miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met
Abstract
:1. Introduction
2. Materials and Methods
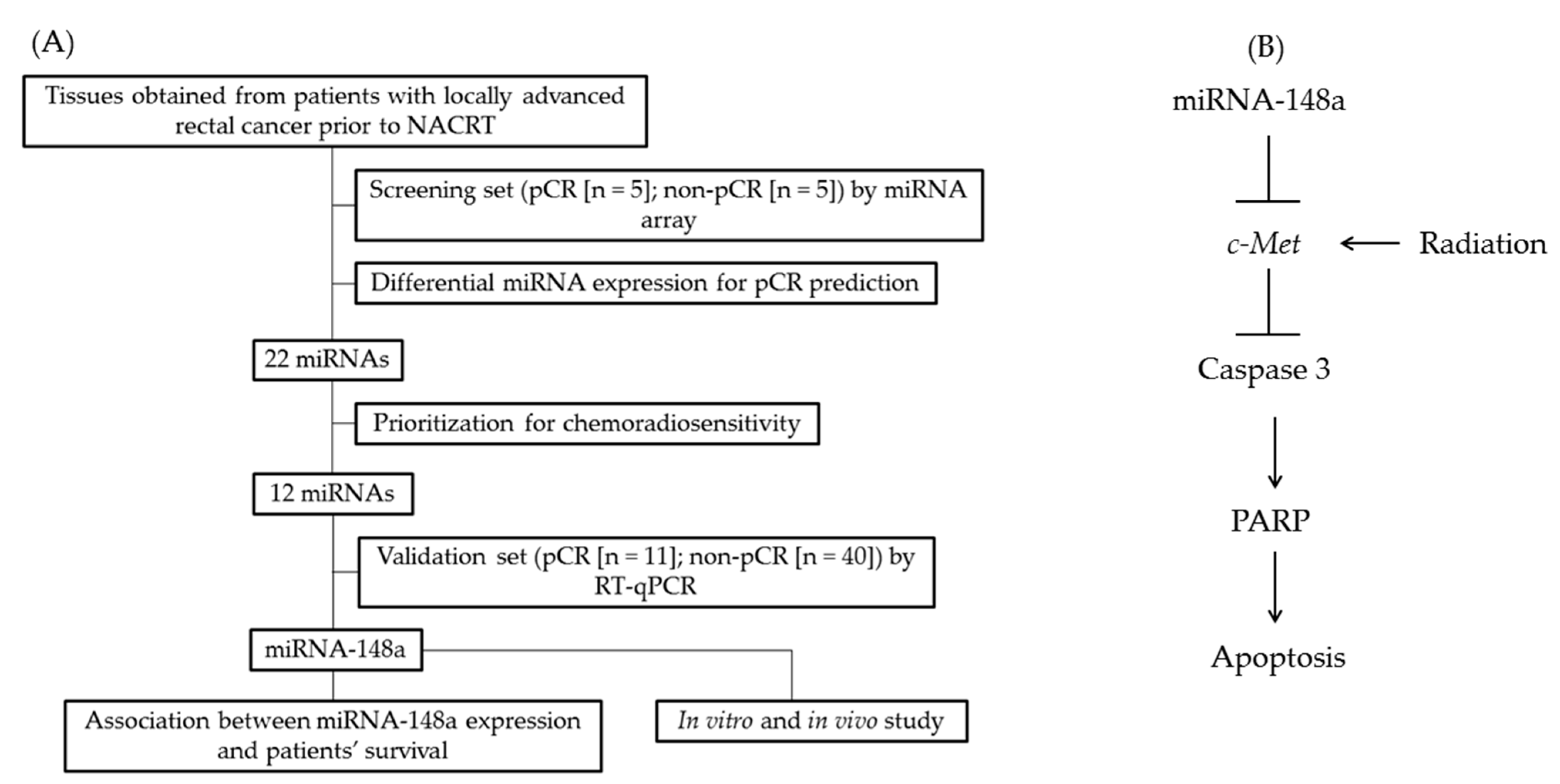
2.1. Patients and Tissue Specimens
2.2. miRNA Microarray
2.3. miRNA Expression by RT-qPCR
2.4. Putative Target Genes of miRNA-148a
2.5. Cell Culture and Irradiation
2.6. Cell Transfection
2.7. Cell Viability Assay
2.8. Colony Formation Assay
2.9. Cell Cycle and Apoptosis Analysis by Flow Cytometry
2.10. Western Blotting
2.11. Luciferase Reporter Assay
2.12. Animal Studies
2.13. Statistical Analysis
3. Results
3.1. Demographic Data
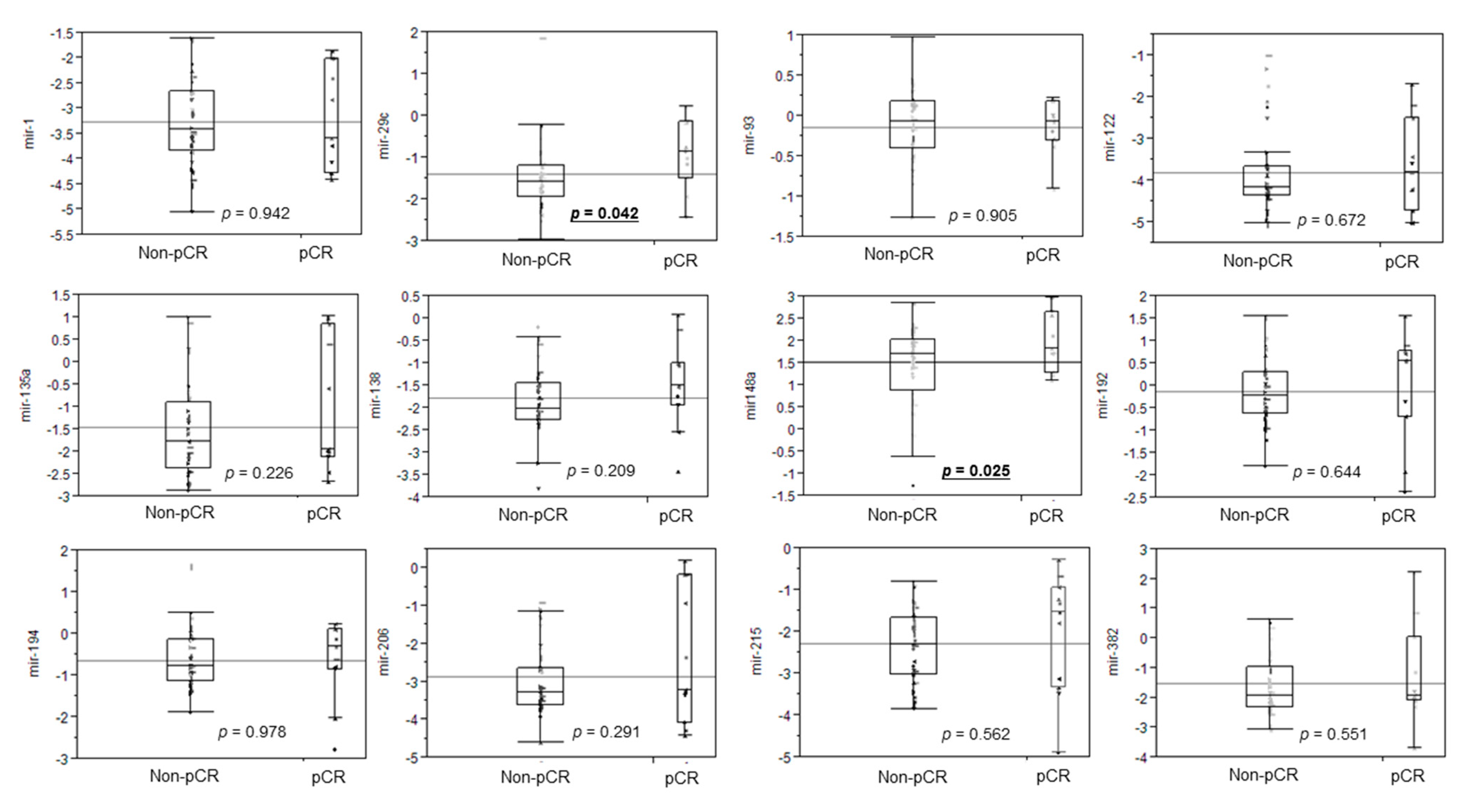
3.2. Differential miRNA Expression for pCR Prediction
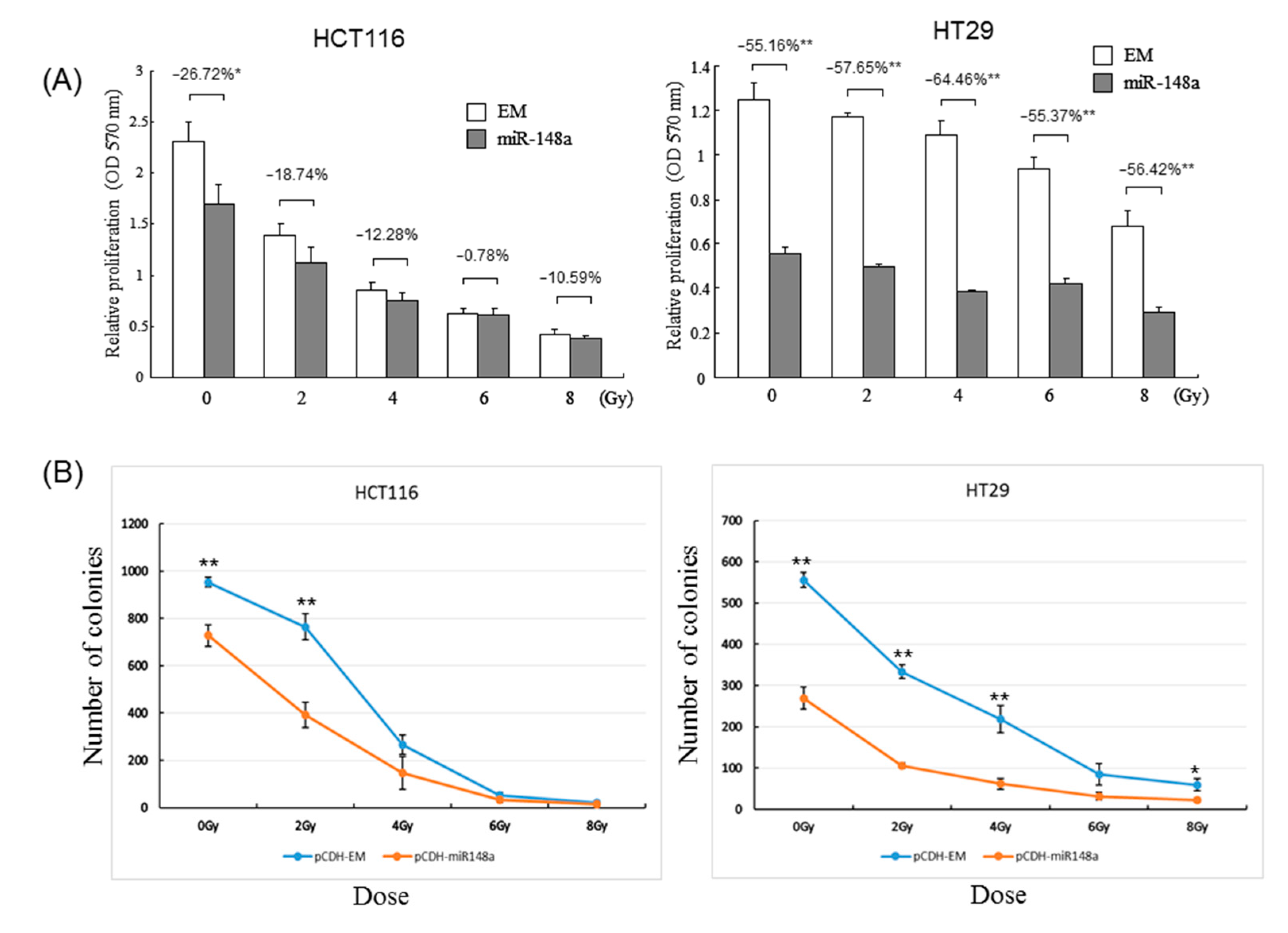
3.3. miRNA-148a Overexpression Promoted Radiosensitivity in CRC Cell Lines
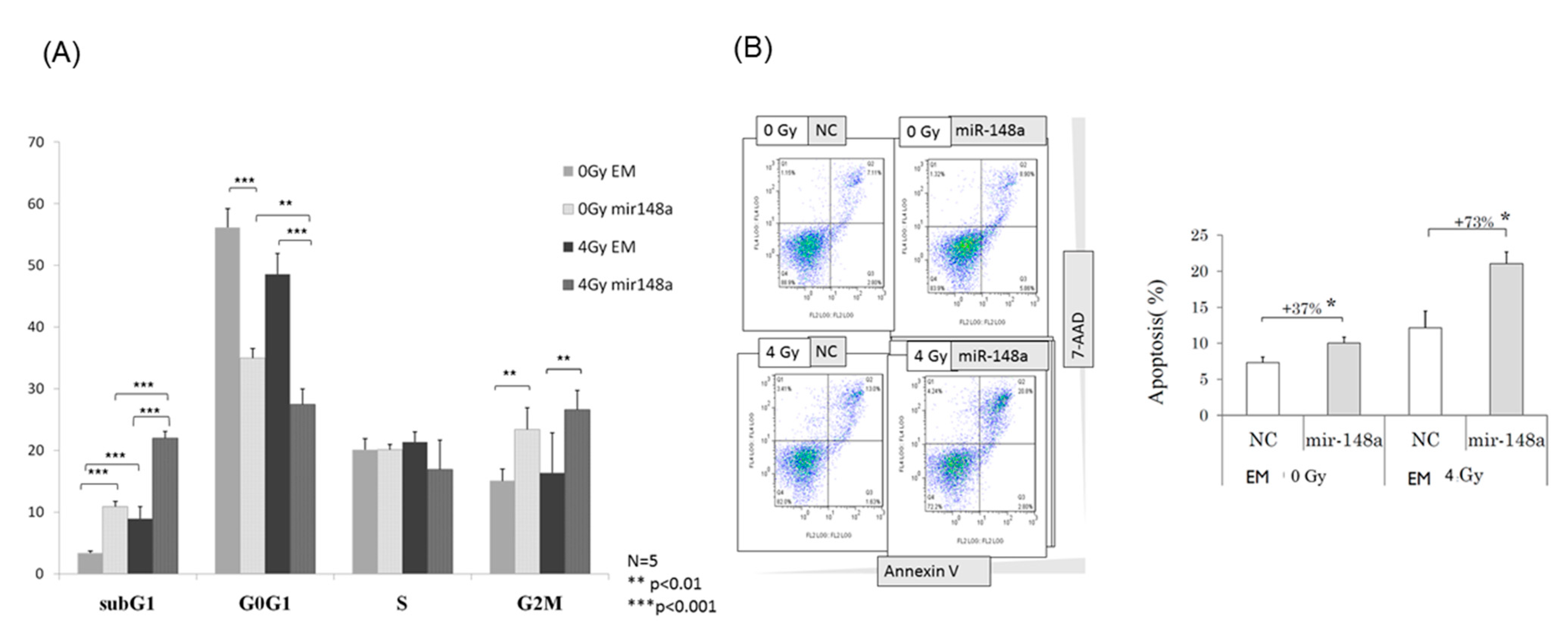
3.4. miRNA-148a Overexpression Led to Cell Cycle Changes in Irradiated CRC Cells
3.5. miRNA-148a Overexpression Enhanced Radiation-Induced Apoptosis in CRC Cells
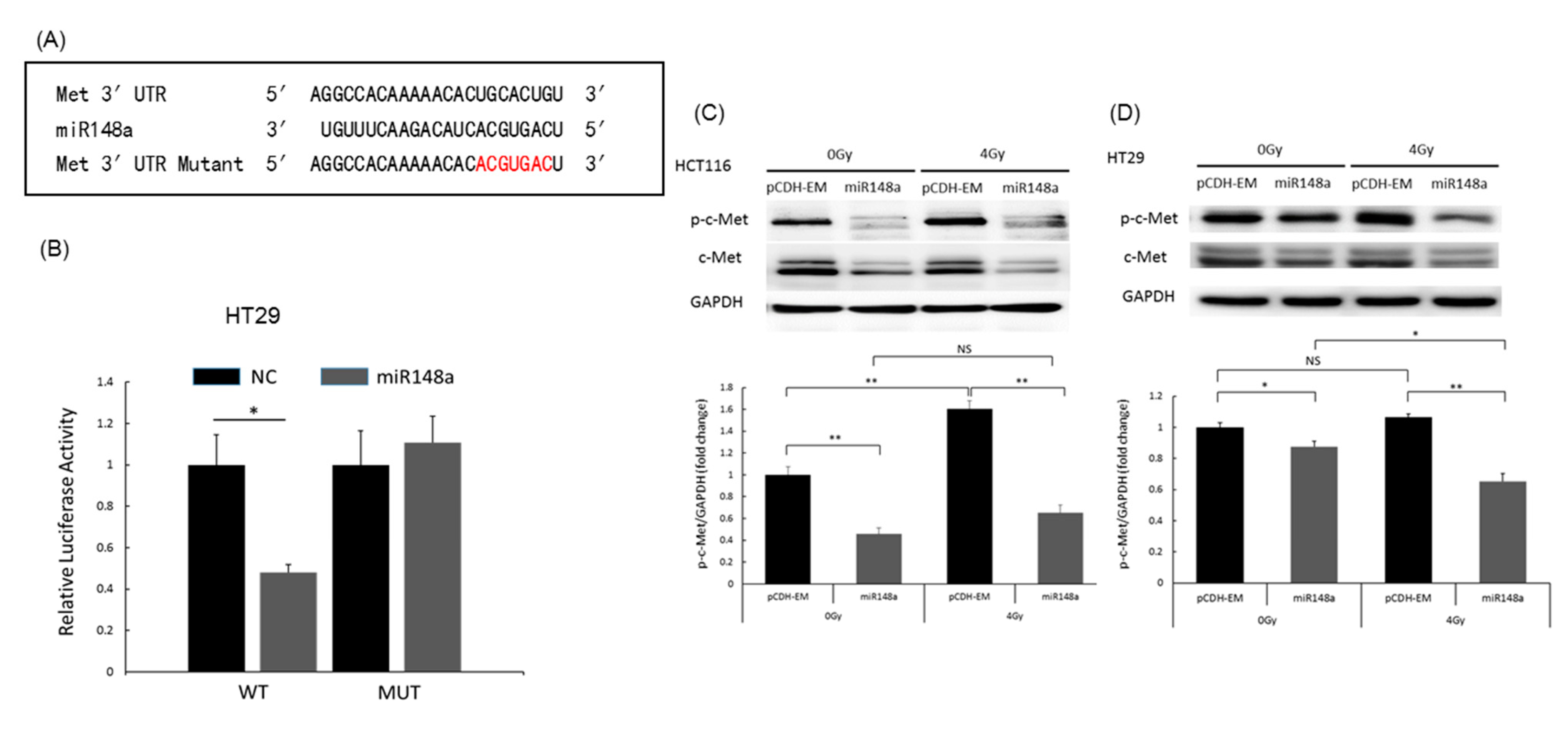
3.6. miRNA-148a Overexpression Promoted Apoptosis in CRC Cells by Directly Targeting c-Met
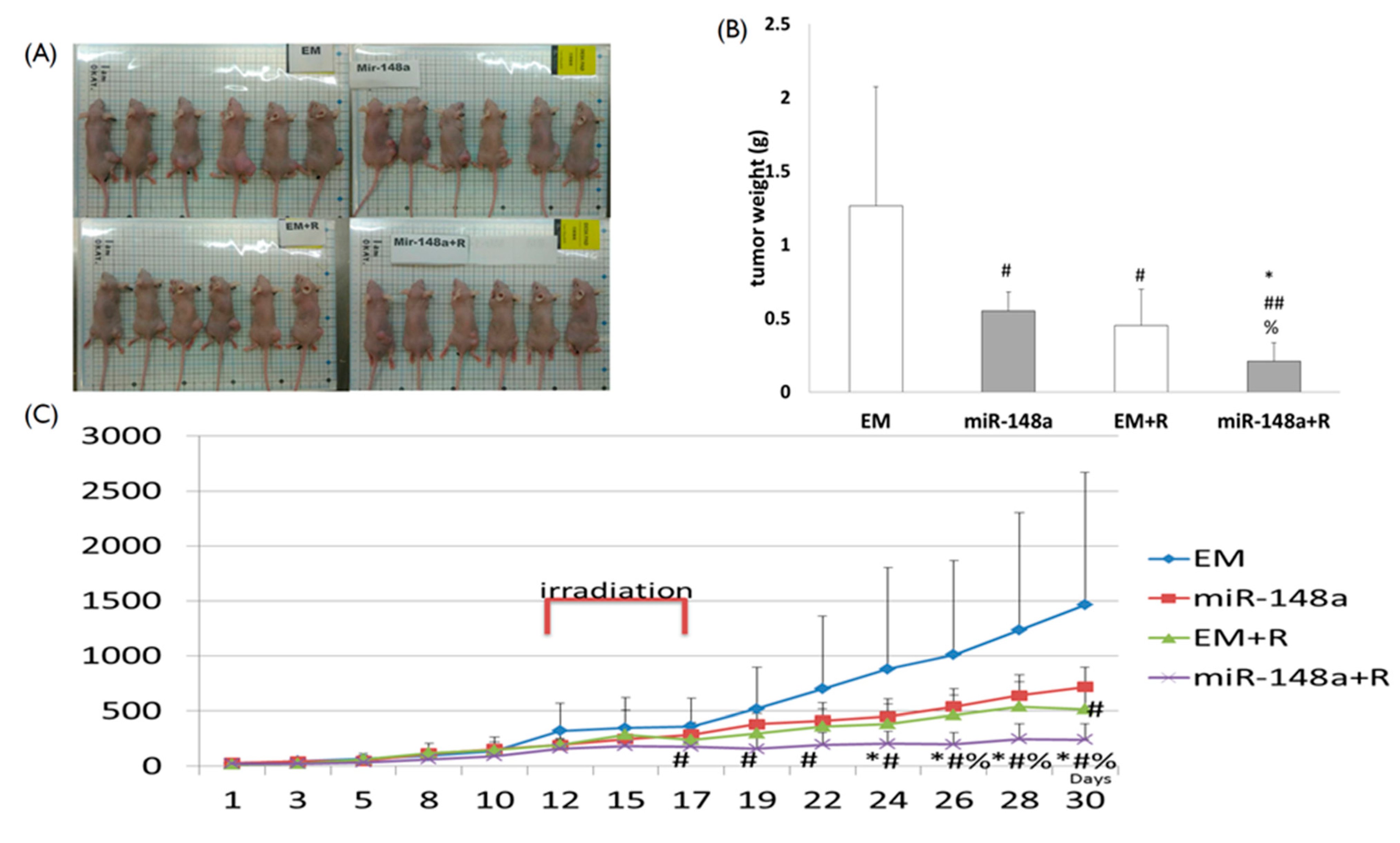
3.7. miRNA-148a Overexpression Enhanced Tumor Response to Radiation in Nude Mice
3.8. miRNA-148a Overexpression Is Associated with a Favorable Prognosis in Patients with LARC Following NACRT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics. CA A Cancer J. Clin. 2017, 67, 177–193. [Google Scholar]
