Splice and Dice: Intronic microRNAs, Splicing and Cancer
Abstract
:1. Introduction
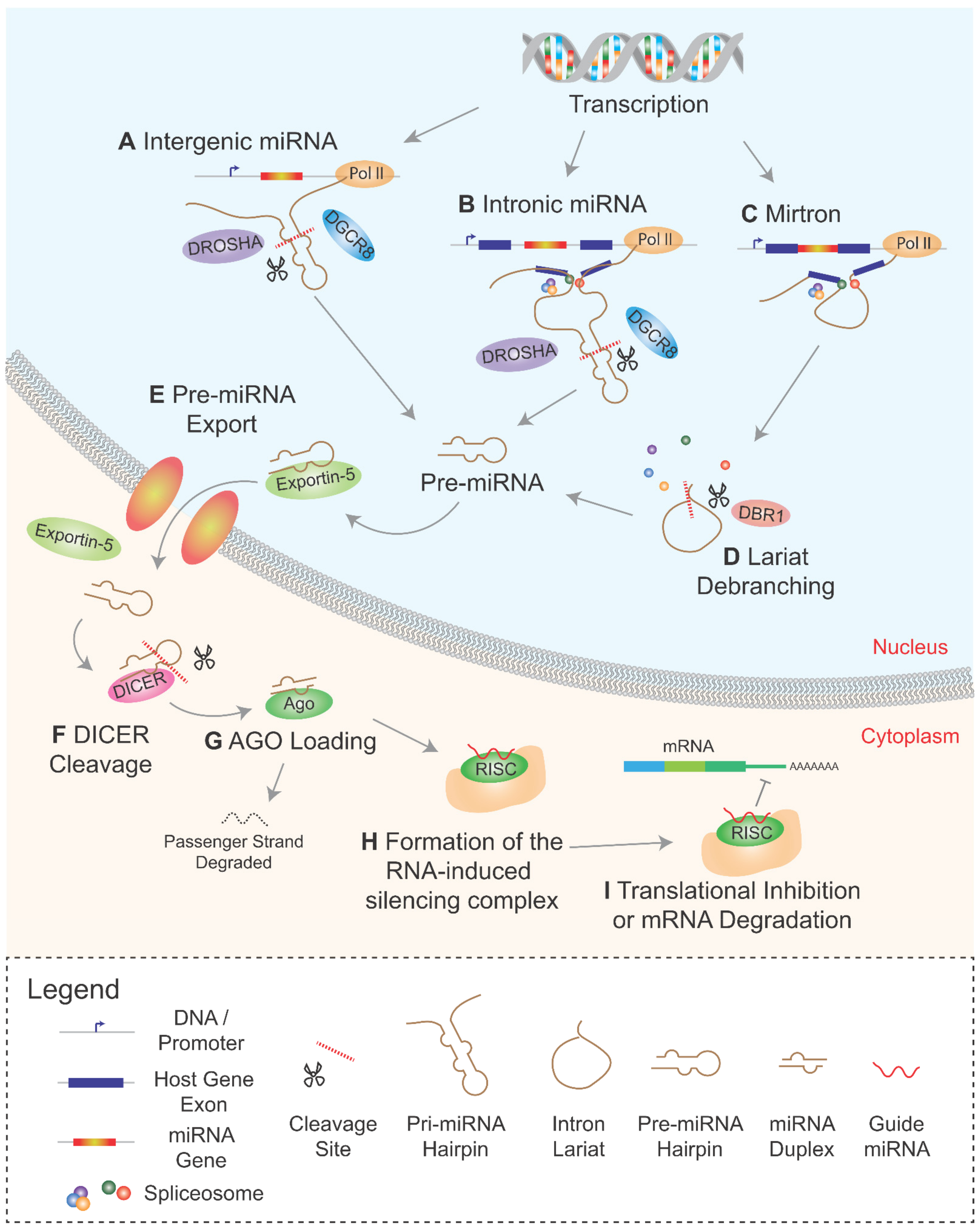
2. Biogenesis of Intronic miRNAs
2.1. Co-Transcriptional Processing of Intronic miRNAs
2.2. Mirtrons and the Non-Canonical miRNA Pathway
2.3. Comparisons and Contrasts between Mirtrons and Canonical Intronic miRNAs
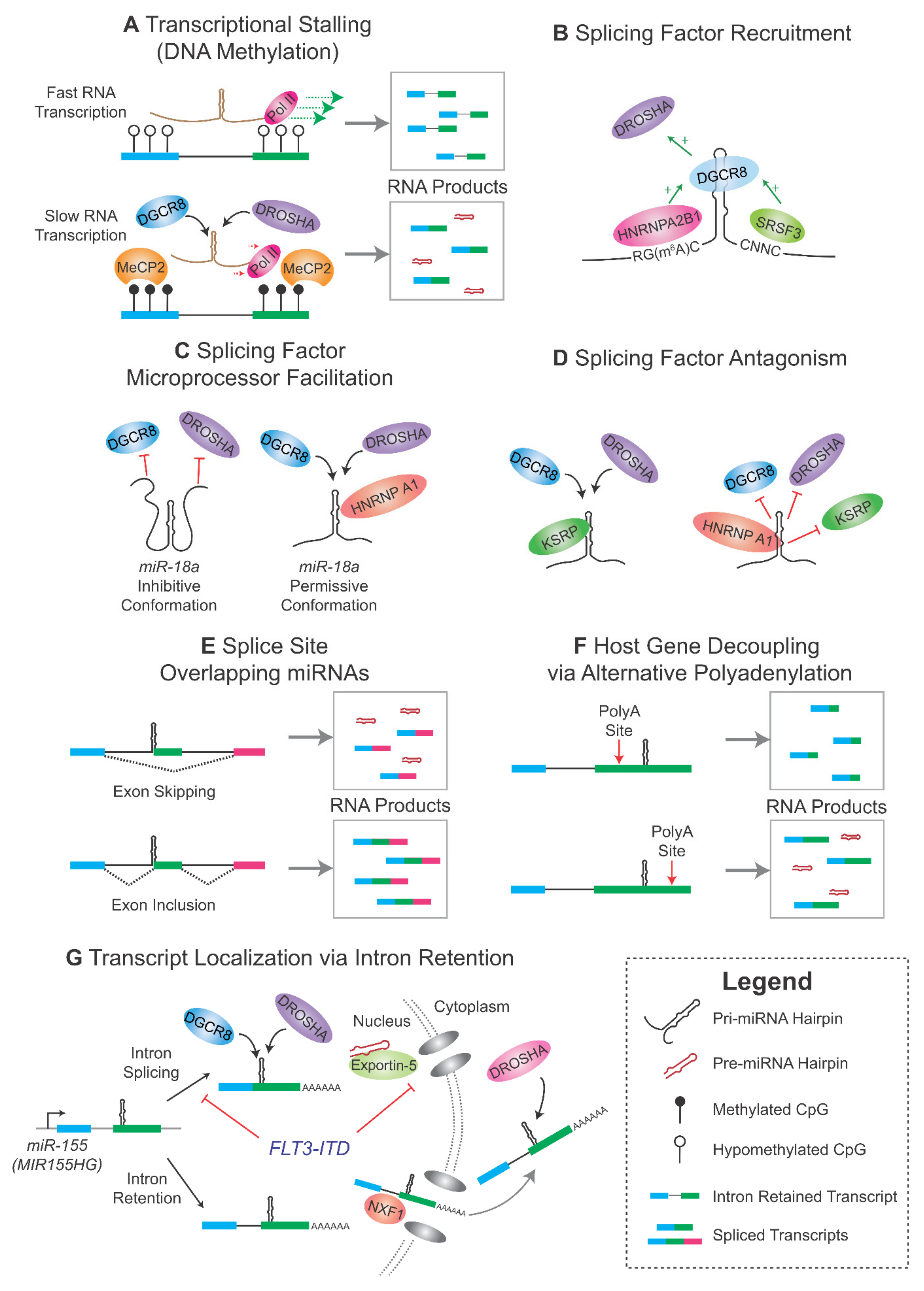
3. Interplay between miRNA Processing and the Spliceosome
3.1. Kinetic Regulation of Microprocessing and Splicing
3.2. Splicing Factors Regulating Microprocessing
3.3. Role of RNA m6A Modification
3.4. The Microprocessor as a Splicing Factor
3.5. Alternative Splicing and miRNA Biogenesis
4. Intronic miRNAs—Host Gene Interactions and Their Roles in Cancer
4.1. mir-615-3p/HOXC5 Axis and Telomerase Regulation
4.2. miR-374b/miR-545/FTX
4.3. miR-944/TP63 Axis and p53 Maintenance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Förstemann, K.; Horwich, M.D.; Wee, L.; Tomari, Y.; Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Linsen, S.E.; Tops, B.B.; Cuppen, E. miRNAs: Small changes, widespread effects. Cell Res. 2008, 18, 1157–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef]
- Wong, G.K.; Passey, D.A.; Huang, Y.; Yang, Z.; Yu, J. Is “junk” DNA mostly intron DNA? Genome Res. 2000, 10, 1672–1678. [Google Scholar] [CrossRef] [Green Version]
- Hubé, F.; Francastel, C. Mammalian introns: When the junk generates molecular diversity. Int. J. Mol. Sci. 2015, 16, 4429–4452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, U.; Pinello, N.; Jia, F.; Alasmari, S.; Ritchie, W.; Keightley, M.C.; Shini, S.; Lieschke, G.J.; Wong, J.J.; Rasko, J.E.J. Intron retention enhances gene regulatory complexity in vertebrates. Genome Biol. 2017, 18, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.J.; Au, A.Y.; Ritchie, W.; Rasko, J.E. Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. Bioessays 2016, 38, 41–49. [Google Scholar] [CrossRef]
- Boutz, P.L.; Bhutkar, A.; Sharp, P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015, 29, 63–80. [Google Scholar] [CrossRef] [Green Version]
- Rearick, D.; Prakash, A.; McSweeny, A.; Shepard, S.S.; Fedorova, L.; Fedorov, A. Critical association of ncRNA with introns. Nucleic Acids Res. 2011, 39, 2357–2366. [Google Scholar] [CrossRef] [Green Version]
- Kwok, Z.H.; Zhang, B.; Chew, X.H.; Chan, J.J.; Teh, V.; Yang, H.; Kappei, D.; Tay, Y. Systematic analysis of intronic miRNAs reveals cooperativity within the multicomponent FTX locus to promote colon cancer development. Cancer Res. 2021, 81, 1308–1320. [Google Scholar] [CrossRef]
- Jo, B.S.; Choi, S.S. Introns: The functional benefits of introns in genomes. Genom. Inf. 2015, 13, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [Green Version]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef]
- Gao, X.; Qiao, Y.; Han, D.; Zhang, Y.; Ma, N. Enemy or partner: Relationship between intronic micrornas and their host genes. IUBMB Life 2012, 64, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Lutter, D.; Marr, C.; Krumsiek, J.; Lang, E.W.; Theis, F.J. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genom. 2010, 11, 224. [Google Scholar] [CrossRef] [Green Version]
- Shomron, N.; Levy, C. MicroRNA-biogenesis and pre-mRNA splicing crosstalk. J. Biomed. Biotechnol. 2009, 2009, 594678. [Google Scholar] [CrossRef]
- Faller, M.; Guo, F. MicroRNA biogenesis: There’s more than one way to skin a cat. Biochim. Biophys. Acta 2008, 1779, 663–667. [Google Scholar] [CrossRef] [Green Version]
- Monteys, A.M.; Spengler, R.M.; Wan, J.; Tecedor, L.; Lennox, K.A.; Xing, Y.; Davidson, B.L. Structure and activity of putative intronic miRNA promoters. RNA 2010, 16, 495–505. [Google Scholar] [CrossRef] [Green Version]
- Roden, C.; Gaillard, J.; Kanoria, S.; Rennie, W.; Barish, S.; Cheng, J.; Pan, W.; Liu, J.; Cotsapas, C.; Ding, Y.; et al. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017, 27, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zeng, Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010, 38, 7689–7697. [Google Scholar] [CrossRef] [PubMed]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. microRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsico, A.; Huska, M.R.; Lasserre, J.; Hu, H.; Vucicevic, D.; Musahl, A.; Orom, U.; Vingron, M. PROmiRNA: A new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 2013, 14, R84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.L.; Miller, J.D.; Ying, S.Y. Intronic microRNA (miRNA). J. Biomed. Biotechnol. 2006, 2006, 26818. [Google Scholar] [CrossRef]
- Kataoka, N.; Fujita, M.; Ohno, M. Functional association of the microprocessor complex with the spliceosome. Mol. Cell. Biol. 2009, 29, 3243–3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Ladewig, E.; Shenker, S.; Mohammed, J.; Lai, E.C. Analysis of nearly one thousand mammalian mirtrons reveals novel features of Dicer substrates. PLoS Comput. Biol. 2015, 11, e1004441. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westholm, J.O.; Lai, E.C. Mirtrons: microRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef] [Green Version]
- Flynt, A.S.; Greimann, J.C.; Chung, W.J.; Lima, C.D.; Lai, E.C. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell 2010, 38, 900–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Greimann, J.C.; Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006, 127, 1223–1237. [Google Scholar] [CrossRef] [Green Version]
- Rorbach, G.; Unold, O.; Konopka, B.M. Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods. Sci. Rep. 2018, 8, 7560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef] [Green Version]
- Donayo, A.O.; Johnson, R.M.; Tseng, H.W.; Izreig, S.; Gariepy, A.; Mayya, V.K.; Wu, E.; Alam, R.; Lussier, C.; Jones, R.G.; et al. Oncogenic biogenesis of pri-miR-17~92 reveals hierarchy and competition among polycistronic MicroRNAs. Mol. Cell 2019, 75, 340–356. [Google Scholar] [CrossRef]
