Clinical Characterization of a 6-Year-Old Patient with Autism and Two Adjacent Duplications on 10q11.22q11.23. A Case Report
Abstract
1. Introduction
Clinical History
2. Cognitive and Behavioral Assessment
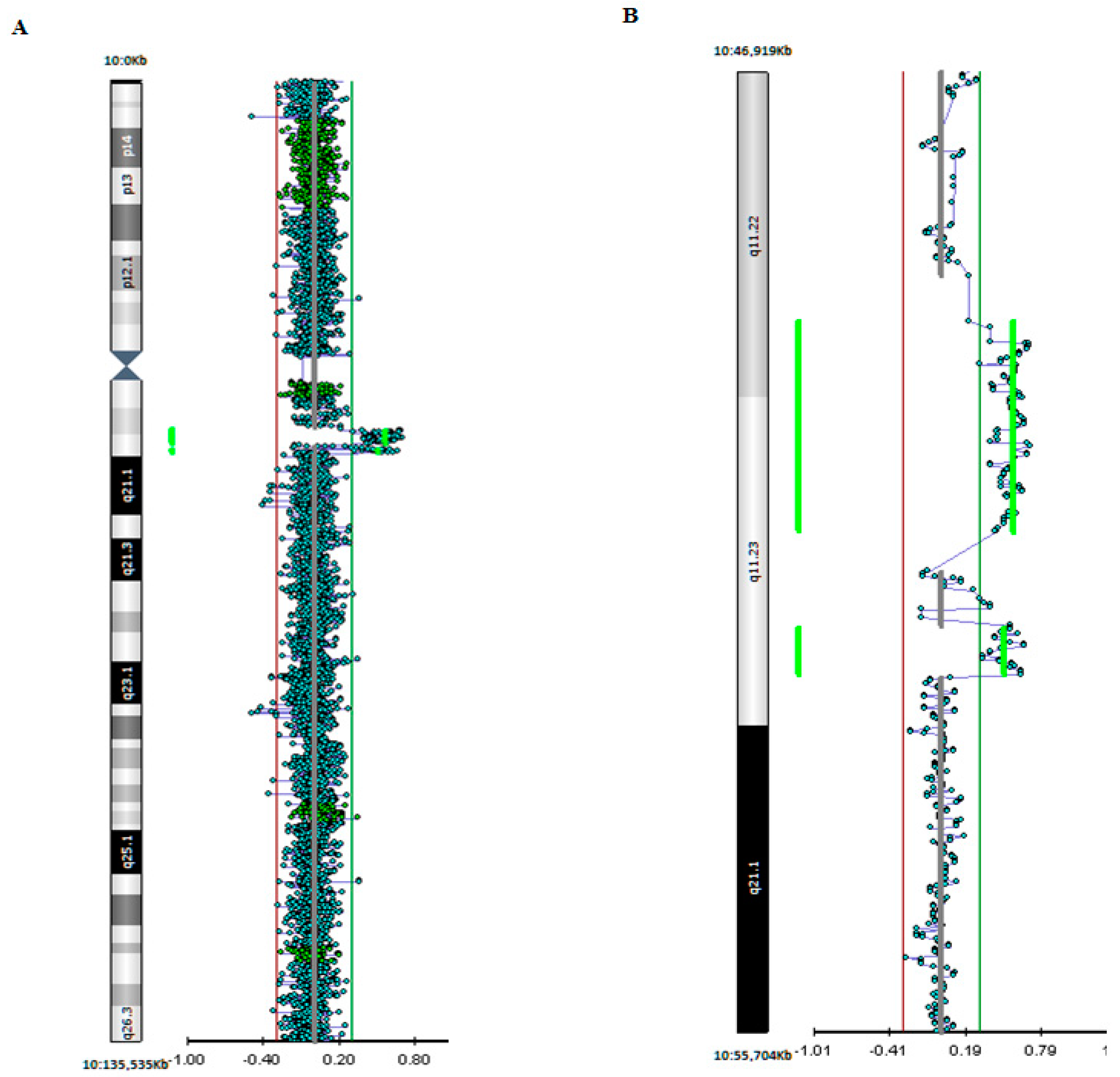
3. Genetic Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elsabbagh, M.; Divan, G.; Koh, Y.J.; Kim, Y.S.; Kauchali, S.; Marcin, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef]
- Lai, M.C.; Lombardo, M.V.; Baron-Cohen, S. Autism. Lancet 2014, 383, 896–910. [Google Scholar] [CrossRef]
- Chaste, P.; Leboyer, M. Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar] [PubMed]
- Bailey, A.; Le Couteur, A.; Gottesman, I.; Bolton, P.; Simonoff, E.; Yuzda, E.; Rutter, M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol. Med. 1995, 25, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Gillberg, C.; Steffenburg, S.; Wahlstrom, J.; Gillberg, I.C.; Sjostedt, A.; Martinsson, T.; Liedgren, S.; Eeg-Olofsson, O. Autism associated with marker chromosome. J. Am. Acad. Child. Adolesc. Psychiatry 1991, 30, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Colvert, E.; Tick, B.; McEwen, F.; Stewart, C.; Curran, S.R.; Woodhouse, E.; Gillan, N.; Hallett, V.; Lietz, S.; Garnett, T.; et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry 2015, 72, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Folstein, S.; Rutter, M. Genetic influences and infantile autism. Nature 1977, 265, 726–728. [Google Scholar] [CrossRef]
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K.; et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Hultman, C.; Larsson, H.; Reichenberg, A. The heritability of autism spectrum disorder. JAMA 2017, 318, 1182–1184. [Google Scholar] [CrossRef]
- Ozonoff, S.; Young, G.S.; Carter, A.; Messinger, D.; Yirmiya, N.; Zwaigenbaum, L.; Bryson, S.; Carver, L.J.; Constantino, J.N.; Dobkins, K.; et al. Recurrence risk for autism spectrum disorders: A baby siblings research consortium study. Pediatrics 2011, 128, e488–e495. [Google Scholar] [CrossRef]
- Werling, D.M.; Geschwind, D.H. Recurrence rates provide evidence for sex-differential, familial genetic liability for autism spectrum disorders in multiplex families and twins. Mol. Autism. 2015, 6, 27. [Google Scholar] [CrossRef]
- Buxbaum, J.D. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin. Neurosci. 2009, 11, 35–43. [Google Scholar]
- Yu, L.; Wu, Y.; Wu, B.L. Genetic architecture, epigenetic influence and environment exposure in the pathogenesis of Autism. Sci. China Life Sci. 2015, 58, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.; Beck, J.C.; Bernier, R.; Bisson, E.; Braun, T.A.; Casavant, T.L.; Childress, D.; Folstein, S.E.; Garcia, M.; Gardiner, M.B.; et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am. J. Med. Genet. 1999, 88, 609–615. [Google Scholar] [CrossRef]
- IMGSAC. A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International molecular genetic study of autism consortium. Hum. Mol. Genet. 1998, 7, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Risch, N.; Spiker, D.; Lotspeich, L.; Nouri, N.; Hinds, D.; Hallmayer, J.; Kalaydjieva, L.; McCague, P.; Dimiceli, S.; Pitts, T.; et al. A genomic screen of autism: Evidence for a multilocus etiology. Am. J. Med. Genet. 1999, 65, 493–507. [Google Scholar] [CrossRef]