- Sauer, R.; Becker, H.; Hohenberger, W.; Rödel, C.; Wittekind, C.; Fietkau, R.; Martus, P.; Tschmelitsch, J.; Hager, E.; Hess, C.F.; et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N. Engl. J. Med. 2004, 351, 1731–1740. [Google Scholar] [CrossRef] [Green Version]
- van Gijn, W.; Marijnen, C.A.; Nagtegaal, I.D.; Kranenbarg, E.M.; Putter, H.; Wiggers, T.; Rutten, H.J.; Påhlman, L.; Glimelius, B.; van de Velde, C.J. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011, 12, 575–582. [Google Scholar] [CrossRef]
- Huang, C.M.; Huang, M.Y.; Tsai, H.L.; Huang, C.W.; Ma, C.J.; Yeh, Y.S.; Juo, S.H.; Huang, C.J.; Wang, J.Y. An observational study of extending FOLFOX chemotherapy, lengthening the interval between radiotherapy and surgery, and enhancing pathological complete response rates in rectal cancer patients following preoperative chemoradiotherapy. Ther. Adv. Gastroenterol. 2016, 9, 702–712. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.W.; Su, W.C.; Yin, T.C.; Chen, P.J.; Chang, T.K.; Chen, Y.C.; Li, C.C.; Hsieh, Y.C.; Tsai, H.L.; Wang, J.Y. Time interval between the completion of radiotherapy and robotic-assisted surgery among patients with stage I-III rectal cancer undergoing preoperative chemoradiotherapy. PLoS ONE 2020, 15, e0240742. [Google Scholar] [CrossRef]