- Mohammed, J.; Flynt, A.S.; Siepel, A.; Lai, E.C. The impact of age, biogenesis, and genomic clustering on Drosophila microRNA evolution. RNA 2013, 19, 1295–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; He, C.; Fu, B.; Kong, X.; Bu, J.; Zhu, K.; Zheng, W.; Zhou, F.; Ni, B. microRNA-877 contributes to decreased non-small cell lung cancer cell growth via the PI3K/AKT pathway by targeting tartrate resistant acid phosphatase 5 activity. Cell Cycle 2020, 19, 3260–3276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, Y.; Kang, M.; Zhang, B. MiR-877 suppresses gastric cancer progression by downregulating AQP3. J. Int. Med. Res. 2020, 48, 300060520903661. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhang, S.; Wang, W.; Xu, Y.; Kawuli, A.; Lu, J.; Xiu, X. Long non-coding RNA DSCAM-AS1 contributes to the tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3 axis. Biosci. Rep. 2020, 40, BSR20192061. [Google Scholar] [CrossRef] [Green Version]
- Du, L.J.; Mao, L.J.; Jing, R.J. Long noncoding RNA DNAH17-AS1 promotes tumorigenesis and metastasis of non-small cell lung cancer via regulating miR-877-5p/CCNA2 pathway. Biochem. Biophys. Res. Commun. 2020, 533, 565–572. [Google Scholar] [CrossRef]
- Guo, T.; Wang, W.; Ji, Y.; Zhang, M.; Xu, G.; Lin, S. LncRNA PROX1-AS1 facilitates gastric cancer progression via miR-877-5p/PD-L1 Axis. Cancer Manag. Res. 2021, 13, 2669–2680. [Google Scholar] [CrossRef]
- Yan, T.H.; Qiu, C.; Sun, J.; Li, W.H. MiR-877-5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3038–3046. [Google Scholar] [CrossRef]
- Li, S.; Zhu, Y.; Liang, Z.; Wang, X.; Meng, S.; Xu, X.; Xu, X.; Wu, J.; Ji, A.; Hu, Z.; et al. Up-regulation of p16 by miR-877-3p inhibits proliferation of bladder cancer. Oncotarget 2016, 7, 51773–51783. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Li, K. HOXD-AS1 facilitates cell migration and invasion as an oncogenic lncRNA by competitively binding to miR-877-3p and upregulating FGF2 in human cervical cancer. BMC Cancer 2020, 20, 924. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qin, J.; Lu, S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int. J. Clin. Exp. Pathol. 2015, 8, 1515–1524. [Google Scholar]
- Wang, X.; Liu, L.; Zhao, W.; Li, Q.; Wang, G.; Li, H. LncRNA SNHG16 promotes the progression of laryngeal squamous cell carcinoma by mediating miR-877-5p/FOXP4 Axis. OncoTargets Ther. 2020, 13, 4569–4579. [Google Scholar] [CrossRef]
- Zhou, G.; Xie, J.; Gao, Z.; Yao, W. MicroRNA-877 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting IGF-1R. Exp. Ther. Med. 2019, 18, 1449–1457. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Ou, J.; Liu, J.; Li, X.; Meng, Y.; Yan, L.; Deng, P.; Sun, B. MicroRNA-877 is downregulated in cervical cancer and directly targets MACC1 to inhibit cell proliferation and invasion. Exp. Ther. Med. 2019, 18, 3650–3658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, C.; Cao, L.; Li, H.; Zou, H.; Li, H.; Pei, H. microRNA-877 inhibits malignant progression of colorectal cancer by directly targeting MTDH and regulating the PTEN/Akt pathway. Cancer Manag. Res. 2019, 11, 2769–2781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Bian, L.; Liu, R.; Wang, Y.; Xiao, X. Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 2021, 21, 10. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Sun, N.; Zhang, X.; Liang, G.; Zhu, J.; Xia, L.; Kou, Y.; Lu, J. Linc00941 regulates esophageal squamous cell carcinoma via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression. Aging 2021, 13, 17830–17846. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zheng, S. MiR-887-3p negatively regulates STARD13 and promotes pancreatic cancer progression. Cancer Manag. Res. 2020, 12, 6137–6147. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Shi, S.; Chen, Q.; Chen, Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol. Res. Pract. 2019, 215, 152476. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Hu, G.Q.; Da Costa, C.; Tang, J.H.; Li, Q.R.; Du, L.; Pan, Y.W.; Lv, S.Q. Long noncoding RNA UBE2R2-AS1 promotes glioma cell apoptosis via targeting the miR-877-3p/TLR4 axis. OncoTargets Ther. 2019, 12, 3467–3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Wang, Y.H.; Yoon, C.; Huang, X.Y.; Xu, Y.; Xie, J.W.; Wang, J.B.; Lin, J.X.; Chen, Q.Y.; Cao, L.L.; et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020, 471, 38–48. [Google Scholar] [CrossRef]
- Qian, J.; Li, R.; Wang, Y.Y.; Shi, Y.; Luan, W.K.; Tao, T.; Zhang, J.X.; Xu, Y.C.; You, Y.P. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol. Cell. Biochem. 2015, 403, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Liu, P.; Zheng, M.; Wang, Z.; Gao, Y.; Liang, K.; Wang, H.; Tan, X. The miR-1224-5p/ELF3 axis regulates malignant behaviors of pancreatic cancer via PI3K/AKT/Notch signaling pathways. OncoTargets Ther. 2020, 13, 3449–3466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Y.; Shen, W.; Wang, C.; Niu, N.; Pu, J. Circular RNA circ-ZNF609 promotes lung adenocarcinoma proliferation by modulating miR-1224-3p/ETV1 signaling. Cancer Manag. Res. 2020, 12, 2471–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wen, T.; Li, Z.; Che, X.; Gong, L.; Yang, X.; Zhang, J.; Tang, H.; He, L.; Qu, X.; et al. MicroRNA-1224 inhibits tumor metastasis in intestinal-type gastric cancer by directly targeting FAK. Front. Oncol. 2019, 9, 222. [Google Scholar] [CrossRef]
- Yu, P.F.; Wang, Y.; Lv, W.; Kou, D.; Hu, H.L.; Guo, S.S.; Zhao, Y.J. LncRNA NEAT1/miR-1224/KLF3 contributes to cell proliferation, apoptosis and invasion in lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8403–8410. [Google Scholar] [CrossRef]
- Ran, F.; Zhang, Y.; Shi, Y.; Liu, J.; Li, H.; Ding, L.; Ye, Q. miR-1224-3p promotes breast cancer cell proliferation and migration through PGM5-mediated aerobic glycolysis. J. Oncol. 2021, 2021, 5529770. [Google Scholar] [CrossRef]
- Jin, B.; Jin, D.; Zhuo, Z.; Zhang, B.; Chen, K. miR-1224-5p activates autophagy, cell invasion and inhibits epithelial-to-mesenchymal transition in osteosarcoma cells by directly targeting PLK1 through PI3K/AKT/mTOR signaling pathway. OncoTargets Ther. 2020, 13, 11807–11818. [Google Scholar] [CrossRef]
