Novel Mutations Detection with Next-Generation Sequencing and Its Association with Clinical Outcome in Unilateral Primary Aldosteronism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Mutation Analysis
2.3. Bioinformatics Analysis
2.4. Pathogenicity Criteria for Filtered Variants
2.5. Clinical Parameters
2.6. Clinical and Biochemical Success after Unilateral Adrenalectomy
2.7. Tissue Immunohistochemistry
2.8. Statistical Analysis
3. Results
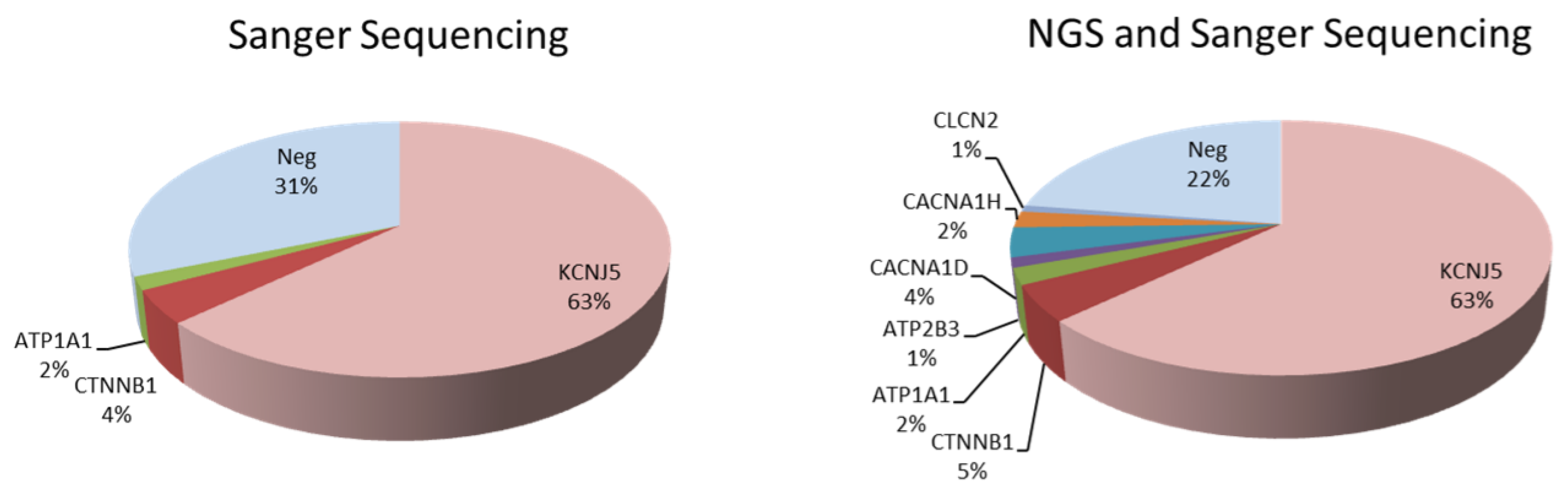
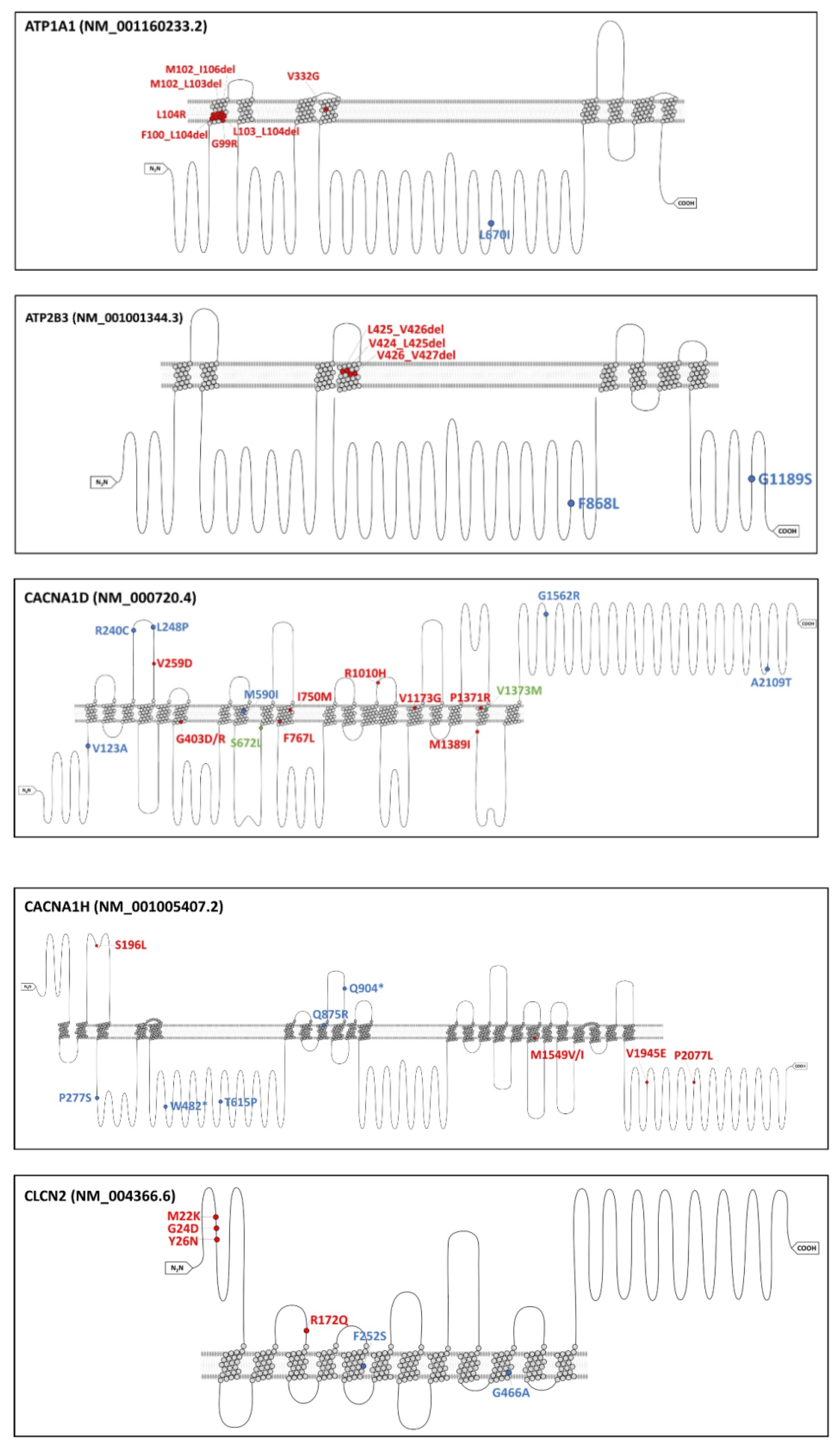
3.1. Identification of Somatic Mutations in uPA
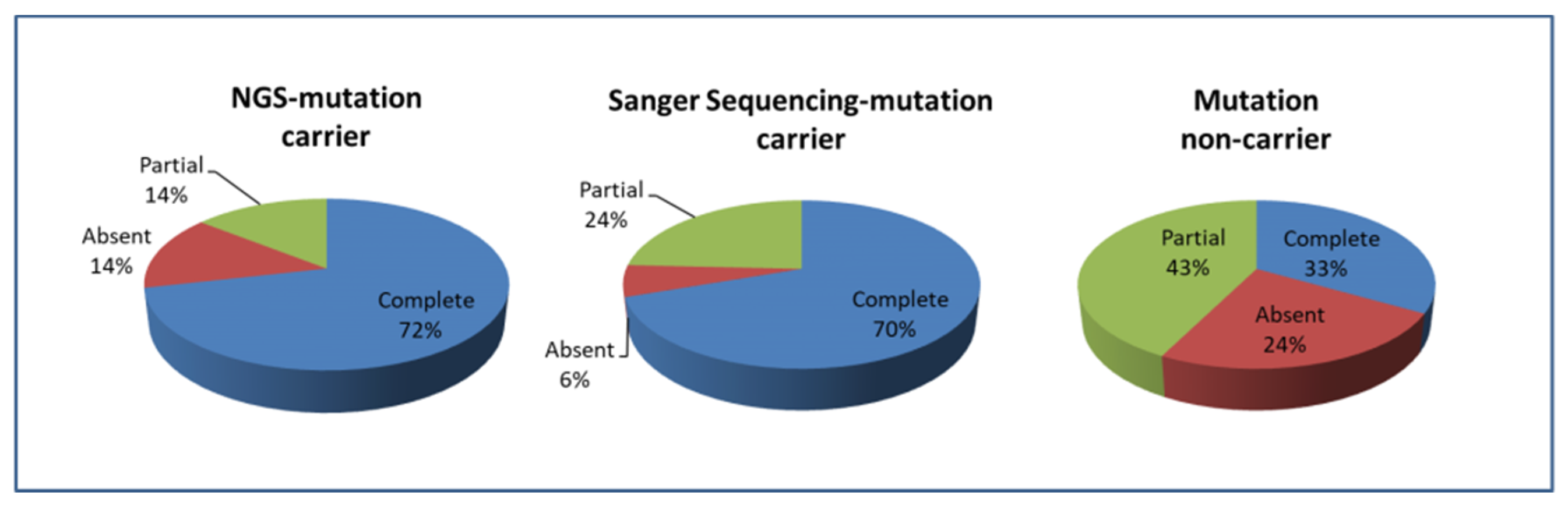
3.2. Phenotypic Characteristics of uPA Patients with Negative Sanger Sequencing Results
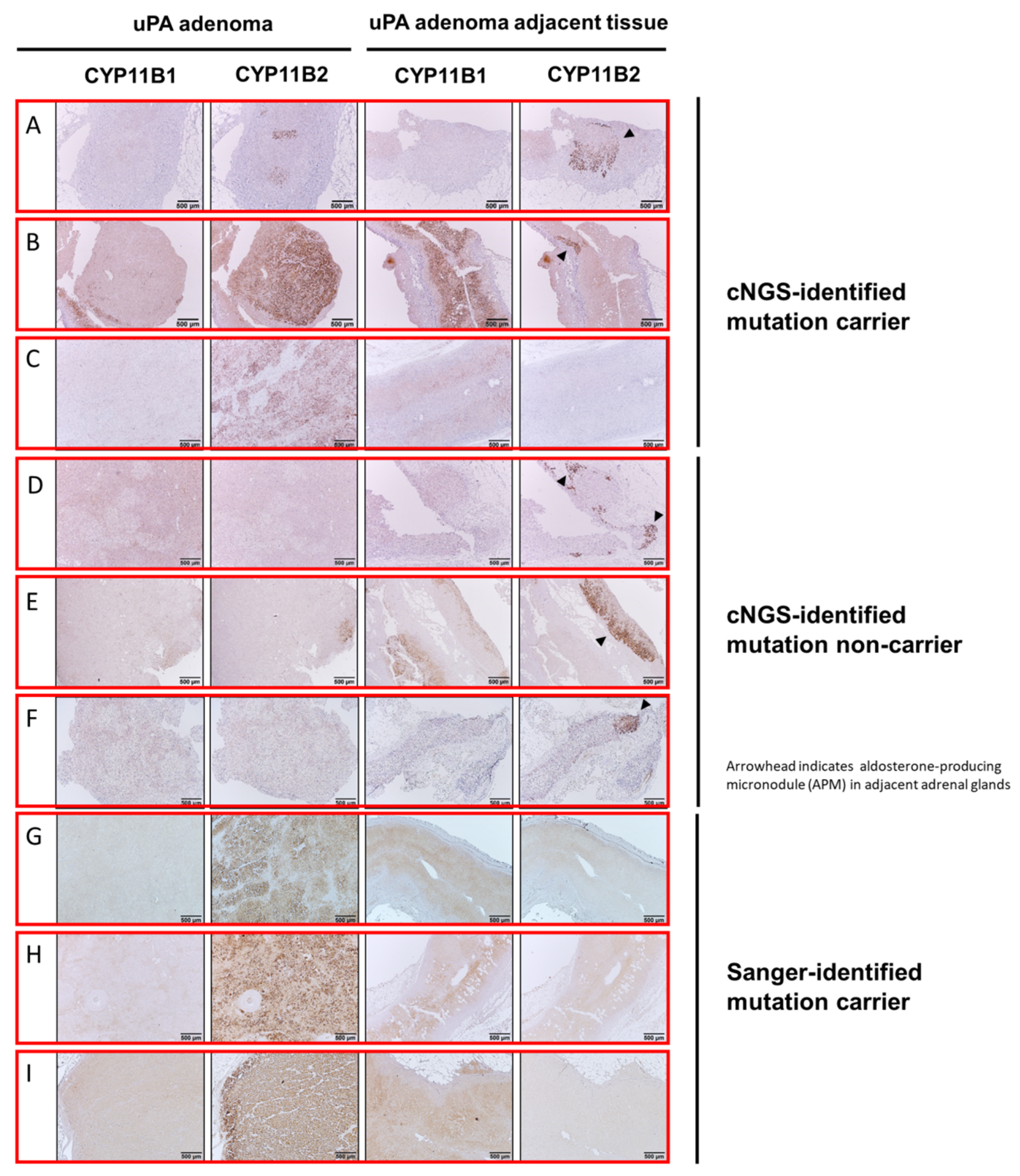
3.3. Histopathologic Characteristics of Adrenal Tumoral Tissues of uAPA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, W.F., Jr. Minireview: Primary aldosteronism—Changing concepts in diagnosis and treatment. Endocrinology 2003, 144, 2208–2213. [Google Scholar] [CrossRef]
- Fagugli, R.M.; Taglioni, C. Changes in the perceived epidemiology of primary hyperaldosteronism. Int. J. Hypertens. 2011, 2011, 162804. [Google Scholar] [CrossRef] [Green Version]
- Choi, M.; Scholl, U.I.; Yue, P.; Bjorklund, P.; Zhao, B.; Nelson-Williams, C.; Ji, W.; Cho, Y.; Patel, A.; Men, C.J.; et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 2011, 331, 768–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholl, U.I.; Goh, G.; Stolting, G.; de Oliveira, R.C.; Choi, M.; Overton, J.D.; Fonseca, A.L.; Korah, R.; Starker, L.F.; Kunstman, J.W.; et al. Somatic and germline cacna1d calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat. Genet. 2013, 45, 1050–1054. [Google Scholar] [CrossRef]
- Azizan, E.A.; Poulsen, H.; Tuluc, P.; Zhou, J.; Clausen, M.V.; Lieb, A.; Maniero, C.; Garg, S.; Bochukova, E.G.; Zhao, W.; et al. Somatic mutations in atp1a1 and cacna1d underlie a common subtype of adrenal hypertension. Nat. Genet. 2013, 45, 1055–1060. [Google Scholar] [CrossRef]