- Zorcolo, L.; Rosman, A.S.; Restivo, A.; Pisano, M.; Nigri, G.R.; Fancellu, A.; Melis, M. Complete pathologic response after combined modality treatment for rectal cancer and long-term survival: A meta-analysis. Ann. Surg. Oncol. 2012, 19, 2822–2832. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.Q.; Ma, X.J.; Zhang, L.; Cai, S.J.; Peng, J.J. Adjuvant chemotherapy for rectal cancer with complete pathological response (pCR) may not be necessary: A pooled analysis of 5491 patients. Cancer Cell Int. 2019, 19, 127. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The Role of Mir-148a in Cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.H.; Wang, P.P.; Chen, C.T.; Li, D.; Liu, Q.Y.; Lv, L.; Liu, X.; Wang, L.N.; Li, B.X.; Weng, C.Y.; et al. MicroRNA-148b enhances the radiosensitivity of B-cell lymphoma cells by targeting Bcl-w to promote apoptosis. Int. J. Biol. Sci. 2020, 16, 935–946. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015, 13, 252. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.S.; Kim, J.J.; Byun, J.Y.; Kim, I.A. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J. Radiat. Oncol. Biol. Phys. 2010, 76, 5–8. [Google Scholar] [CrossRef]
- Tsai, H.L.; Miao, Z.F.; Chen, Y.T.; Huang, C.W.; Yeh, Y.S.; Yang, I.P.; Wang, J.Y. MiR-148a inhibits early relapsed colorectal cancers and the secretion of VEGF by indirectly targeting HIF-1α under non-hypoxia/hypoxia conditions. J. Cell Mol. Med. 2019, 23, 3572–3582. [Google Scholar] [CrossRef]
