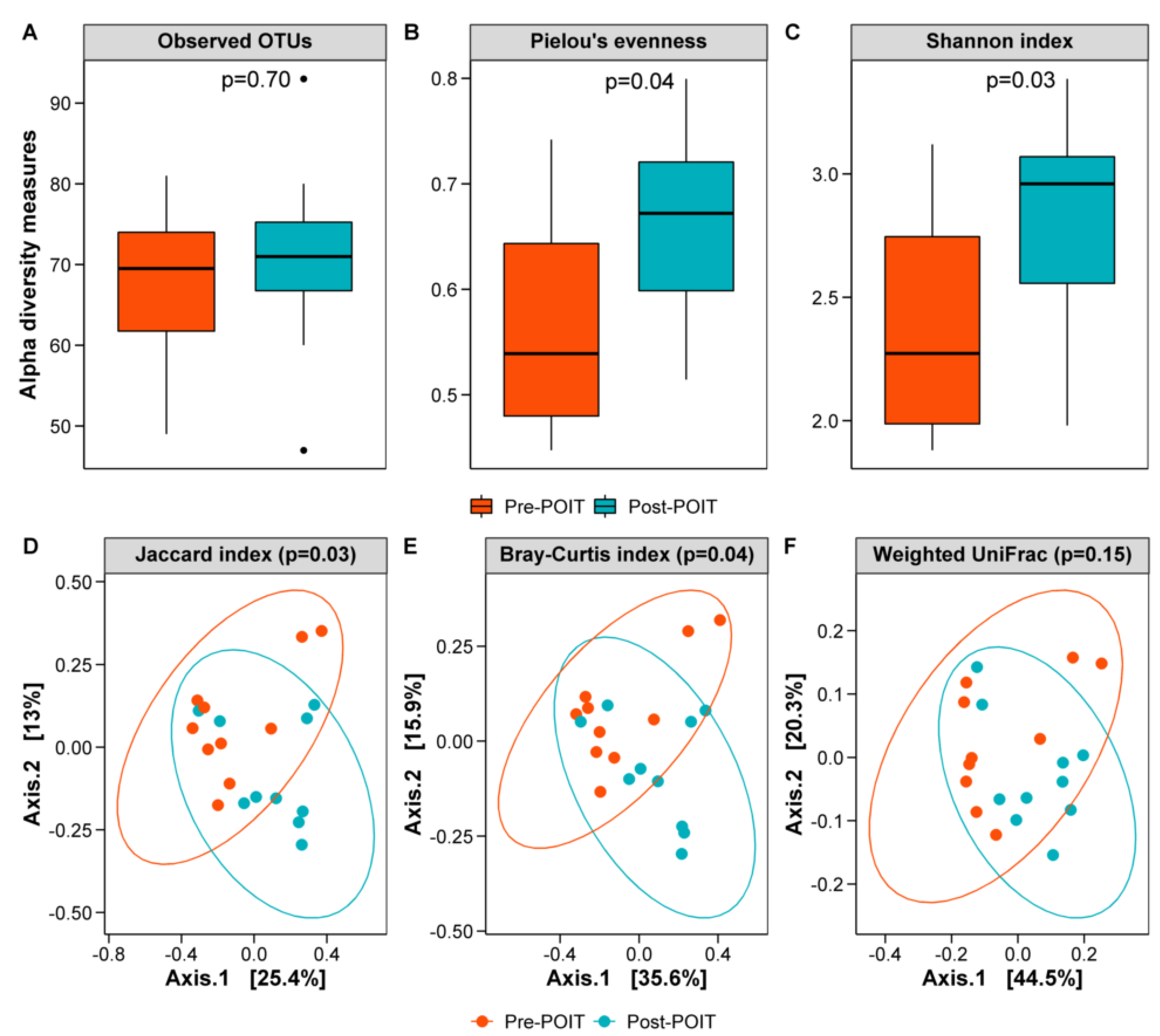
3.5. Gut Microbiome before and after POIT
3.6. Fecal SCFAs Profiles in Pre-POIT and Post-POIT Samples
3.7. Microbiome and SCFAs Levels in Patients Taking Probiotics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IFN-γ | Interferon gamma |
| IL-10 | Interleukin-10 |
| POIT | peanut oral immunotherapy |
| SCFAs | short chain fatty acids |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| Treg | T regulatory |
References
- Gupta, R.S.; Springston, E.E.; Warrier, M.R.; Smith, B.; Kumar, R.; Pongracic, J.; Holl, J.L. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics 2011, 128, e9–e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, N.J.; Koplin, J.J.; Martin, P.E.; Gurrin, L.C.; Lowe, A.J.; Matheson, M.C.; Ponsonby, A.-L.; Wake, M.; Tang, M.L.; Dharmage, S.C. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J. Allergy Clin. Immunol. 2011, 127, 668–676.e2. [Google Scholar] [CrossRef] [PubMed]
- Avery, N.J.; King, R.M.; Knight, S.; Hourihane, J.O. Assessment of quality of life in children with peanut allergy. Pediatr. Allergy Immunol. 2003, 14, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Bunyavanich, S.; Rifas-Shiman, S.L.; Platts-Mills, T.A.; Workman, L.; Sordillo, J.E.; Gillman, M.W.; Gold, D.R.; Litonjua, A.A. Peanut allergy prevalence among school-age children in a US cohort not selected for any disease. J. Allergy Clin. Immunol. 2014, 134, 753–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capucilli, P.; Wang, K.Y.; Spergel, J.M. Food reactions during avoidance: Focus on peanut. Ann. Allergy Asthma Immunol. 2020, 124, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Stensgaard, A.; Bindslev-Jensen, C.; Nielsen, D.; Munch, M.; Dunngalvin, A. Quality of life in childhood, adolescence and adult food allergy: Patient and parent perspectives. Clin. Exp. Allergy 2017, 47, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Greenhawt, M. Food Allergy Quality of Life. Ann. Allergy Asthma Immunol. 2016, 113, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Herbert, L.; Shemesh, E.; Bender, B. Clinical Management of Psychosocial Concerns Related to Food Allergy. J. Allergy Clin. Immunol. Pract. 2016, 4, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, R.A.; Rubes, M.; Ambrose, M.A.; Mullarkey, C.; Shemesh, E.; Sicherer, S.H. Longitudinal evaluation of food allergy-related bullying. J. Allergy Clin. Immunol. Pract. 2014, 2, 639–641. [Google Scholar] [CrossRef]
- Burks, A.W.; Tang, M.; Sicherer, S.; Muraro, A.; Eigenmann, P.A.; Ebisawa, M.; Fiocchi, A.; Chiang, W.; Beyer, K.; Wood, R.; et al. ICON: Food allergy. J. Allergy Clin. Immunol. 2012, 129, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Pons, L.; Roberts, J.L.; Scurlock, A.M.; Perry, T.T.; Kulis, M.; Shreffler, W.G.; Steele, P.; Henry, K.A.; Adair, M.; et al. Clinical efficacy and immune regulation with peanut oral immunotherapy. J. Allergy Clin. Immunol. 2009, 124, 292–300.e97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, P.; Jones, S.M.; Scurlock, A.M.; Perry, T.T.; Kemper, A.; Steele, P.; Hiegel, A.; Kamilaris, J.; Carlisle, S.; Yue, X.; et al. A randomized controlled study of peanut oral immunotherapy: Clinical desensitization and modulation of the allergic response. J. Allergy Clin. Immunol. 