- Bacchelli, E.; Cameli, C.; Viggiano, M.; Igliozzi, R.; Mancini, A.; Tancredi, R.; Battaglia, A.; Maestrini, E. An integrated analysis of rare CNV and exome variation in autism spectrum disorder using the infinium psycharray. Sci. Rep. 2020, 10, 3198. [Google Scholar] [CrossRef] [PubMed]
- O’Roak, B.J.; Vives, L.; Girirajan, S.; Karakoc, E.; Krumm, N.; Coe, B.P.; Levy, R.; Ko, A.; Lee, C.; Smith, J.D.; et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012, 485, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.J.; Murtha, M.T.; Gupta, A.R.; Murdoch, J.D.; Raubeson, M.J.; Willsey, A.J.; Ercan-Sencicek, A.G.; DiLullo, N.M.; Parikshak, N.N.; Stein, J.L.; et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012, 485, 237–241. [Google Scholar] [CrossRef]
- van Bon, B.W.M.; Balciuniene, J.; Fruhman, G.; Nagamani, S.C.S.; Broome, D.L.; Cameron, E.; Martinet, D.; Roulet, E.; Jacquemont, S.; Beckmann, J.S.; et al. The phenotype of recurrent 10q22q23 deletions and duplications. Eur. J. Hum. Genet. 2011, 19, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, S.; Kruer, M.; Bader, P.; Corzo, D.; Schuette, J.; Keegan, C.; Nowakowska, B.; Peacock, S.; Cai, W.; Peiffer, D.; et al. Identification of critical regions for clinical features of distal 10q deletion syndrome. Clin. Genet. 2009, 76, 54–62. [Google Scholar] [CrossRef]
- Aalfs, C.M.; Hoovers, J.M.; Nieste-Otter, M.A.; Mannens, M.M.; Hennekam, R.C.; Leschot, N.J. Further delineation of the partial proximal trisomy 10q syndrome. J. Med. Genet. 1995, 32, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Fryns, J.P.; Kleczkowska, A.; Igodt-Ameye, L.; Van den Berghe, H. Proximal duplication of the long arm of chromosome 10 (10q11.2—10q22): A distinct clinical entity. Clin. Genet. 1987, 32, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.W.; Chan, W.K.; Lam, S.T.; Chu, W.P.; Kwong, N.S. Proximal 10q trisomy: A new case with anal atresia. J. Med. Genet. 2000, 37, E24. [Google Scholar] [CrossRef]
- Lysy, P.A.; Sibille, C.; Gillerot, Y.; Smets, F.; Sokal, E.M. Partial proximal 10q trisomy: A new case associated with biliary atresia. Hereditas 2007, 144, 191–194. [Google Scholar] [CrossRef]
- Nucaro, A.; Faedda, A.; Cao, A.; Boccone, L. Partial proximal trisomy 10q syndrome: A new case. Genet. Couns. 2002, 13, 411–416. [Google Scholar] [PubMed]
- van Buggenhout, G.; Decock, P.; Fryns, J.P. A distinct phenotype associated with partial trisomy 10q due to proximal direct duplication 10q11 --> q223? Genet. Couns. 1996, 7, 53–59. [Google Scholar] [PubMed]
- Vogel, W.; Back, E.; Imm, W. Serial duplication of 10 (q11 leads to q22) in a patient with minor congenital malformations. Clin. Genet. 1978, 13, 159–163. [Google Scholar] [CrossRef]
- Cheung, S.W.; Shaw, C.A.; Yu, W.; Li, J.; Ou, Z.; Patel, A.; Yatsenko, S.A.; Cooper, M.L.; Furman, P.; Stankiewicz, P.; et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet. Med. 2005, 7, 422–432. [Google Scholar] [CrossRef]
- Kearney, H.M.; Thorland, E.C.; Brown, K.K.; Quintero-Rivera, F.; South, S.T.; Working Group of the American College of Medical Genetics Laboratory Quality Assurance C. American college of medical genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011, 13, 680–685. [Google Scholar] [CrossRef]