- Hao, J.; Magnelli, A.; Godley, A.; Yu, J.S. Use of a Linear Accelerator for Conducting In Vitro Radiobiology Experiments. J. Vis. Exp. 2019, 147. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Qvarnström, O.F.; Simonsson, M.; Eriksson, V.; Turesson, I.; Carlsson, J. GammaH2AX and cleaved PARP-1 as apoptotic markers in irradiated breast cancer BT474 cellular spheroids. Int. J. Oncol. 2009, 35, 41–47. [Google Scholar] [CrossRef]
- Martens, M.H.; Maas, M.; Heijnen, L.A.; Lambregts, D.M.; Leijtens, J.W.; Stassen, L.P.; Breukink, S.O.; Hoff, C.; Belgers, E.J.; Melenhorst, J.; et al. Long-term Outcome of an Organ Preservation Program After Neoadjuvant Treatment for Rectal Cancer. J. Natl. Cancer Inst. 2016, 108, djw171. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.M.; Huang, C.W.; Ma, C.J.; Yeh, Y.S.; Su, W.C.; Chang, T.K.; Tsai, H.L.; Juo, S.H.; Huang, M.Y.; Wang, J.Y. Predictive Value of FOLFOX-Based Regimen, Long Interval, Hemoglobin Levels and Clinical Negative Nodal Status, and Postchemoradiotherapy CEA Levels for Pathological Complete Response in Patients with Locally Advanced Rectal Cancer after Neoadjuvant Chemoradiotherapy. J. Oncol. 2020, 2020, 9437684. [Google Scholar]
- Huang, C.M.; Huang, M.Y.; Huang, C.W.; Tsai, H.L.; Su, W.C.; Chang, W.C.; Wang, J.Y.; Shi, H.Y. Machine learning for predicting pathological complete response in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy. Sci. Rep. 2020, 10, 12555. [Google Scholar] [CrossRef]