- Yang, L.; Wang, M.; He, P. LncRNA NEAT1 promotes the progression of gastric cancer through modifying the miR-1224-5p/RSF1 signaling axis. Cancer Manag. Res. 2020, 12, 11845–11855. [Google Scholar] [CrossRef]
- Song, N.S.; Pei, Z.D.; Fu, G. miR-1224-5p acts as a tumor suppressor via inhibiting the malignancy of rectal cancer through targeting SLC29A3. IUBMB Life 2020, 72, 2204–2213. [Google Scholar] [CrossRef]
- Li, J.; Peng, W.; Yang, P.; Chen, R.; Gu, Q.; Qian, W.; Ji, D.; Wang, Q.; Zhang, Z.; Tang, J.; et al. microRNA-1224-5p inhibits metastasis and epithelial-mesenchymal transition in colorectal cancer by targeting SP1-mediated NF-κB signaling pathways. Front. Oncol. 2020, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, Y.; Ye, C.; Liu, J. miR-1224-5p inhibits the proliferation and invasion of ovarian cancer via targeting SND1. Hum. Cell 2020, 33, 780–789. [Google Scholar] [CrossRef]
- Chen, W.; Yu, Z.; Huang, W.; Yang, Y.; Wang, F.; Huang, H. LncRNA LINC00665 promotes prostate cancer progression via miR-1224-5p/SND1 axis. OncoTargets Ther. 2020, 13, 2527–2535. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Zhang, B.; Yang, Y.; Li, Z.; Zhao, P.; Wu, W.; Zhang, H.; Mao, J. LncRNA MIR4435-2HG potentiates the proliferation and invasion of glioblastoma cells via modulating miR-1224-5p/TGFBR2 axis. J. Cell. Mol. Med. 2020, 24, 6362–6372. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.Z.; Wang, W.J.; Chen, Y.X.; Fan, Z.W.; Xie, X.F.; Yang, L.Y.; Chang, C.; Cai, Y.; Hao, J.J.; Wang, M.R.; et al. The miR-1224-5p/TNS4/EGFR axis inhibits tumour progression in oesophageal squamous cell carcinoma. Cell Death Dis. 2020, 11, 597. [Google Scholar] [CrossRef] [PubMed]
- Mohamadzade, Z.; Soltani, B.M.; Ghaemi, Z.; Hoseinpour, P. Cell specific tumor suppressor effect of hsa-miR-1226-3p through downregulation of HER2, PIK3R2, and AKT1 genes. Int. J. Biochem. Cell Biol. 2021, 134, 105965. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Jung, H.J.; Choi, H.J.; Jang, H.J.; Park, H.J.; Nejsum, L.N.; Kwon, T.H. Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells. FASEB J. 2020, 34, 3379–3398. [Google Scholar] [CrossRef]
- Chen, X.; Tan, W.; Li, W.; Li, W.; Zhu, S.; Zhong, J.; Shang, C.; Chen, Y. miR-1226-3p promotes sorafenib sensitivity of hepatocellular carcinoma via downregulation of DUSP4 expression. J. Cancer 2019, 10, 2745–2753. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhang, G.Q.; Zhu, D.Y.; Wang, L.J.; Li, G.T.; Xu, J.G.; Jin, X.L.; Zhu, Y.M.; Yang, X.Y. Long noncoding RNA ZFPM2-AS1 regulates ITGB1 by miR-1226-3p to promote cell proliferation and invasion in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7612–7620. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Rajabi, H.; Kufe, D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int. J. Oncol. 2010, 37, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Chen, D. Circular RNA hsa_circ_0000658 inhibits osteosarcoma cell proliferation and migration via the miR-1227/IRF2 axis. J. Cell. Mol. Med. 2021, 25, 510–520. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, Y.; Cai, Y. circTNFRSF21, a newly identified circular RNA promotes endometrial carcinoma pathogenesis through regulating miR-1227-MAPK13/ATF2 axis. Aging 2020, 12, 6774–6792. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Jiao, X.; Tian, B.; Zhang, M.; Zhou, C.; Wang, R.; Chen, H.; Wang, B.; Li, J.; et al. Suppressor of Ty 16 promotes lung cancer malignancy and is negatively regulated by miR-1227-5p. Cancer Sci. 2020, 111, 4075–4087. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Wu, J.; Zhang, L.; Chen, J.; Zhong, D.; Xu, S.; Xie, C.; Cai, J. Restoration of miR-1228* expression suppresses epithelial-mesenchymal transition in gastric cancer. PLoS ONE 2013, 8, e58637. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Chen, J.; Xie, C.; Shao, L.; Xu, Z.; Zhang, L. microRNA-1228(*) impairs the pro-angiogenic activity of gastric cancer cells by targeting macrophage migration inhibitory factor. Life Sci. 2017, 180, 9–16. [Google Scholar] [CrossRef]
- Chang, L.; Gao, H.; Wang, L.; Wang, N.; Zhang, S.; Zhou, X.; Yang, H. Exosomes derived from miR-1228 overexpressing bone marrow-mesenchymal stem cells promote growth of gastric cancer cells. Aging 2021, 13, 11808–11821. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, D.; Liang, H.; Xue, L.; Su, C.; Liu, M. miR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int. J. Clin. Exp. Pathol. 2015, 8, 6646–6655. [Google Scholar] [PubMed]
- Wang, J.W.; Wu, X.F.; Gu, X.J.; Jiang, X.H. Exosomal miR-1228 from cancer-associated fibroblasts promotes cell migration and invasion of osteosarcoma by directly targeting SCAI. Oncol. Res. 2019, 27, 979–986. [Google Scholar] [CrossRef]
- Sun, J.; Li, X.; Yu, E.; Liu, J.; Sun, L.; He, Q.; Lu, Q. A novel tumor suppressor ASMTL-AS1 regulates the miR-1228-3p/SOX17/β-catenin axis in triple-negative breast cancer. Diagn. Pathol. 2021, 16, 45. [Google Scholar] [CrossRef]
- Chen, D.; Ma, W.; Ke, Z.; Xie, F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 2018, 17, 2080–2090. [Google Scholar] [CrossRef]
- Li, X.; Lin, S.; Mo, Z.; Jiang, J.; Tang, H.; Wu, C.; Song, J. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J. Cancer 2020, 11, 599–609. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, J.; Deng, H.; Wan, H.; Liu, M.; Wang, J.; Li, S.; Li, X.; Tang, H. miR-1228 promotes the proliferation and metastasis of hepatoma cells through a p53 forward feedback loop. Br. J. Cancer 2015, 112, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Z.; Zheng, H.; Liu, X.; Zhang, W.; Zhu, J.; Wu, G.; Cao, L.; Song, J.; Wu, S.; Song, L.; et al. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/β-catenin signaling in breast cancer. Oncotarget 2016, 7, 24076–24087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.Y.; Yu, C.H.; Zhang, H.H.; Zhang, S.Z.; Yu, W.Y.; Yang, Y.; Chen, Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Cao, Y.; Cheng, K.; Li, C.; Zhu, F.; Yang, S.; Jin, M.; Song, S. Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR-1229-3p/ITGB8 axis. Open Life Sci. 2021, 16, 442–454. [Google Scholar] [CrossRef]