- Beuschlein, F.; Boulkroun, S.; Osswald, A.; Wieland, T.; Nielsen, H.N.; Lichtenauer, U.D.; Penton, D.; Schack, V.R.; Amar, L.; Fischer, E.; et al. Somatic mutations in atp1a1 and atp2b3 lead to aldosterone-producing adenomas and secondary hypertension. Nat. Genet. 2013, 45, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Akerstrom, T.; Maharjan, R.; Sven Willenberg, H.; Cupisti, K.; Ip, J.; Moser, A.; Stalberg, P.; Robinson, B.; Alexander Iwen, K.; Dralle, H.; et al. Activating mutations in ctnnb1 in aldosterone producing adenomas. Sci. Rep. 2016, 6, 19546. [Google Scholar] [CrossRef] [Green Version]
- Scholl, U.I.; Healy, J.M.; Thiel, A.; Fonseca, A.L.; Brown, T.C.; Kunstman, J.W.; Horne, M.J.; Dietrich, D.; Riemer, J.; Kucukkoylu, S.; et al. Novel somatic mutations in primary hyperaldosteronism are related to the clinical, radiological and pathological phenotype. Clin. Endocrinol. 2015, 83, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Scholl, U.I.; Stolting, G.; Schewe, J.; Thiel, A.; Tan, H.; Nelson-Williams, C.; Vichot, A.A.; Jin, S.C.; Loring, E.; Untiet, V.; et al. Clcn2 chloride channel mutations in familial hyperaldosteronism type II. Nat. Genet. 2018, 50, 349–354. [Google Scholar] [CrossRef]
- Dutta, R.K.; Arnesen, T.; Heie, A.; Walz, M.; Alesina, P.; Soderkvist, P.; Gimm, O. A somatic mutation in clcn2 identified in a sporadic aldosterone-producing adenoma. Eur. J. Endocrinol. 2019, 181, K37–K41. [Google Scholar] [CrossRef]
- Nanba, K.; Blinder, A.R.; Rege, J.; Hattangady, N.G.; Else, T.; Liu, C.J.; Tomlins, S.A.; Vats, P.; Kumar-Sinha, C.; Giordano, T.J.; et al. Somatic cacna1h mutation as a cause of aldosterone-producing adenoma. Hypertension 2020, 75, 645–649. [Google Scholar] [CrossRef]
- Fernandes-Rosa, F.L.; Williams, T.A.; Riester, A.; Steichen, O.; Beuschlein, F.; Boulkroun, S.; Strom, T.M.; Monticone, S.; Amar, L.; Meatchi, T.; et al. Genetic spectrum and clinical correlates of somatic mutations in aldosterone-producing adenoma. Hypertension 2014, 64, 354–361. [Google Scholar] [CrossRef]
- Zheng, F.F.; Zhu, L.M.; Nie, A.F.; Li, X.Y.; Lin, J.R.; Zhang, K.; Chen, J.; Zhou, W.L.; Shen, Z.J.; Zhu, Y.C.; et al. Clinical characteristics of somatic mutations in chinese patients with aldosterone-producing adenoma. Hypertension 2015, 65, 