- Manolakos, E.; Vetro, A.; Garas, A.; Thomaidis, L.; Kefalas, K.; Kitsos, G.; Ziegler, M.; Liehr, T.; Zuffardi, O.; Papoulidis, I. Proximal 10q duplication in a child with severe central hypotonia characterized by array-comparative genomic hybridization: A case report and review of the literature. Exp. Ther. Med. 2014, 7, 953–957. [Google Scholar] [CrossRef]
- Girirajan, S.; Johnson, R.L.; Tassone, F.; Balciuniene, J.; Katiyar, N.; Fox, K.; Baker, C.; Srikanth, A.; Yeoh, K.H.; Khoo, S.J.; et al. Global increases in both common and rare copy number load associated with autism. Hum. Mol. Genet. 2013, 22, 2870–2880. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Kulkarni, S.; Dharmadhikari, A.V.; Sampath, S.; Bhatt, S.S.; Shaikh, T.H.; Xia, Z.; Pursley, A.N.; Cooper, M.L.; Shinawi, M.; et al. Recurrent deletions and reciprocal duplications of 10q11.21q11.23 including CHAT and SLC18A3 are likely mediated by complex low-copy repeats. Hum. Mutat. 2012, 33, 165–179. [Google Scholar] [CrossRef][Green Version]
- Meguid, N.A.; Eid, M.M.; Mohamed, A.M.; Ghanoum, H.; Helmy, N.A.; Eid, O.M. Contribution of chromosomal abnormalities at 10q and 22q to autism. Res. Autism Spectr. Disord. 2018, 50, 43–50. [Google Scholar] [CrossRef]
- Bertino, E.; Spada, E.; Occhi, L.; Coscia, A.; Giuliani, F.; Gagliardi, L.; Gilli, G.; Bona, G.; Fabris, C.; De Curtis, M.; et al. Neonatal anthropometric charts: The Italian neonatal study compared with other European studies. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Cacciari, E.; Milani, S.; Balsamo, A.; Spada, E.; Bona, G.; Cavallo, L.; Cerutti, F.; Gargantini, L.; Greggio, N.; Tonini, G.; et al. Italian cross-sectional growth charts for height, weight and BMI (2 to 20 yr). J. Endocrinol. Invest. 2006, 29, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.D.; Collins, J.S.; Holden, K.R. United States head circumference growth reference charts: Birth to 21 years. J. Pediatr. 2010, 156, 907–913.e902. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; DiLavore, P.C.; Risi, S.; Gotham, K.; Bishop, S.L.; Luyster, R.J.; Guthrie, W. Autism Diagnostic Observation Schedule, Second Edition: ADOS-2; Services, T.W.P., Ed.; Massachusetts General Hospital: Boston, MA, USA, 2012. [Google Scholar]
- Griffiths, R. The Griffiths Mental Developmental Scales, Extended Revised; Association for Research in Infant and Child Development, the Test Agency: Oxford, UK, 2006. [Google Scholar]
- Roid, G.M.; Miller, L.J. Leiter International Performance Scale—Revised: Examiners Manual; Wood Dale, I.S.C., Ed.; Springer: Cham, Switzerland, 1997. [Google Scholar]
- Fenson, L. MacArthur-Bates Communicative Development Inventories: User’s Guide and Technical Manual; Paul, H., Ed.; Brookes Pub. Co.: Baltimore, MD, USA, 2007. [Google Scholar]
- Coonrod, E.; Marcus, L. Psychoeducational profile—Revised (PEP-3). In Encyclopedia of Autism Spectrum Disorders; Volkmar, F.R., Ed.; Springer: New York, NY, USA, 2013; pp. 2439–2444. [Google Scholar] [CrossRef]
- American Association on Intellectual and Developmental Disabilities. Intellectual Disability: Definition, Classification, and Systems of Supports; American Association on Intellectual and Developmental Disabilities: Washington, DC, USA, 2010. [Google Scholar]