- Cao, Q.; Shi, Y.; Wang, X.; Yang, J.; Mi, Y.; Zhai, G.; Zhang, M. Circular METRN RNA hsa_circ_0037251 promotes glioma progression by sponging miR-1229-3p and regulating mTOR expression. Sci. Rep. 2019, 9, 19791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Cai, C.; Gan, J.; Yang, X.; Shuang, Z.; Liu, M.; Li, S.; Tang, H. miR-1236 down-regulates alpha-fetoprotein, thus causing PTEN accumulation, which inhibits the PI3K/Akt pathway and malignant phenotype in hepatoma cells. Oncotarget 2015, 6, 6014–6028. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, M.; Ding, D.; Yang, F.; Chen, Z. Long non-coding RNA NNT-AS1 contributes to cisplatin resistance via miR-1236-3p/ATG7 axis in lung cancer cells. OncoTargets Ther. 2020, 13, 3641–3652. [Google Scholar] [CrossRef]
- Hao, J.; Du, X.; Lv, F.; Shi, Q. Knockdown of circ_0006528 suppresses cell proliferation, migration, invasion, and adriamycin chemoresistance via regulating the miR-1236-3p/CHD4 axis in breast cancer. J. Surg. Res. 2021, 260, 104–115. [Google Scholar] [CrossRef]
- Wang, C.; Tang, K.; Li, Z.; Chen, Z.; Xu, H.; Ye, Z. Targeted p21(WAF1/CIP1) activation by miR-1236 inhibits cell proliferation and correlates with favorable survival in renal cell carcinoma. Urol. Oncol. 2016, 34, 59.e23-34. [Google Scholar] [CrossRef]
- Chen, S.Y.; Teng, S.C.; Cheng, T.H.; Wu, K.J. miR-1236 regulates hypoxia-induced epithelial-mesenchymal transition and cell migration/invasion through repressing SENP1 and HDAC3. Cancer Lett. 2016, 378, 59–67. [Google Scholar] [CrossRef]
- Chen, L.; Chi, K.; Xiang, H.; Yang, Y. Circ_0032821 facilitates gastric cancer cell proliferation, migration, invasion and glycolysis by regulating MiR-1236-3p/HMGB1 axis. Cancer Manag. Res. 2020, 12, 9965–9976. [Google Scholar] [CrossRef]
- Feng, W.; Gong, H.; Wang, Y.; Zhu, G.; Xue, T.; Wang, Y.; Cui, G. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol. Ther. Nucleic Acids 2019, 18, 375–387. [Google Scholar] [CrossRef] [Green Version]
- Duan, X.; Liu, D.; Wang, Y.; Chen, Z. Circular RNA hsa_circ_0074362 promotes glioma cell proliferation, migration, and invasion by attenuating the inhibition of miR-1236-3p on HOXB7 expression. DNA Cell Biol. 2018, 37, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Bian, T.; Jiang, D.; Liu, J.; Yuan, X.; Feng, J.; Li, Q.; Zhang, Q.; Li, X.; Liu, Y.; Zhang, J. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem. Biophys. Res. Commun. 2017, 492, 461–467. [Google Scholar] [CrossRef] [PubMed]
- An, J.X.; Ma, M.H.; Zhang, C.D.; Shao, S.; Zhou, N.M.; Dai, D.Q. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018, 18, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Zhu, H.; Sun, H.; Hua, Y.; Zhang, G.; Jiang, J.; Wang, X. Adipose mesenchymal stem cell-derived exosomal microRNA-1236 reduces resistance of breast cancer cells to cisplatin by suppressing SLC9A1 and the Wnt/β-catenin signaling. Cancer Manag. Res. 2020, 12, 8733–8744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, L.; Guo, X.; Guo, C.; Wang, W. microRNA-1236-3p regulates DDP resistance in lung cancer cells. Open Med. 2018, 14, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Song, T.F.; Xu, A.L.; Chen, X.H.; Gao, J.Y.; Gao, F.; Kong, X.C. Circular RNA circRNA_101996 promoted cervical cancer development by regulating miR-1236-3p/TRIM37 axis. Kaohsiung J. Med. Sci. 2021, 37, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, S.; Liu, X.; Zhang, W.; Li, Y.; Dong, R.; Zhang, Q.; Yang, Q.; Yuan, C.; Shen, K.; et al. miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncol. Rep. 2014, 31, 1905–1910. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.C.; Fu, W.G.; Zhong, Y.S. MicroRNA-1236-3p inhibits proliferation and invasion of breast cancer cells by targeting ZEB1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9988–9995. [Google Scholar] [CrossRef]
- Fang, H.; Jiang, W.; Jing, Z.; Mu, X.; Xiong, Z. miR-937 regulates the proliferation and apoptosis via targeting APAF1 in breast cancer. OncoTargets Ther. 2019, 12, 5687–5699. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Zhong, J.; Zhang, G.; Lin, L.; Liu, Z. Oncogenic role and prognostic value of microRNA-937-3p in patients with breast cancer. OncoTargets Ther. 2019, 12, 11045–11056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Guo, X.; Zhang, W.; Cong, Q. MicroRNA-937 inhibits the malignant phenotypes of breast cancer by directly targeting and downregulating forkhead box Q1. OncoTargets Ther. 2019, 12, 4813–4824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zeng, D.; Chen, Y.; Li, N.; Lv, Y.; Li, Y.; Xu, X.; Xu, G. miR-937 contributes to the lung cancer cell proliferation by targeting INPP4B. Life Sci. 2016, 155, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Xu, W.; Gong, J.; Wang, L.; Dai, M.; Chen, G.; Yuan, L. miR-937-5p targets SOX17 to modulate breast cancer cell cycle and cell proliferation through the Wnt signaling pathway. Cell. Signal. 2021, 77, 109818. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, Y.; Li, W.T.; Li, Y.; Zhu, J.F. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol. Oncol. 2020, 14, 2960–2984. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Li-ya, Q.; Feng, Z.; Yin, W.; Ji-hong, L. miR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed. Pharmacother. 2015, 71, 64–69. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, G.; Gao, H.; Li, Y.; Zhuang, L.; Meng, Z.; Liu, L. miR-939-5p contributes to the migration and invasion of pancreatic cancer by targeting ARHGAP4. OncoTargets Ther. 2020, 13, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.; Yu, J.; Huang, S.; Zhu, H.; Huang, Z. Transcriptional regulation of gene expression by microRNAs as endogenous decoys of transcription factors. Cell. Physiol. Biochem. 2014, 33, 1698–1714. [Google Scholar] [CrossRef]