622–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Sousa, K.; Boulkroun, S.; Baron, S.; Nanba, K.; Wack, M.; Rainey, W.E.; Rocha, A.; Giscos-Douriez, I.; Meatchi, T.; Amar, L.; et al. Genetic, cellular, and molecular heterogeneity in adrenals with aldosterone-producing adenoma. Hypertension 2020, 75, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Nanba, K.; Omata, K.; Gomez-Sanchez, C.E.; Stratakis, C.A.; Demidowich, A.P.; Suzuki, M.; Thompson, L.D.R.; Cohen, D.L.; Luther, J.M.; Gellert, L.; et al. Genetic characteristics of aldosterone-producing adenomas in blacks. Hypertension 2019, 73, 885–892. [Google Scholar] [CrossRef]
- Wu, C.H.; Yang, Y.W.; Hu, Y.H.; Tsai, Y.C.; Kuo, K.L.; Lin, Y.H.; Hung, S.C.; Wu, V.C.; Wu, K.D.; Taiwan Primary Aldosteronism Investigation Study Group. Comparison of 24-h urinary aldosterone level and random urinary aldosterone-to-creatinine ratio in the diagnosis of primary aldosteronism. PLoS ONE 2013, 8, e67417. [Google Scholar] [CrossRef] [Green Version]
- Wu, V.C.; Hu, Y.H.; Wu, C.H.; Kao, C.C.; Wang, C.Y.; Yang, W.S.; Lee, H.H.; Chang, Y.S.; Lin, Y.H.; Wang, S.M.; et al. Administrative data on diagnosis and mineralocorticoid receptor antagonist prescription identified patients with primary aldosteronism in taiwan. J. Clin. Epidemiol. 2014, 67, 1139–1149. [Google Scholar] [CrossRef]
- Wu, V.C.; Hu, Y.H.; Er, L.K.; Yen, R.F.; Chang, C.H.; Chang, Y.L.; Lu, C.C.; Chang, C.C.; Lin, J.H.; Lin, Y.H.; et al. Case detection and diagnosis of primary aldosteronism—The consensus of Taiwan society of aldosteronism. J. Formos. Med. Assoc. 2017, 116, 993–1005. [Google Scholar] [CrossRef]
- Yen, R.F.; Wu, V.C.; Liu, K.L.; Cheng, M.F.; Wu, Y.W.; Chueh, S.C.; Lin, W.C.; Wu, K.D.; Tzen, K.Y.; Lu, C.C. 131i-6beta-iodomethyl-19-norcholesterol spect/ct for primary aldosteronism patients with inconclusive adrenal venous sampling and ct results. J. Nucl. Med. 2009, 50, 1631–1637. [Google Scholar] [CrossRef] [Green Version]
