Triazole-Modified Nucleic Acids for the Application in Bioorganic and Medicinal Chemistry
Abstract
1. Introduction
1.1. A Brief Introduction to DNA and RNA World
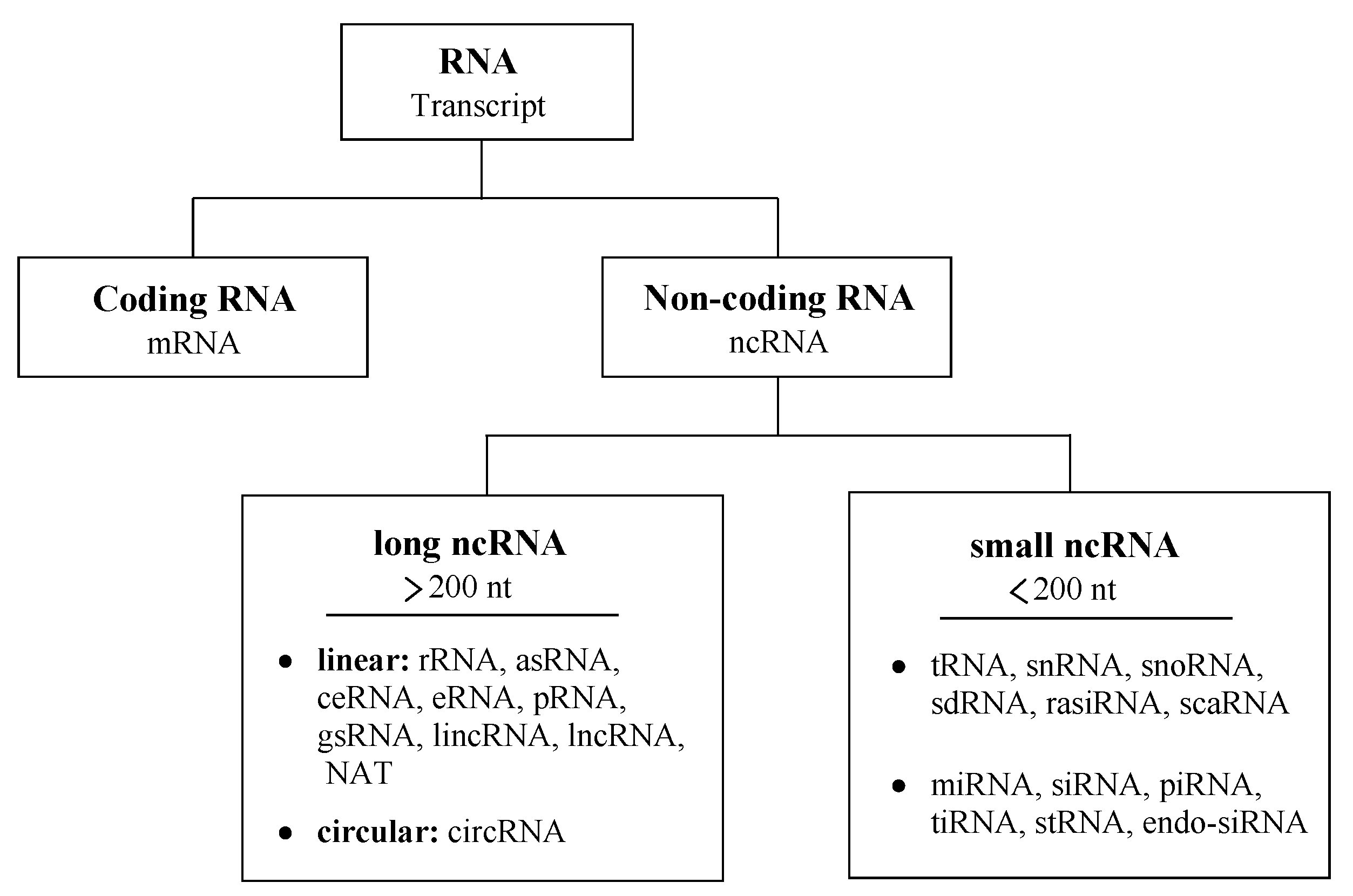
1.2. Non-Coding RNA
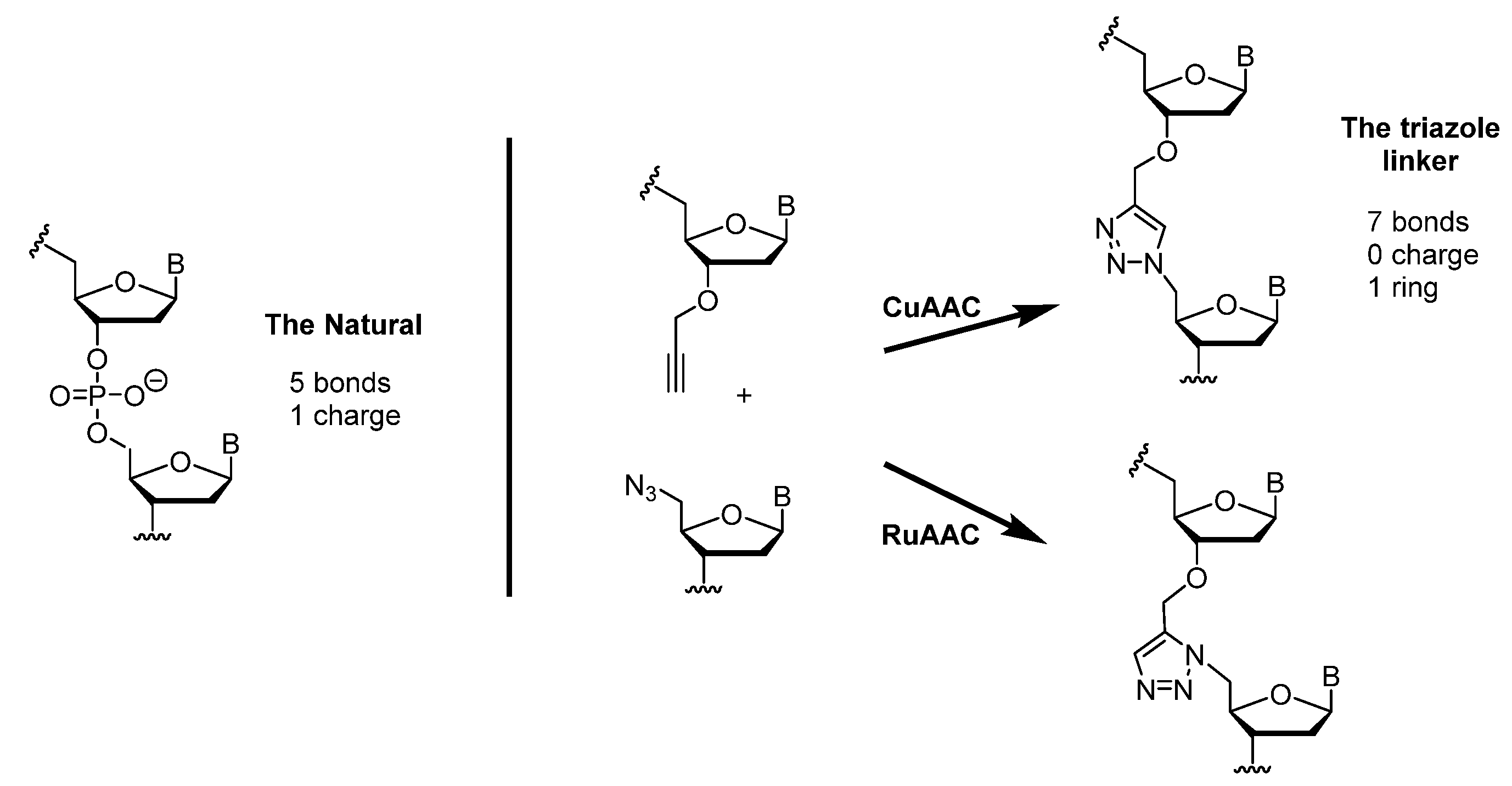