2011, 127, 654–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickery, B.P.; Scurlock, A.M.; Kulis, M.; Steele, P.H.; Kamilaris, J.; Berglund, J.P.; Burk, C.; Hiegel, A.; Carlisle, S.; Christie, L.; et al. Sustained unresponsiveness to peanut in subjects who have completed peanut oral immunotherapy. J. Allergy Clin. Immunol. 2014, 133, 468–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickery, B.P.; Berglund, J.P.; Burk, C.M.; Fine, J.P.; Kim, E.H.; Kim, J.I.; Keet, C.A.; Kulis, M.; Orgel, K.G.; Guo, R.; et al. Early oral immunotherapy in peanut-allergic preschool children is safe and highly effective. J. Allergy Clin. Immunol. 2017, 139, 173–181.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickery, B.P.; Vereda, A.; Casale, T.B.; Beyer, K.; du Toit, G.; Hourihane, J.O.; Jones, S.M.; Shreffler, W.G.; Marcantonio, A.; Zawadzki, R.; et al. AR101 Oral Immunotherapy for Peanut Allergy. N. Engl. J. Med. 2018, 379, 1991–2001. [Google Scholar]
- Clark, A.T.; Islam, S.; King, Y.; Deighton, J.; Anagnostou, K.; Ewan, P.W. Successful oral tolerance induction in severe peanut allergy. Allergy 2009, 64, 1218–1220. [Google Scholar] [CrossRef]
- Anagnostou, K.; Islam, S.; King, Y.; Foley, L.; Pasea, L.; Bond, S.; Palmer, C.; Deighton, J.; Ewan, P.; Clark, A. Assessing the efficacy of oral immunotherapy for the desensitisation of peanut allergy in children (STOP II): A phase 2 randomised controlled trial. Lancet 2014, 383, 1297–1304. [Google Scholar] [CrossRef] [Green Version]
- Vickery, B.P.; Vereda, A.; Nilsson, C.; du Toit, G.; Shreffler, W.G.; Burks, A.W.; Jones, S.M.; Fernández-Rivas, M.; Blümchen, K.; Hourihane, J.O.; et al. Continuous and Daily Oral Immunotherapy for Peanut Allergy: Results from a 2-Year Open-Label Follow-On Study. J. Allergy Clin. Immunol. Pract. 2021, 9, 1879–1889.e14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ho, H.; Bunyavanich, S. The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 2019, 122, 276–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Blackman, A.C.; Staggers, K.A.; Kronisch, L.; Davis, C.M.; Anagnostou, A. Quality of life improves significantly after real-world oral immunotherapy for children with peanut allergy. Ann. Allergy Asthma Immunol. 2020, 125, 196–201.e1. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate With Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2016, 3, 218–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapa, S.; Runge, J.K.; Venkatachalam, A.; Denne, C.; Luna, R.A.; Anon, J.B. The Nasopharyngeal and Gut Microbiota in Children in a Pediatric Otolaryngology Practice. Pediatr. Infect. Dis. J. 2020, 39, e226–e233. [Google Scholar] [CrossRef]
- Hildebrand, F.; Tadeo, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 30. [Google Scholar] [CrossRef] [Green Version]