- Lopes-Ramos, C.M.; Habr-Gama, A.; Quevedo Bde, S.; Felício, N.M.; Bettoni, F.; Koyama, F.C.; Asprino, P.F.; Galante, P.A.; Gama-Rodrigues, J.; Camargo, A.A.; et al. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med. Genom. 2014, 7, 68. [Google Scholar] [CrossRef] [Green Version]
- Kral, J.; Korenkova, V.; Novosadova, V.; Langerova, L.; Schneiderova, M.; Liska, V.; Levy, M.; Veskrnova, V.; Spicak, J.; Opattova, A.; et al. Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 2018, 39, 1359–1367. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, M.; Peng, F.; Fang, L.; Zhang, Y.; Liang, H.; Zhou, W.; Ao, L.; Guo, Z. The BRCA1/2-directed miRNA signature predicts a good prognosis in ovarian cancer patients with wild-type BRCA1/2. Oncotarget 2015, 6, 2397–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.L.; Yang, I.P.; Huang, C.W.; Ma, C.J.; Kuo, C.H.; Lu, C.Y.; Juo, S.H.; Wang, J.Y. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl. Res. 2013, 162, 258–268. [Google Scholar] [CrossRef]
- Ikediobi, O.N.; Davies, H.; Bignell, G.; Edkins, S.; Stevens, C.; O’Meara, S.; Santarius, T.; Avis, T.; Barthorpe, S.; Brackenbury, L.; et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol. Cancer Ther. 2006, 5, 2606–2612. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.R.; Zhang, Y.; Zhou, H.; Gridley, D.S.; Koch, C.J.; Slater, J.M.; Little, J.B. Overview of radiosensitivity of human tumor cells to low-dose-rate irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2008, 72, 909–917. [Google Scholar] [CrossRef]