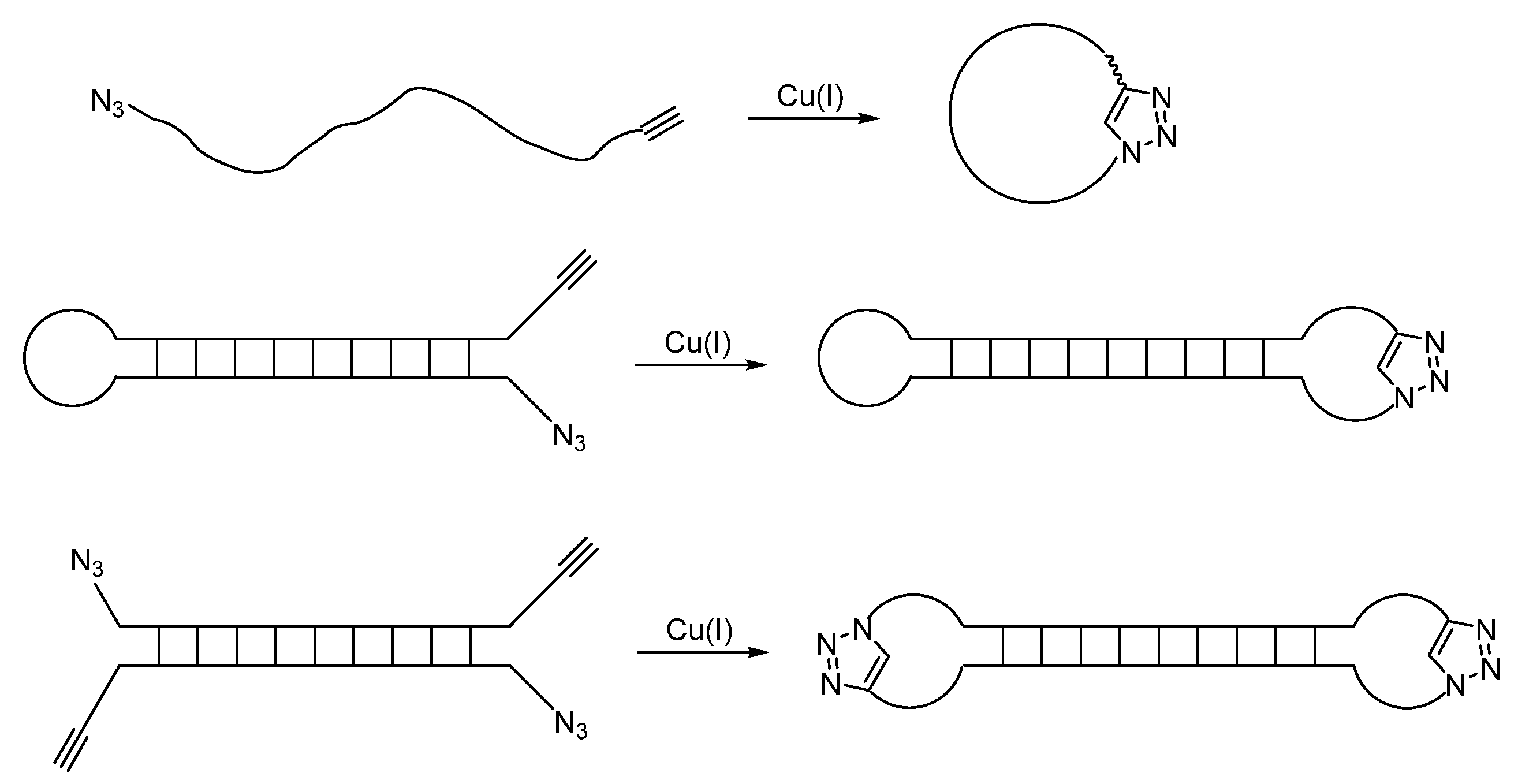
1.3. Click Chemistry
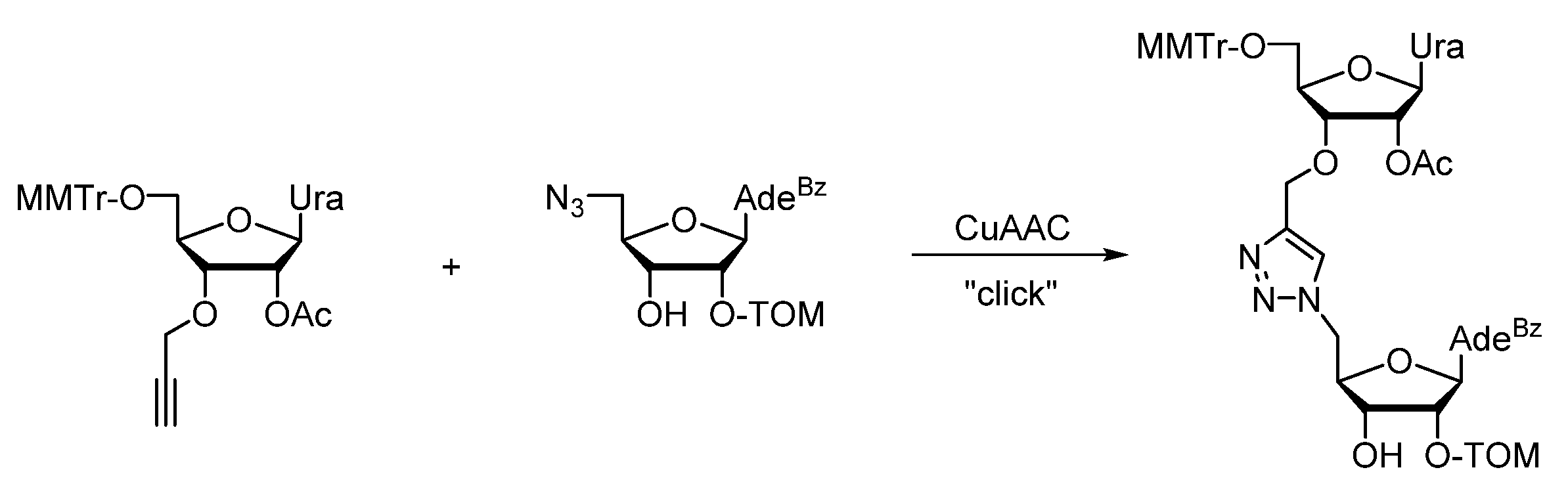