- Pammi, M.; Thapa, S.; Balderas, M.; Runge, J.K.; Venkatachalam, A.; Luna, R.A. Microbiome signatures in neonatal central line associated bloodstream infections. PLoS ONE 2020, 15, e0227967. [Google Scholar]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Engevik, M.A.; Luck, B.; Visuthranukul, C.; Ihekweazu, F.D.; Engevik, A.C.; Shi, Z.; Danhof, H.A.; Chang-Graham, A.L.; Hall, A.; Endres, B.T.; et al. Human-Derived Bifidobacterium dentium Modulates the Mammalian Serotonergic System and Gut-Brain Axis. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Seguel, M.; Gottdenker, N. The diversity and impact of hookworm infections in wildlife. Int. J. Parasitol. Parasites Wildl. 2017, 6, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Package “Vegan”. 2022. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 30 May 2022).
- Dzidic, M.; Abrahamsson, T.R.; Artacho, A.; Collado, M.C.; Mira, A.; Jenmalm, M.C. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 2000–2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Vadali, V.G.; Szabady, R.L.; Zhang, W.; Norman, J.M.; Roberts, B.; Tibshirani, R.; Desai, M.; Chinthrajah, R.S.; Galli, S.J.; et al. Increased diversity of gut microbiota during active oral immunotherapy in peanut-allergic adults. Allergy 2020, 76, 927–930. [Google Scholar] [CrossRef]
- Abrahamsson, T.; Jakobsson, H.E.; Andersson, A.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2014, 44, 842–850. [Google Scholar] [CrossRef] [Green Version]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012, 129, 434–440.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Hong, S.M.C.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Feehley, T.; Stefka, A.T.; Nagler, C.R.; Zhou, L.; Mazmanian, S.K.; Seo, G.-Y.; Tjota, M.Y.; Antonopoulos, A.D.; Fu, Y.-X.; Tripathi, P.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.M.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities—2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [PubMed] [Green Version]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Investig. 2021, 131, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kourosh, A.; Luna, R.A.; Nance, C.; Anagnostou, A.; Devaraj, S.; Davis, C.M. Fecal Microbiome Signatures are Different in Food Allergic Children Compared to Siblings and Healthy Children. J. Allergy Clin. Immunol. 2018, 141, AB183. [Google Scholar] [CrossRef]
- Sudo, N.; Sawamura, S.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 1997, 159, 1739–1745. [Google Scholar] [PubMed]
- Rodriguez, B.; Prioult, G.; Hacini-Rachinel, F.; Moine, D.; Bruttin, A.; Ngom-Bru, C.; Labellie, C.; Nicolis, I.; Berger, B.; Mercenier, A.; et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol. Ecol. 2012, 79, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, C.H.; Nagler, C.R. The Influence of the Microbiome on Allergic Sensitization to Food. J. Immunol. 2017, 198, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Song, J.; Wang, C.; Fu, S.; Wang, Y. Bifidobacterium infantis Potentially Alleviates Shrimp Tropomyosin-Induced Allergy by Tolerogenic Dendritic Cell-Dependent Induction of Regulatory T Cells and Alterations in Gut Microbiota. Front. Immunol. 2017, 8, 1536. [Google Scholar] [CrossRef]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef] [Green Version]
- Venegas, D.P.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Bakdash, G.; Vogelpoel, L.T.C.; Van Capel, T.M.M.; Kapsenberg, M.L.; De Jong, E.C. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. 2015, 8, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. Available online: https://pubmed.ncbi.nlm.nih.gov/26963409 (accessed on 3 May 2016). [CrossRef] [PubMed] [Green Version]