- Cui, C.; Zhai, D.; Cai, L.; Duan, Q.; Xie, L.; Yu, J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939‒mediated transcriptional repression of Bcl-xL. Cancer Res. Treat. 2018, 50, 992–1008. [Google Scholar] [CrossRef] [Green Version]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Situ, J.; Zhang, H.; Jin, Z.; Li, K.; Mao, Y.; Huang, W. MicroRNA-939 directly targets HDGF to inhibit the aggressiveness of prostate cancer via deactivation of the WNT/β-catenin pathway. OncoTargets Ther. 2020, 13, 4257–4270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Yu, D. MicroRNA-939-5p directly targets IGF-1R to inhibit the aggressive phenotypes of osteosarcoma through deactivating the PI3K/Akt pathway. Int. J. Mol. Med. 2019, 44, 1833–1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbin, A.; Lovisa, F.; Holmes, A.B.; Damanti, C.C.; Gallingani, I.; Carraro, E.; Accordi, B.; Veltri, G.; Pizzi, M.; d’Amore, E.S.G.; et al. miR-939 acts as tumor suppressor by modulating JUNB transcriptional activity in pediatric anaplastic large cell lymphoma. Haematologica 2021, 106, 610–613. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, X.; Li, Q.; Zhang, Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manag. Res. 2019, 11, 1779–1789. [Google Scholar] [CrossRef] [Green Version]
- Aghdaei, F.H.; Soltani, B.M.; Dokanehiifard, S.; Mowla, S.J.; Soleimani, M. Overexpression of hsa-miR-939 follows by NGFR down-regulation and apoptosis reduction. J. Biosci. 2017, 42, 23–30. [Google Scholar] [CrossRef]
- Zhang, J.X.; Xu, Y.; Gao, Y.; Chen, C.; Zheng, Z.S.; Yun, M.; Weng, H.W.; Xie, D.; Ye, S. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol. Cancer 2017, 16, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Liu, S.; Lu, X.; Wei, L.; Chen, Y. Inhibition of microRNA-939 suppresses the development of human non-small cell lung cancer via the upregulation of tissue inhibitor of metalloproteinases 2. Mol. Med. Rep. 2018, 18, 4831–4838. [Google Scholar] [CrossRef] [Green Version]
- Hui, W.; Ma, X.; Zan, Y.; Song, L.; Zhang, S.; Dong, L. MicroRNA-1292-5p inhibits cell growth, migration and invasion of gastric carcinoma by targeting DEK. Am. J. Cancer Res. 2018, 8, 1228–1238. [Google Scholar]
- Wang, J.; Li, M.; Han, X.; Wang, H.; Wang, X.; Ma, G.; Xia, T.; Wang, S. miR-1976 knockdown promotes epithelial-mesenchymal transition and cancer stem cell properties inducing triple-negative breast cancer metastasis. Cell Death Dis. 2020, 11, 500. [Google Scholar] [CrossRef]
- Chen, G.; Hu, J.; Huang, Z.; Yang, L.; Chen, M. MicroRNA-1976 functions as a tumor suppressor and serves as a prognostic indicator in non-small cell lung cancer by directly targeting PLCE1. Biochem. Biophys. Res. Commun. 2016, 473, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Pekow, J.; Hutchison, A.L.; Meckel, K.; Harrington, K.; Deng, Z.; Talasila, N.; Rubin, D.T.; Hanauer, S.B.; Hurst, R.; Umanskiy, K.; et al. miR-4728-3p functions as a tumor suppressor in ulcerative colitis-associated colorectal neoplasia through regulation of focal adhesion signaling. Inflamm. Bowel Dis. 2017, 23, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yuan, Y.; Li, L.; Wang, X.; Quan, Y.; Liu, C.; Yu, M.; Hu, X.; Meng, X.; Zhou, Z.; et al. HER2-intronic miR-4728-5p facilitates HER2 expression and accelerates cell proliferation and migration by targeting EBP1 in breast cancer. PLoS ONE 2021, 16, e0245832. [Google Scholar] [CrossRef]
- Newie, I.; Søkilde, R.; Persson, H.; Grabau, D.; Rego, N.; Kvist, A.; von Stedingk, K.; Axelson, H.; Borg, Å.; Vallon-Christersson, J.; et al. The HER2-encoded miR-4728-3p regulates ESR1 through a non-canonical internal seed interaction. PLoS ONE 2014, 9, e97200. [Google Scholar] [CrossRef]
- Schmitt, D.C.; Madeira da Silva, L.; Zhang, W.; Liu, Z.; Arora, R.; Lim, S.; Schuler, A.M.; McClellan, S.; Andrews, J.F.; Kahn, A.G.; et al. ErbB2-intronic microRNA-4728: A novel tumor suppressor and antagonist of oncogenic MAPK signaling. Cell Death Dis. 2015, 6, e1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newie, I.; Søkilde, R.; Persson, H.; Jacomasso, T.; Gorbatenko, A.; Borg, Å.; de Hoon, M.; Pedersen, S.F.; Rovira, C. HER2-encoded mir-4728 forms a receptor-independent circuit with miR-21-5p through the non-canonical poly(A) polymerase PAPD5. Sci. Rep. 2016, 6, 35664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Zhang, X.; Gong, C.; Li, J. Aberrant expression of miR-4728 in patients with non-small cell lung cancer and its regulatory effects on tumor progression in tumor cells. Exp. Ther. Med. 2020, 20, 15. [Google Scholar] [CrossRef]
- Zhou, W.; Ding, X.; Jin, P.; Li, P. miR-6838-5p affects cell growth, migration, and invasion by targeting GPRIN3 via the Wnt/β-catenin signaling pathway in gastric cancer. Pathobiology 2020, 87, 327–337. [Google Scholar] [CrossRef]
- Liu, G.; Wang, P.; Zhang, H. miR-6838-5p suppresses cell metastasis and the EMT process in triple-negative breast cancer by targeting WNT3A to inhibit the WNT pathway. J. Gene Med. 2019, 21, e3129. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, J.; Wen, Q.; Dai, Y.; Zhang, W.; Li, F.; Li, J. MicroRNA-6852 suppresses cell proliferation and invasion via targeting forkhead box J1 in gastric cancer. Exp. Ther. Med. 2018, 16, 3249–3255. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, H.; Zheng, K.; Zhang, S.; Dong, W. MicroRNA-6852 suppresses glioma A172 cell proliferation and invasion by targeting LEF1. Exp. Ther. Med. 2019, 18, 1877–1883. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Li, Z.Y.; Zhang, L.M.; Wu, X.Y.; Xiang, S.H.; Wang, Y.G.; Zhang, Y.Q. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021, 12, 94. [Google Scholar] [CrossRef]
- Cui, B.H.; Hong, X. miR-6852 serves as a prognostic biomarker in colorectal cancer and inhibits tumor growth and metastasis by targeting TCF7. Exp. Ther. Med. 2018, 16, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 1996, 6, 215–220. [Google Scholar] [CrossRef]
- Fu, X.D.; Ares, M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Agranat-Tamir, L.; Shomron, N.; Sperling, J.; Sperling, R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014, 42, 4640–4651. [Google Scholar] [CrossRef] [PubMed]