- Hwang, D.Y.; Dworschak, G.C.; Kohl, S.; Saisawat, P.; Vivante, A.; Hilger, A.C.; Reutter, H.M.; Soliman, N.A.; Bogdanovic, R.; Kehinde, E.O.; et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014, 85, 1429–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, C.A.; Petrovski, S.; Oliver, K.L.; Berkovic, S.F. Exactly zero or once: A clinically helpful guide to assessing genetic variants in mild epilepsies. Neurol. Genet. 2017, 3, e163. [Google Scholar] [CrossRef] [Green Version]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Ng, P.C.; Henikoff, S. Sift: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, E.; Schewe, J.; Scholl, U.I. Genetic causes of primary aldosteronism. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wu, V.C.; Lo, S.C.; Chen, Y.L.; Huang, P.H.; Tsai, C.T.; Liang, C.J.; Kuo, C.C.; Kuo, Y.S.; Lee, B.C.; Wu, E.L.; et al. Endothelial progenitor cells in primary aldosteronism: A biomarker of severity for aldosterone vasculopathy and prognosis. J. Clin. Endocrinol. Metab. 2011, 96, 3175–3183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.A.; Lenders, J.W.M.; Mulatero, P.; Burrello, J.; Rottenkolber, M.; Adolf, C.; Satoh, F.; Amar, L.; Quinkler, M.; Deinum, J.; et al. Outcomes after adrenalectomy for unilateral primary aldosteronism: An international consensus on outcome measures and analysis of remission rates in an international cohort. Lancet Diabetes Endocrinol. 2017, 5, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Sanchez, C.E.; Qi, X.; Velarde-Miranda, C.; Plonczynski, M.W.; Parker, C.R.; Rainey, W.; Satoh, F.; Maekawa, T.; Nakamura, Y.; Sasano, H.; et al. Development of monoclonal antibodies against human cyp11b1 and cyp11b2. Mol. Cell Endocrinol. 2014, 383, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Williams, T.A.; Gomez-Sanchez, C.E.; Rainey, W.E.; Giordano, T.J.; Lam, A.K.; Marker, A.; Mete, O.; Yamazaki, Y.; Zerbini, M.C.N.; Beuschlein, F.; et al. International histopathology consensus for unilateral primary aldosteronism. J. Clin. Endocrinol. Metab. 2021, 106, 42–54. [Google Scholar] [CrossRef]
- Nanba, K.; Yamazaki, Y.; Bick, N.; Onodera, K.; Tezuka, Y.; Omata, K.; Ono, Y.; Blinder, A.R.; Tomlins, S.A.; Rainey, W.E.; et al. Prevalence of somatic mutations in aldosterone-producing adenomas in japanese patients. J. Clin. Endocrinol. Metab. 2020, 105, e4066–e4073. [Google Scholar] [CrossRef] [PubMed]