- Varshney, P.; Jones, S.; Pons, L.; Kulis, M.; Steele, P.; Kemper, A.; Scurlock, A.; Perry, T.; Burks, A. Peanut Oral Immunotherapy (OIT) Induces Immunologic Changes Supporting the Development of Tolerance. J. Allergy Clin. Immunol. 2010, 125 (Suppl. 1), AB59. [Google Scholar] [CrossRef]
- Canani, R.B.; De Filippis, F.; Nocerino, R.; Paparo, L.; Di Scala, C.; Cosenza, L.; Gatta, G.D.; Calignano, A.; De Caro, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 12500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vonk, M.M.; Blokhuis, B.R.J.; Diks, M.A.P.; Wagenaar, L.; Smit, J.J.; Pieters, R.H.H.; Garssen, J.; Knippels, L.M.J.; Van Esch, B.C.A.M. Butyrate Enhances Desensitization Induced by Oral Immunotherapy in Cow’s Milk Allergic Mice. Mediat. Inflamm. 2019, 2019, 9062537. [Google Scholar] [CrossRef] [Green Version]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez–Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Chan, J.C.; Kioh, D.Y.; Yap, G.C.; Lee, B.W.; Chan, E.C. A novel LCMSMS method for quantitative measurement of short-chain fatty acids in human stool derivatized with 12C- and 13C-labelled aniline. J. Pharm. Biomed. Anal. 2017, 138, 43–53. [Google Scholar] [CrossRef]



| Variable | Peanut Allergic Cohort (n = 17) |
|---|---|
| Demographics | |
| Age (years) | 7 ± 3 |
| History of atopy | |
| Atopic dermatitis | 13 (76%) |
| Allergic rhinitis | 12 (71%) |
| Asthma | 8 (47%) |
| Multiple food allergies | 12 (71%) |
| Mode of delivery | |
| Vaginal | 5 (29%) |
| Cesarean-section | 12 (71%) |
| Feeding pattern | |
| Breastfed | 12 (71%) |
| Formula | 5 (29%) |
| Medications | |
| Antibiotics | 5 (29%) |
| Probiotics | 3 (18%) |
| Birth place-Urban | 15 (88%) |
| Pets | 9 (53%) |
| Dog | 7 |
| Cats and dogs | 1 |
| Rabbit | 1 |
| History of food allergies in sibling(s) | 7 (41%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blackman, A.C.; Thapa, S.; Venkatachalam, A.; Horvath, T.D.; Runge, J.K.; Haidacher, S.J.; Hoch, K.M.; Haag, A.M.; Luna, R.A.; Anagnostou, A. Insights into Microbiome and Metabolic Signatures of Children Undergoing Peanut Oral Immunotherapy. Children 2022, 9, 1192. https://doi.org/10.3390/children9081192
Blackman AC, Thapa S, Venkatachalam A, Horvath TD, Runge JK, Haidacher SJ, Hoch KM, Haag AM, Luna RA, Anagnostou A. Insights into Microbiome and Metabolic Signatures of Children Undergoing Peanut Oral Immunotherapy. Children. 2022; 9(8):1192. https://doi.org/10.3390/children9081192
Chicago/Turabian StyleBlackman, Andrea C., Santosh Thapa, Alamelu Venkatachalam, Thomas D. Horvath, Jessica K. Runge, Sigmund J. Haidacher, Kathleen M. Hoch, Anthony M. Haag, Ruth Ann Luna, and Aikaterini Anagnostou. 2022. "Insights into Microbiome and Metabolic Signatures of Children Undergoing Peanut Oral Immunotherapy" Children 9, no. 8: 1192. https://doi.org/10.3390/children9081192
APA StyleBlackman, A. C., Thapa, S., Venkatachalam, A., Horvath, T. D., Runge, J. K., Haidacher, S. J., Hoch, K. M., Haag, A. M., Luna, R. A., & Anagnostou, A. (2022). Insights into Microbiome and Metabolic Signatures of Children Undergoing Peanut Oral Immunotherapy. Children, 9(8), 1192. https://doi.org/10.3390/children9081192