- Ip, J.Y.; Schmidt, D.; Pan, Q.; Ramani, A.K.; Fraser, A.G.; Odom, D.T.; Blencowe, B.J. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011, 21, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Pawlicki, J.M.; Steitz, J.A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008, 182, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.L.; Wang, Z.; Liao, Q.; Zhu, Y.; Zhou, W.H.; Xu, W.; Qiu, Z. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev. Cell 2014, 28, 547–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shayevitch, R.; Askayo, D.; Keydar, I.; Ast, G. The importance of DNA methylation of exons on alternative splicing. RNA 2018, 24, 1351–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, J.J.; Gao, D.; Nguyen, T.V.; Kwok, C.T.; van Geldermalsen, M.; Middleton, R.; Pinello, N.; Thoeng, A.; Nagarajah, R.; Holst, J.; et al. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 2017, 8, 15134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schor, I.E.; Gómez Acuña, L.I.; Kornblihtt, A.R. Coupling between transcription and alternative splicing. Cancer Treat. Res. 2013, 158, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Janas, M.M.; Khaled, M.; Schubert, S.; Bernstein, J.G.; Golan, D.; Veguilla, R.A.; Fisher, D.E.; Shomron, N.; Levy, C.; Novina, C.D. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011, 7, e1002330. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.J.; van der Weyden, L.; Mayeda, A.; Stamm, S.; Morris, B.J.; Rasko, J.E. ZNF265--a novel spliceosomal protein able to induce alternative splicing. J. Cell Biol. 2001, 154, 25–32. [Google Scholar] [CrossRef]
- Wu, H.; Sun, S.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.R.; Zhu, J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell 2010, 38, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Hao, Q.; Lin, Y.C.; Song, Y.J.; Bangru, S.; Arif, W.; Tripathi, V.; Zhang, Y.; Cho, J.H.; Freier, S.M.; et al. Antagonism between splicing and microprocessor complex dictates the serum-induced processing of lnc-MIRHG for efficient cell cycle reentry. RNA 2020, 26, 1603–1620. [Google Scholar] [CrossRef]
- Butkytė, S.; Čiupas, L.; Jakubauskienė, E.; Vilys, L.; Mocevicius, P.; Kanopka, A.; Vilkaitis, G. Splicing-dependent expression of microRNAs of mirtron origin in human digestive and excretory system cancer cells. Clin. Epigenetics 2016, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Nguyen, T.D.; Li, S.; Nguyen, T.A. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA 2018, 24, 892–898. [Google Scholar] [CrossRef] [Green Version]
- Ajiro, M.; Jia, R.; Yang, Y.; Zhu, J.; Zheng, Z.M. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016, 44, 1854–1870. [Google Scholar] [CrossRef]
- Guil, S.; Cáceres, J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007, 14, 591–596. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Ota, A.; Tagawa, H.; Karnan, S.; Tsuzuki, S.; Karpas, A.; Kira, S.; Yoshida, Y.; Seto, M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004, 64, 3087–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Kooshapur, H.; Choudhury, N.R.; Simon, B.; Mühlbauer, M.; Jussupow, A.; Fernandez, N.; Jones, A.N.; Dallmann, A.; Gabel, F.; Camilloni, C.; et al. Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. Nat. Commun. 2018, 9, 2479. [Google Scholar] [CrossRef] [Green Version]
- Gherzi, R.; Lee, K.Y.; Briata, P.; Wegmüller, D.; Moroni, C.; Karin, M.; Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [Green Version]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michlewski, G.; Cáceres, J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010, 17, 1011–1018. [Google Scholar] [CrossRef] [Green Version]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Church, V.A.; Pressman, S.; Isaji, M.; Truscott, M.; Cizmecioglu, N.T.; Buratowski, S.; Frolov, M.V.; Carthew, R.W. Microprocessor recruitment to elongating RNA polymerase II is required for differential expression of microRNAs. Cell Rep. 2017, 20, 3123–3134. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.M.; Hwang, S.J.; Masuda, K.; Choi, K.M.; Jeong, M.R.; Nam, D.H.; Gorospe, M.; Kim, H.H. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol. Cell. Biol. 2012, 32, 4237–4244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havens, M.A.; Reich, A.A.; Hastings, M.L. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014, 10, e1004312. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Nam, J.W.; Shin, C. DROSHA targets its own transcript to modulate alternative splicing. RNA 2017, 23, 1035–1047. [Google Scholar] [CrossRef] [Green Version]
- Link, S.; Grund, S.E.; Diederichs, S. Alternative splicing affects the subcellular localization of Drosha. Nucleic Acids Res. 2016, 44, 5330–5343. [Google Scholar] [CrossRef]
- Nguyen, L.X.T.; Zhang, B.; Hoang, D.H.; Zhao, D.; Wang, H.; Wu, H.; Su, Y.L.; Dong, H.; Rodriguez-Rodriguez, S.; Armstrong, B.; et al. Cytoplasmic DROSHA and non-canonical mechanisms of MiR-155 biogenesis in FLT3-ITD acute myeloid leukemia. Leukemia 2021, 35, 2285–2298. [Google Scholar] [CrossRef]
- Dai, L.; Hallmark, L.; Bofill De Ros, X.; Crouch, H.; Chen, S.; Shi, T.; Yang, A.; Lian, C.; Zhao, Y.; Tran, B.; et al. Novel, abundant Drosha isoforms are deficient in miRNA processing in cancer cells. RNA Biol. 2020, 17, 1603–1612. [Google Scholar] [CrossRef]
- Mattioli, C.; Pianigiani, G.; Pagani, F. A competitive regulatory mechanism discriminates between juxtaposed splice sites and pri-miRNA structures. Nucleic Acids Res. 2013, 41, 8680–8691. [Google Scholar] [CrossRef] [Green Version]
- Melamed, Z.; Levy, A.; Ashwal-Fluss, R.; Lev-Maor, G.; Mekahel, K.; Atias, N.; Gilad, S.; Sharan, R.; Levy, C.; Kadener, S.; et al. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol. Cell 2013, 50, 869–881. [Google Scholar] [CrossRef] [Green Version]
- Pianigiani, G.; Licastro, D.; Fortugno, P.; Castiglia, D.; Petrovic, I.; Pagani, F. Microprocessor-dependent processing of splice site overlapping microRNA exons does not result in changes in alternative splicing. RNA 2018, 24, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Bell, M.L.; Buvoli, M.; Leinwand, L.A. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol. Cell. Biol. 2010, 30, 1937–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef] [Green Version]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V. FLT3-ITD and its current role in acute myeloid leukaemia. Med. Oncol. 2017, 34, 114. [Google Scholar] [CrossRef] [PubMed]