- Vilela, L.A.P.; Rassi-Cruz, M.; Guimaraes, A.G.; Moises, C.C.S.; Freitas, T.C.; Alencar, N.P.; Petenuci, J.; Goldbaum, T.S.; Maciel, A.A.W.; Pereira, M.A.A.; et al. Kcnj5 somatic mutation is a predictor of hypertension remission after adrenalectomy for unilateral primary aldosteronism. J. Clin. Endocrinol. Metab. 2019, 104, 4695–4702. [Google Scholar] [CrossRef]
- Ip, J.C.; Pang, T.C.; Pon, C.K.; Zhao, J.T.; Sywak, M.S.; Gill, A.J.; Soon, P.S.; Sidhu, S.B. Mutations in kcnj5 determines presentation and likelihood of cure in primary hyperaldosteronism. ANZ J. Surg. 2015, 85, 279–283. [Google Scholar] [CrossRef]
- Okamura, T.; Nakajima, Y.; Katano-Toki, A.; Horiguchi, K.; Matsumoto, S.; Yoshino, S.; Yamada, E.; Tomaru, T.; Ishii, S.; Saito, T.; et al. Characteristics of japanese aldosterone-producing adenomas with kcnj5 mutations. Endocr. J. 2017, 64, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gomez-Sanchez, C.E.; Jaquin, D.; Aristizabal Prada, E.T.; Meyer, L.S.; Knosel, T.; Schneider, H.; Beuschlein, F.; Reincke, M.; Williams, T.A. Primary aldosteronism: Kcnj5 mutations and adrenocortical cell growth. Hypertension 2019, 74, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, C.E.; Gomez-Sanchez, E.P. Mutations of the potassium channel kcnj5 causing aldosterone-producing adenomas: One or two hits? Hypertension 2012, 59, 196–197. [Google Scholar] [CrossRef] [PubMed]
- Nanba, K.; Chen, A.X.; Omata, K.; Vinco, M.; Giordano, T.J.; Else, T.; Hammer, G.D.; Tomlins, S.A.; Rainey, W.E. Molecular heterogeneity in aldosterone-producing adenomas. J. Clin. Endocrinol. Metab. 2016, 101, 999–1007. [Google Scholar] [CrossRef]
- Jamuar, S.S.; Lam, A.T.; Kircher, M.; D’Gama, A.M.; Wang, J.; Barry, B.J.; Zhang, X.; Hill, R.S.; Partlow, J.N.; Rozzo, A.; et al. Somatic mutations in cerebral cortical malformations. N. Engl. J. Med. 2014, 371, 733–743. [Google Scholar] [CrossRef] [Green Version]
- Rivas, M.A.; Beaudoin, M.; Gardet, A.; Stevens, C.; Sharma, Y.; Zhang, C.K.; Boucher, G.; Ripke, S.; Ellinghaus, D.; Burtt, N.; et al. Deep resequencing of gwas loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011, 43, 1066–1073. [Google Scholar] [CrossRef] [Green Version]