- Shen, Y.N.; Bae, I.S.; Park, G.H.; Choi, H.S.; Lee, K.H.; Kim, S.H. MicroRNA-196b enhances the radiosensitivity of SNU-638 gastric cancer cells by targeting RAD23B. Biomed. Pharmacother. 2018, 105, 362–369. [Google Scholar] [CrossRef]
- De Bacco, F.; Luraghi, P.; Medico, E.; Reato, G.; Girolami, F.; Perera, T.; Gabriele, P.; Comoglio, P.M.; Boccaccio, C. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J. Natl. Cancer Inst. 2011, 103, 645–661. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.; Zheng, X.W.; Li, C.; Chen, Q.; Ye, Y.B. KRAS mutation and NF-κB activation indicates tolerance of chemotherapy and poor prognosis in colorectal cancer. Dig. Dis. Sci. 2012, 57, 2325–2333. [Google Scholar] [CrossRef]
- Wang, C.; Shao, S.; Deng, L.; Wang, S.; Zhang, Y. LncRNA SNHG12 regulates the radiosensitivity of cervical cancer through the miR-148a/CDK1 pathway. Cancer Cell Int. 2020, 20, 554. [Google Scholar] [CrossRef]
- Lopez-Bertoni, H.; Kozielski, K.L.; Rui, Y.; Lal, B.; Vaughan, H.; Wilson, D.R.; Mihelson, N.; Eberhart, C.G.; Laterra, J.; Green, J.J. Bioreducible Polymeric Nanoparticles Containing Multiplexed Cancer Stem Cell Regulating miRNAs Inhibit Glioblastoma Growth and Prolong Survival. Nano Lett. 2018, 18, 4086–4094. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.; Trusolino, L.; Comoglio, P.M. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008, 27, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Parizadeh, S.M.; Jafarzadeh-Esfehani, R.; Fazilat-Panah, D.; Hassanian, S.M.; Shahidsales, S.; Khazaei, M.; Parizadeh, S.M.R.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. The potential therapeutic and prognostic impacts of the c-MET/HGF signaling pathway in colorectal cancer. IUBMB Life 2019, 71, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Dai, G.; Wang, J.; Gao, X.; Zhao, Z.; Duan, Z.; Gu, B.; Yang, W.; Wu, J.; Ju, Y.; et al. C-MET inhibition enhances the response of the colorectal cancer cells to irradiation in vitro and in vivo. Oncol. Lett. 2016, 11, 2879–2885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, B.; Xia, S.; Abounader, R.; Laterra, J. Targeting the c-Met pathway potentiates glioblastoma responses to gamma-radiation. Clin. Cancer Res. 2005, 11, 4479–4486. [Google Scholar] [CrossRef] [Green Version]
- Cuneo, K.C.; Mehta, R.K.; Kurapati, H.; Thomas, D.G.; Lawrence, T.S.; Nyati, M.K. Enhancing the Radiation Response in KRAS Mutant Colorectal Cancers Using the c-Met Inhibitor Crizotinib. Transl. Oncol. 2019, 12, 209–216. [Google Scholar] [CrossRef]
- Kawamura, M.; Saigusa, S.; Toiyama, Y.; Tanaka, K.; Okugawa, Y.; Hiro, J.; Uchida, K.; Mohri, Y.; Inoue, Y.; Kusunoki, M. Correlation of MACC1 and MET expression in rectal cancer after neoadjuvant chemoradiotherapy. Anticancer Res. 2012, 32, 1527–1531. [Google Scholar]


| Variables | Numbers (%) |
|---|---|