- Ivanov, H.Y.; Stoyanova, V.K.; Popov, N.T.; Vachev, T.I. Autism spectrum disorder—A complex genetic disorder. Folia. Medica. 2015, 57, 19–28. [Google Scholar] [CrossRef]
- Reiff, M.; Giarelli, E.; Bernhardt, B.A.; Easley, E.; Spinner, N.B.; Sankar, P.L.; Mulchandani, S. Parents’ perceptions of the usefulness of chromosomal microarray analysis for children with autism spectrum disorders. J. Autism Dev. Disord. 2015, 45, 3262–3275. [Google Scholar] [CrossRef]
- de Michelena, M.I.; Campos, P.J. A new case of proximal 10q partial trisomy. J. Med. Genet. 1991, 28, 205–206. [Google Scholar] [CrossRef]
- Lyall, K.; Song, L.; Botteron, K.; Croen, L.A.; Dager, S.R.; Fallin, M.D.; Hazlett, H.C.; Kauffman, E.; Landa, R.; Ladd-Acosta, C.; et al. The association between parental age and autism-related outcomes in children at high familial risk for autism. Autism Res. 2020, 13, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Rosenfeld, J.A.; Coe, B.P.; Parikh, S.; Friedman, N.; Goldstein, A.; Filipink, R.A.; McConnell, J.S.; Angle, B.; Meschino, W.S.; et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012, 367, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
| Scales | Percentile | Developmental Age (Months) |
|---|---|---|
| Cognitive verbal/pre-verbal (CVP) | 7 | 27 |
| Expressive language (EL) | 2 | <12 |
| Receptive language (RL) | 7 | 18 |
| Fine motor (FM) | 9 | 29 |
| Gross motor (GM) | 13 | 29 |
| Visual-motor imitation (VMI) | 9 | 24 |
| Affective expression (AE) | 15 | |
| Social reciprocity (SR), | 41 | |
| Characteristic motor behaviour (CMB) | 9 | |
| Characteristic verbal behaviour (CVB) | 11 |
| Subscale | Equivalent Age (Months) | |
|---|---|---|
| Communication | Receptive | 13 |
| Expressive | 18 | |
| Written | 43 | |
| Daily living skills | Personal | 27 |
| Domestic | 33 | |
| Community | 18 | |
| Socialization | Interpersonal | 17 |
| Play and leisure time | 24 | |
| Coping skills | 13 | |
| Motor skills | Gross | 42 |
| Fine | 31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tritto, G.; Ricca, I.; Turi, M.; Gemma, A.; Muratori, F.; Scarano, G.; Lonardo, F. Clinical Characterization of a 6-Year-Old Patient with Autism and Two Adjacent Duplications on 10q11.22q11.23. A Case Report. Children 2021, 8, 518. https://doi.org/10.3390/children8060518
Tritto G, Ricca I, Turi M, Gemma A, Muratori F, Scarano G, Lonardo F. Clinical Characterization of a 6-Year-Old Patient with Autism and Two Adjacent Duplications on 10q11.22q11.23. A Case Report. Children. 2021; 8(6):518. https://doi.org/10.3390/children8060518
Chicago/Turabian StyleTritto, Giovanna, Ivana Ricca, Marco Turi, Andrea Gemma, Filippo Muratori, Gioacchino Scarano, and Fortunato Lonardo. 2021. "Clinical Characterization of a 6-Year-Old Patient with Autism and Two Adjacent Duplications on 10q11.22q11.23. A Case Report" Children 8, no. 6: 518. https://doi.org/10.3390/children8060518
APA StyleTritto, G., Ricca, I., Turi, M., Gemma, A., Muratori, F., Scarano, G., & Lonardo, F. (2021). Clinical Characterization of a 6-Year-Old Patient with Autism and Two Adjacent Duplications on 10q11.22q11.23. A Case Report. Children, 8(6), 518. https://doi.org/10.3390/children8060518