- Konig, K.; Peifer, M.; Fassunke, J.; Ihle, M.A.; Kunstlinger, H.; Heydt, C.; Stamm, K.; Ueckeroth, F.; Vollbrecht, C.; Bos, M.; et al. Implementation of amplicon parallel sequencing leads to improvement of diagnosis and therapy of lung cancer patients. J. Thorac. Oncol. 2015, 10, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Nanba, K.; Omata, K.; Else, T.; Beck, P.C.C.; Nanba, A.T.; Turcu, A.F.; Miller, B.S.; Giordano, T.J.; Tomlins, S.A.; Rainey, W.E. Targeted molecular characterization of aldosterone-producing adenomas in white americans. J. Clin. Endocrinol. Metab. 2018, 103, 3869–3876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| No. | NGS-Identified Mutated Gene | Chr | Ref | Alt | Amino Acid Change | VAF | CADD Score | M-C | M-C Pred | PP2 Score | PP2_ Pred | MSC-PP2 Score | MSC-PP2 Pred | SIFT Score | SIFT Pred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ATP1A1 | 1 | C | A | Leu670Ile | 5.07 | 25.9 | 17.8 | high | 0.978 | P | 0.239 | high | 0.01 | D |
| 2 | ATP2B3 | X | G | A | Gly1189Ser | 12.56 | 25.7 | 3.313 | high | 0.901 | P | 0.239 | high | 0 | n/a |
| 3 | ATP2B3 | X | G | A | Gly1189Ser | 12.56 | 25.7 | 3.313 | high | 0.901 | P | 0.239 | high | 0 | n/a |
| 4 | ATP2B3 | X | T | C | Phe868Leu | 5.15 | 24.7 | 3.313 | high | 0.415 | benign | 0.239 | high | n/a | n/a |
| 5 | CACNA1D | 3 | G | A | Val1373Met | 5.59 | 33 | 16.36 | high | 1 | P | 0.57 | high | 0 | D |
| 6 | CACNA1D | 3 | G | A | Val1373Met | 20.35 | 33 | 16.36 | high | 1 | P | 0.57 | high | 0 | D |
| 7 | CACNA1D | 3 | C | T | Ser672Leu | 39.75 | 35 | 16.36 | high | 0.992 | P | 0.57 | high | 0 | D |
| 8 | CACNA1D | 3 | C | T | Arg240Cys | 5.58 | 35 | 16.36 | high | 1 | P | 0.57 | high | 0 | D |
| 9 | CACNA1D | 3 | G | A | Met590Ile | 5.56 | 30 | 16.36 | high | 0.917 | P | 0.57 | high | 0 | D |
| 10 | CACNA1D | 3 | T | C | Leu248Pro | 5.14 | 28 | 16.36 | high | 1 | P | 0.57 | high | 0 | D |
| 11 | CACNA1D | 3 | G | A | Ala2109Thr | 8.25 | 28.7 | 16.36 | high | 0.999 | P | 0.57 | high | 0 | D |
| 12 | CACNA1D | 3 | T | C | Val123Ala | 5.45 | 26.4 | 16.36 | high | 0.999 | P | 0.57 | high | 0.02 | D |
| 13 | CACNA1D | 3 | G | A | Gly1562Arg | 5.60 | 34 | 16.36 | high | 0.986 | P | 0.57 | high | 0 | D |
| 14 | CACNA1H | 16 | C | T | Pro277Ser | 48.37 | 24.4 | 0.001 | high | 0.971 | P | 0.001 | high | 0.03 | D |