- Narayan, N.; Bracken, C.P.; Ekert, P.G. MicroRNA-155 expression and function in AML: An evolving paradigm. Exp. Hematol. 2018, 62, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Shyr, Y.; Cai, J.; Liu, Q. Interplay between miRNAs and host genes and their role in cancer. Brief. Funct. Genom. 2018, 18, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steiman-Shimony, A.; Shtrikman, O.; Margalit, H. Assessing the functional association of intronic miRNAs with their host genes. RNA 2018, 24, 991–1004. [Google Scholar] [CrossRef] [Green Version]
- Hinske, L.C.; Galante, P.A.; Kuo, W.P.; Ohno-Machado, L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genom. 2010, 11, 533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.; Quarton, T.; Nowak, C.M.; Ehrhardt, K.; Singh, A.; Li, Y.; Bleris, L. Robust filtering and noise suppression in intragenic miRNA-mediated host regulation. iScience 2020, 23, 101595. [Google Scholar] [CrossRef]
- Campo-Paysaa, F.; Sémon, M.; Cameron, R.A.; Peterson, K.J.; Schubert, M. microRNA complements in deuterostomes: Origin and evolution of microRNAs. Evol. Dev. 2011, 13, 15–27. [Google Scholar] [CrossRef] [PubMed]
- França, G.S.; Vibranovski, M.D.; Galante, P.A. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 2016, 7, 11438. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Song, Y.J.; Prasanth, K.V. One locus with two roles: MicroRNA-independent functions of microRNA-host-gene locus-encoded long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2021, 12, e1625. [Google Scholar] [CrossRef]
- Li, Z.; Huang, H.; Chen, P.; He, M.; Li, Y.; Arnovitz, S.; Jiang, X.; He, C.; Hyjek, E.; Zhang, J.; et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012, 3, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, D.P.; Cost, N.G.; Hegde, S.; Kellner, E.; Mikhaylova, O.; Stratton, Y.; Ehmer, B.; Abplanalp, W.A.; Pandey, R.; Biesiada, J.; et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell 2014, 26, 738–753. [Google Scholar] [CrossRef] [Green Version]
- Miele, E.; Po, A.; Mastronuzzi, A.; Carai, A.; Besharat, Z.M.; Pediconi, N.; Abballe, L.; Catanzaro, G.; Sabato, C.; De Smaele, E.; et al. Downregulation of miR-326 and its host gene β-arrestin1 induces pro-survival activity of E2F1 and promotes medulloblastoma growth. Mol. Oncol. 2021, 15, 523–542. [Google Scholar] [CrossRef] [PubMed]
- Herbst, F.; Lang, T.J.L.; Eckert, E.S.P.; Wünsche, P.; Wurm, A.A.; Kindinger, T.; Laaber, K.; Hemmati, S.; Hotz-Wagenblatt, A.; Zavidij, O.; et al. The balance between the intronic miR-342 and its host gene Evl determines hematopoietic cell fate decision. Leukemia 2021. [Google Scholar] [CrossRef]
- Peperstraete, E.; Lecerf, C.; Collette, J.; Vennin, C.; Raby, L.; Völkel, P.; Angrand, P.O.; Winter, M.; Bertucci, F.; Finetti, P.; et al. Enhancement of breast cancer cell aggressiveness by lncRNA H19 and its mir-675 derivative: Insight into shared and different actions. Cancers 2020, 12, 1730. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, R.; Wu, Y.; Liu, Y.; Su, W.; Xiong, W.; Zeng, Z. PVT1 promotes cancer progression via microRNAs. Front. Oncol. 2019, 9, 609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.F.; Li, X.L.; Subramanian, M.; Shabalina, S.A.; Hara, T.; Zhu, Y.; Huang, J.; Yang, Y.; Wakefield, L.M.; Prasanth, K.V.; et al. Growth differentiation factor-15 encodes a novel microRNA 3189 that functions as a potent regulator of cell death. Cell Death Differ. 2015, 22, 1641–1653. [Google Scholar] [CrossRef] [Green Version]
- Floros, K.V.; Lochmann, T.L.; Hu, B.; Monterrubio, C.; Hughes, M.T.; Wells, J.D.; Morales, C.B.; Ghotra, M.S.; Costa, C.; Souers, A.J.; et al. Coamplification of miR-4728 protects HER2-amplified breast cancers from targeted therapy. Proc. Natl. Acad. Sci. USA 2018, 115, E2594–E2603. [Google Scholar] [CrossRef] [Green Version]
- Quah, S.; Holland, P.W. The Hox cluster microRNA miR-615: A case study of intronic microRNA evolution. EvoDevo 2015, 6, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godínez-Rubí, M.; Ortuño-Sahagún, D. miR-615 fine-tunes growth and development and has a role in cancer and in neural repair. Cells 2020, 9, 1566. [Google Scholar] [CrossRef]
- Liu, J.; Jia, Y.; Jia, L.; Li, T.; Yang, L.; Zhang, G. MicroRNA 615-3p inhibits the tumor growth and metastasis of NSCLC via inhibiting IGF2. Oncol. Res. 2019, 27, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, L.; Sun, Y.; Xue, Y.; Qu, J.; Pan, S.; Li, H.; Qu, H.; Wang, J.; Zhang, J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharmacother. 2018, 101, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Ooi, W.F.; Qamra, A.; Cheung, A.; Ma, D.; Sundaram, G.M.; Xu, C.; Xing, M.; Poon, L.; Wang, J.; et al. HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat. Commun. 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, M.; Maeda, T.; Bouillez, A.; Alam, M.; Tagde, A.; Hinohara, K.; Suzuki, Y.; Markert, T.; Miyo, M.; Komura, K.; et al. MUC1-C activates BMI1 in human cancer cells. Oncogene 2017, 36, 2791–2801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; An, O.; Chan, T.H.M.; Ng, V.H.E.; Kwok, H.S.; Lin, J.S.; Qi, L.; Han, J.; Tay, D.J.T.; Tang, S.J.; et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res. 2018, 46, 7953–7969. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Balinth, S.; Mills, A.A. p63-related signaling at a glance. J. Cell Sci. 2020, 133, jcs228015. [Google Scholar] [CrossRef]