| Age, median (range, years) | 63 (28–75) |
| Sex (male/female) | 34 (66.7)/17 (33.3) |
| Histology (WD/MD/PD) | 8 (15.7)/40 (78.4)/3 (5.9) |
| Tumor stage (T2/T3/T4) | 8 (15.7)/32 (62.7)/11 (21.6) |
| Nodal stage (N1/2) | 12 (23.5)/16 (31.4)/23 (45.1) |
| Treatment response (pCR/non-pCR) | 11 (21.6)/40 (78.4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-M.; Huang, M.-Y.; Chen, Y.-C.; Chen, P.-J.; Su, W.-C.; Chang, T.-K.; Li, C.-C.; Huang, C.-W.; Tsai, H.-L.; Wang, J.-Y. miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met. Biomedicines 2021, 9, 1371. https://doi.org/10.3390/biomedicines9101371
Huang C-M, Huang M-Y, Chen Y-C, Chen P-J, Su W-C, Chang T-K, Li C-C, Huang C-W, Tsai H-L, Wang J-Y. miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met. Biomedicines. 2021; 9(10):1371. https://doi.org/10.3390/biomedicines9101371
Chicago/Turabian StyleHuang, Chun-Ming, Ming-Yii Huang, Yen-Cheng Chen, Po-Jung Chen, Wei-Chih Su, Tsung-Kun Chang, Ching-Chun Li, Ching-Wen Huang, Hsiang-Lin Tsai, and Jaw-Yuan Wang. 2021. "miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met" Biomedicines 9, no. 10: 1371. https://doi.org/10.3390/biomedicines9101371
APA StyleHuang, C.-M., Huang, M.-Y., Chen, Y.-C., Chen, P.-J., Su, W.-C., Chang, T.-K., Li, C.-C., Huang, C.-W., Tsai, H.-L., & Wang, J.-Y. (2021). miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met. Biomedicines, 9(10), 1371. https://doi.org/10.3390/biomedicines9101371