| 15 | CACNA1H | 16 | A | G | Gln875Arg | 8.03 | 25.9 | 0.001 | high | 0.830 | P | 0.001 | high | 0 | D |
| 16 | CACNA1H | 16 | A | G | Thr615Pro | 10.45 | 25.3 | 0.001 | high | 0.766 | P | 0.001 | high | 0 | D |
| 17 | CACNA1H | 16 | C | T | Gln904Ter | 10.20 | 38 | 0.001 | high | n/a | n/a | 0.001 | n/a | 0 | D |
| 18 | CACNA1H | 16 | G | A | Trp482Ter | 13.54 | 36 | 0.001 | high | n/a | n/a | 0.001 | n/a | 0.17 | T |
| 19 | CLCN2 | 3 | C | G | Gly466Ala | 37.01 | 25.5 | 0.001 | high | 0.489 | P | 0.02 | high | 0 | D |
| 20 | CLCN2 | 3 | A | G | Phe252Ser | 17.33 | 28.3 | 0.001 | high | 0.997 | P | 0.02 | high | 0 | D |
| 21 | CTNNB1 | 3 | C | T | Gln123Ter | 7.87 | 37 | 16.39 | high | n/a | n/a | 0.239 | n/a | 0.01 | D |
| All | cNGS-Identified Mutation Carrier | Cngs-Identified Mutation Non-Carrier | p Value (Mutation Carrier vs. Non-Carrier) | |
|---|---|---|---|---|
| Number | 75 | 21 | 54 | |
| Female (%) | 43 (57.3%) | 14 (66.7%) | 29 (53.7%) | 0.436 |
| Age (years) | 54.5 (10.6) | 53.5 (10.4) | 54.9 (10.7) | 0.603 |
| BMI (kg/m2) | 26.2 (4.0) | 25.3 (3.4) | 26.5 (4.2) | 0.266 |
| sBP (mmHg) | 152.0 (20.8) | 145.0 (19.9) | 154.6 (20.7) | 0.071 |
| dBP (mmHg) | 90.8 (14.5) | 88.3 (17.4) | 91.8 (13.3) | 0.345 |
| PRA (ng/mL per min)† | 0.83 (2.32) | 1.32 (4.07) | 0.64 (1.06) | 0.455 |
| Plasma aldosterone (ng/dL)† | 49.8 (35.2) | 47.1 (29.9) | 50.8 (37.2) | 0.68 |
| Log ARR† | 2.28 (0.83) | 2.33 (0.96) | 2.26 (0.79) | 0.742 |
| Potassium (mmol/L)† | 3.6 (0.6) | 3.7 (0.6) | 3.5 (0.6) | 0.155 |
| Serum creatinine (mg/dL) | 0.93 (0.36) | 0.85 (0.30) | 0.95 (0.38) | 0.279 |
| Cystatin C (mg/L) | 0.84 (0.23) | 0.75 (0.25) | 0.87 (0.30) | 0.246 |
| Microalbuminuria (g/day) | 0.06 (0.12) | 0.07 (0.13) | 0.05 (0.11) | 0.546 |
| Tumor size (cm) | 2.09 (2.57) | 2.28 (2.98) | 2.02 (2.43) | 0.695 |
| baPWV (cm/s) | 1722.0 (329.2) | 1647.5 (256.4) | 1765.2 (361.8) | 0.231 |
| Clinical success (%) | 0.010 | |||
| Complete | 33 (44%) | 15 (71.4%) | 18 (33.3%) | |
| Absent | 16 (21.3%) | 3 (14.3%) | 13 (24.1%) | |
| Partial | 26 (34.7%) | 3 (14.3%) | 23 (42.6%) | |
| Biochemical success (%) | 0.440 | |||
| Complete | 61 (81.3%) | 18 (85.7%) | 43 (79.6%) | |