- Gatti, V.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019, 13, 981–1001. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Jeong, J.S.; Okamura, J.; Sook-Kim, M.; Zhu, H.; Guerrero-Preston, R.; Ratovitski, E.A. Global tumor protein p53/p63 interactome: Making a case for cisplatin chemoresistance. Cell Cycle 2012, 11, 2367–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.H.; Cho, E.G.; Yu, S.J.; Kang, H.; Kim, Y.J.; Kim, S.H.; Lee, T.R. ΔNp63 intronic miR-944 is implicated in the ΔNp63-mediated induction of epidermal differentiation. Nucleic Acids Res. 2015, 43, 7462–7479. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, J.H.; Jin, S.; Kim, J.H.; Kim, S.H. Primate-specific miR-944 activates p53-dependent tumor suppression in human colorectal cancers. Cancer Lett. 2019, 440–441, 168–179. [Google Scholar] [CrossRef] [PubMed]
| Mirtron | Type | Host Gene and Intron | Validated Gene Targets | Cancer |
|---|---|---|---|---|
| hsa-miR-877 | Mirtron | ABCF1 Intron 12 | ACP5 [52], AQP3 [53], ATXN7L3 [54], CCNA2 [55], CD274 [56], CDK14 [57], CDKN2A (upreg) [58], FGF2 [59], FOXM1 [60], FOXP4 [61], IGF1R [62], MACC1 [63], MTDH [64], PIK3R3 [65], PMEPA1 [66], STARD13 [67], SUZ12 [68], TLR4 [69], VEGFA [70] | Bladder (TS) [58], Cervical (TS) [54,59,63], Colorectal (TS) [64], Gastric (TS) [53,56,70], Glioma (Onc) [69], Glioma (TS) [68], Laryngeal (TS) [61], Liver (TS) [57,60,65], Lung (TS) [52,55,62], Oesophageal (TS) [66], Pancreatic (Onc) [67] |
| hsa-miR-1224 | Mirtron | VWA5B2 Intron 17 | CREB1 [71], ELF3 [72], ETV1 [73], FAK [74], KLF3 [75], PGM5 [76], PLK1 [77], RSF1 [78], SLC29A3 [79], SP1 [80], SND1 [81,82], TGFBR2 [83], TNS4 [84], | Breast (Onc) [76], Colorectal (TS) [80], Gastric (TS) [74,78], Glioma (TS) [71,83], Lung (TS) [73,75], Oesophageal (TS) [84], Osteosarcoma (TS) [77], Ovarian (TS) [81], Pancreatic (TS) [72], Prostate (TS) [82], Rectal (TS) [79] |
| hsa-miR-1226 | Mirtron | DHX30 Intron 20 | AKT1 [85], AQP5 [86], DUSP4 [87], ERBB2 [85], ITGB1 [88], MUC1 [89], PIK3R2 [85] | Breast (TS) [85,86,89], Liver (TS) [87,88], |
| hsa-miR-1227 | Mirtron | PLEKHJ1 Intron 1 | IRF2 [90], MAPK13 [91], SUPT16H [92] | Endometrial (TS) [91], Lung (TS) [92], Osteosarcoma (Onc) [90], |
| hsa-miR-1228 | Mirtron | LRP1 Intron 48 | CSNK2A2 [93], MIF [94], MMP14 [95], SCAI [96,97], SOX17 [98], TCF21 [99], TP53 [100,101], | Breast (Onc) [96,98], Gastric (TS) [93,94,95], Liver (Onc) [101], Lung (Onc) [99], Osteosarcoma (Onc) [97], Ovarian (Onc) [100] |
| hsa-miR-1229 | Mirtron | MGAT4B Intron 1 | APC [102], GSK3B [102], HIPK2 [103], ICAT [102], ITGB8 [104], MTOR [105], | Breast (Onc) [102], Colorectal (Onc) [103], Glioma (TS) [104,105], |
| hsa-miR-1236 | Mirtron | RDBP Intron 7 | AFP [106], ATG7 [107], CHD4 [108], CDKN1A (upreg) [109], HDAC3 [110], HMGB1 [111], HOXB7 [112,113], KLF8 [114], MTA2 [115], SENP1 [110], SLC9A1 [116], TPT1 [117], TRIM37 [118], ZEB1 [119,120], | Breast (TS) [108,116,120], Cervical (TS) [118], Colorectal (TS) [112], Gastric (TS) [111,115], Glioma (TS) [113], Lung (TS) [107,114,117], Liver (TS) [106], Kidney (TS) [109], Ovarian (TS) [119], |
| hsa-miR-937 | 3′-tailed mirtron | SCRIB Intron 29 | APAF1 [121], CCRL2 [122], FOXQ1 [123], INPP4B [124], SOX17 [125], TIMP3 [126], | Breast (Onc) [121,125], Breast (TS) [123], Colorectal (Onc) [126], Lung (Onc) [124], |
| hsa-miR-939 | 5′-tailed mirtron | CPSF1 Intron 4 | APC2 [127], ARHGAP4 [128], BCL2L1 [129,130], CDH5 [131], HDGF [132], IGF1R [133], JUNB [134], LIMK2 [135], NGFR [136], SLC34A2 [137], TIMP2 [138], | Breast (Onc) [131], Colorectal (TS) [130,135], Gastric (TS) [137], Lung (Onc) [138], Lymphoma (TS) [134], Ovarian (Onc) [127], Osteosarcoma (TS) [133], Pancreatic (Onc) [128], Prostate (TS) [132], |
| hsa-miR-1292 | 5′-tailed mirtron | NOP56 Intron 11 | DEK [139] | Gastric (TS) [139], |
| hsa-miR-1976 | 5′-tailed mirtron | RPS6KA1 Intron 6 | PIK3CG [140], PLCE1 [141], | Breast (TS) [140], Lung (TS) [141], |
| hsa-miR-4728 | 5′-tailed mirtron | ERBB2 Intron 25 | CAV1 [142], COL1A2 [142], EBP1 [143], ESR1 [144], MST4 [145], PAPD5 [146], THBS2 [142] | Breast (Onc) [143,146], Breast (TS) [145], Colorectal (TS) [142], Lung (TS) [147], |
| hsa-miR-6838 | 5′-tailed mirtron | POLM Intron 10 | GPRIN3 [148], WNT3A [149], | Breast (TS) [149], Gastric (TS) [148] |
| hsa-miR-6852 | 5′-tailed mirtron | TLN1 Intron 24 | FOXJ1 [150], LEF1 [151], ICAM1 [152], TCF7 [153], | Colorectal (TS) [153], Gastric (TS) [150], Glioma (TS) [151], Liver (TS) [152] |
| miRNA | Host Gene | Cancer | Functional Association |
|---|---|---|---|
| hsa-miR-196b | HOXA9 | Mixed lineage leukemia | Complex: miR-196b directly targets its host gene HOXA9 (oncogene) but also targets FAS (tumor suppressor) [205] |
| hsa-miR-204 | TRPM3 | Renal (Clear cell) | Antagonism: inhibits TRPM3-induced proliferation by directly targeting TRPM3; also inhibits oncogenic autophagy by targeting LC3B [206] |
| hsa-miR-326 | ARBB1 | Medulloblastoma | Synergism: inhibits E2F1-associated pro-survival function by directly targeting E2F1 (miR-326) and E2F1 acetylation (ARBB1) [207] |
| hsa-miR-342 | EVL | Lymphoid leukemia | Antagonism: miR-342/EVL balance determines myeloid (high miR-342) or lymphoid (high EVL) differentiation. miR-342 antagonises EVL-induced lymphoid proliferation [208] |
| hsa-miR-675 | H19 | Breast | Synergism: both promote cell migration, invasiveness and stemness [209] |
| hsa-miR-1204/ 1205/ 1207 | PVT1 | Various | Synergism: PVT1 sponges multiple miRNAs, thereby enhancing tumor proliferation induced by miR-1204, miR-1205 and miR-1207 [210] |
| hsa-miR-3189 | GDF15 | Various | Antagonism: mir-3189 promotes p53-independent apoptosis; GDF15 is implicated in metastasis [211] |
| hsa-miR-4728 | ERBB2 | Breast | Synergism: inhibits apoptosis thereby promoting therapy resistance to HER2(ERBB2) inhibitors [212] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, A.C.H.; Rasko, J.E.J. Splice and Dice: Intronic microRNAs, Splicing and Cancer. Biomedicines 2021, 9, 1268. https://doi.org/10.3390/biomedicines9091268
Wong ACH, Rasko JEJ. Splice and Dice: Intronic microRNAs, Splicing and Cancer. Biomedicines. 2021; 9(9):1268. https://doi.org/10.3390/biomedicines9091268
Chicago/Turabian StyleWong, Alex C. H., and John E. J. Rasko. 2021. "Splice and Dice: Intronic microRNAs, Splicing and Cancer" Biomedicines 9, no. 9: 1268. https://doi.org/10.3390/biomedicines9091268