| Absent | 10 (13.3%) | 3 (14.3%) | 7 (13%) | |
| Partial | 4 (5.3%) | 0 | 4 (7.4%) |
| Model Fitting Criteria | Likelihood Ratio Tests | ||||
|---|---|---|---|---|---|
| AIC of Reduced Model | BIC of Reduced Model | −2 Log Likelihood of Reduced Model | Chi-Square | Sig. | |
| Age(yr) | 165.545 | 216.530 | 121.545 | 1.825 | 0.402 |
| BMI(kg/m2) | 164.742 | 215.727 | 120.742 | 1.022 | 0.600 |
| SBP(mmHg) | 163.969 | 214.954 | 119.969 | 0.249 | 0.883 |
| DBP(mmHg) | 167.660 | 218.645 | 123.660 | 3.940 | 0.139 |
| Aldosterone(ng/dL) † | 167.252 | 218.237 | 123.252 | 3.532 | 0.171 |
| PRA(ng/mL/h) † | 165.395 | 216.380 | 121.395 | 1.675 | 0.433 |
| Potassium(mEq/L) † | 164.553 | 215.538 | 120.553 | 0.833 | 0.659 |
| Cr (mg/dL) | 165.379 | 216.364 | 121.379 | 1.659 | 0.436 |
| Tumor size (cm) | 168.261 | 219.246 | 124.261 | 4.541 | 0.103 |
| cNGS-identified mutation (yes) | 172.646 | 223.631 | 128.646 | 8.926 | 0.012 |
| Sex(Female) | 168.601 | 219.586 | 124.601 | 4.881 | 0.087 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-H.; Peng, K.-Y.; Hwang, D.-Y.; Lin, Y.-H.; Wu, V.-C.; Chueh, J.S. Novel Mutations Detection with Next-Generation Sequencing and Its Association with Clinical Outcome in Unilateral Primary Aldosteronism. Biomedicines 2021, 9, 1167. https://doi.org/10.3390/biomedicines9091167
Wu C-H, Peng K-Y, Hwang D-Y, Lin Y-H, Wu V-C, Chueh JS. Novel Mutations Detection with Next-Generation Sequencing and Its Association with Clinical Outcome in Unilateral Primary Aldosteronism. Biomedicines. 2021; 9(9):1167. https://doi.org/10.3390/biomedicines9091167
Chicago/Turabian StyleWu, Che-Hsiung, Kang-Yung Peng, Daw-Yang Hwang, Yen-Hung Lin, Vin-Cent Wu, and Jeff S. Chueh. 2021. "Novel Mutations Detection with Next-Generation Sequencing and Its Association with Clinical Outcome in Unilateral Primary Aldosteronism" Biomedicines 9, no. 9: 1167. https://doi.org/10.3390/biomedicines9091167
APA StyleWu, C.-H., Peng, K.-Y., Hwang, D.-Y., Lin, Y.-H., Wu, V.-C., & Chueh, J. S. (2021). Novel Mutations Detection with Next-Generation Sequencing and Its Association with Clinical Outcome in Unilateral Primary Aldosteronism. Biomedicines, 9(9), 1167. https://doi.org/10.3390/biomedicines9091167