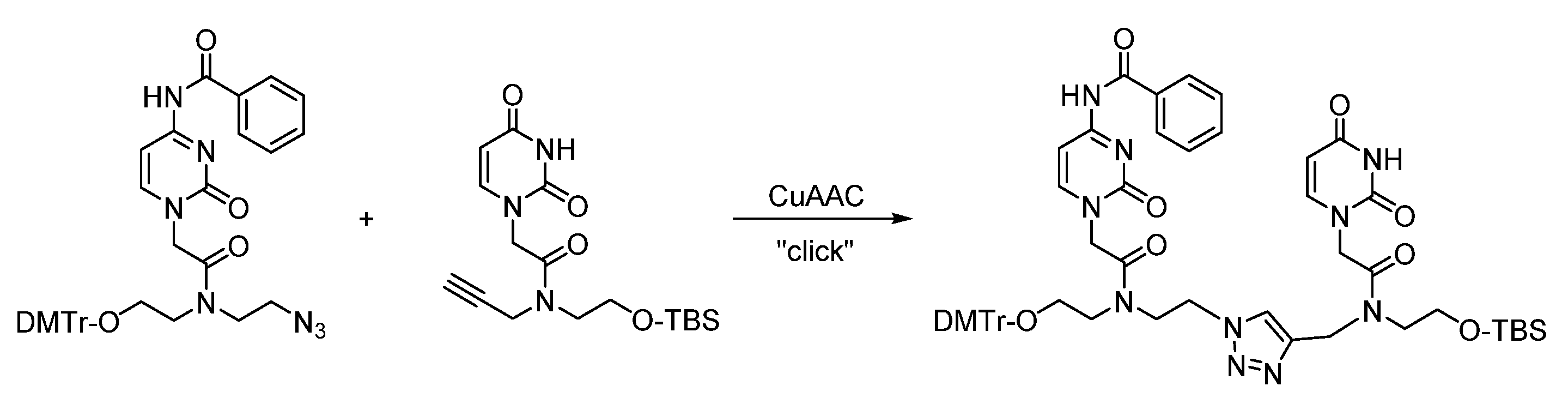
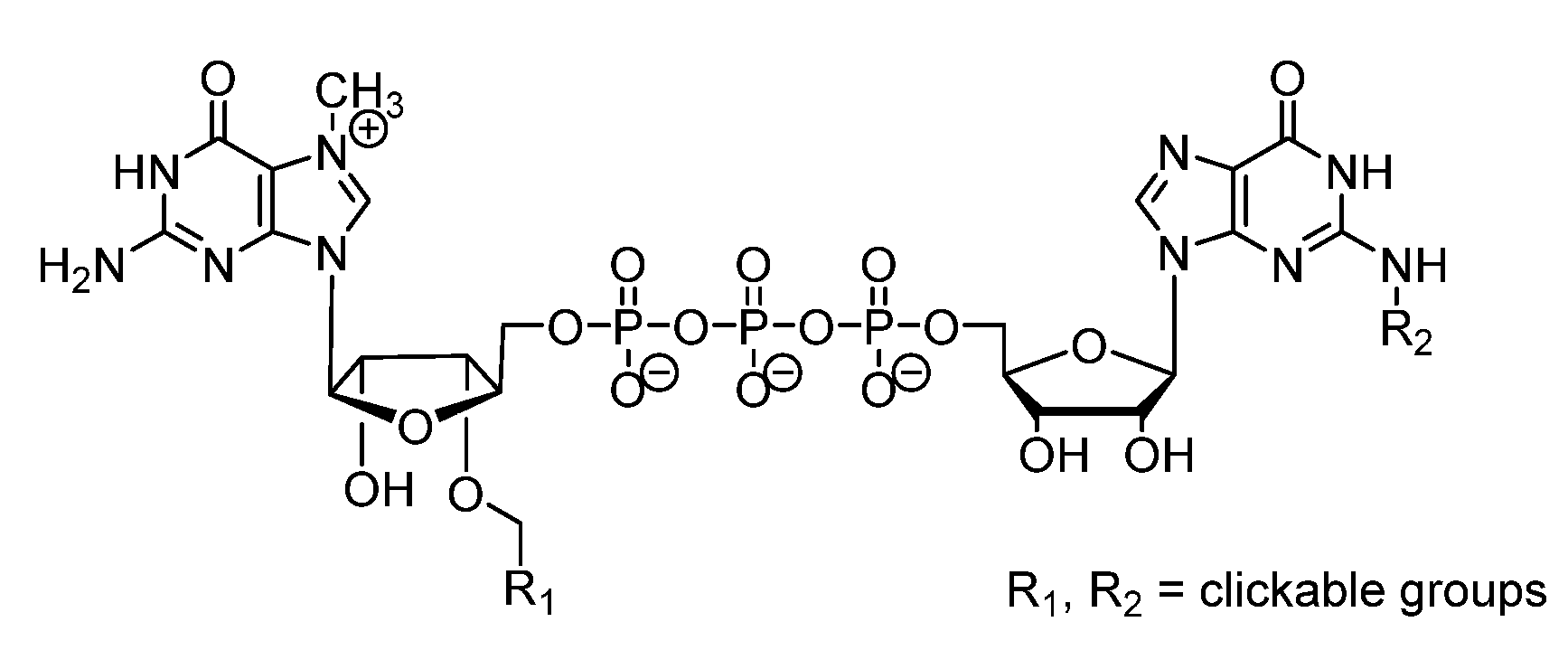
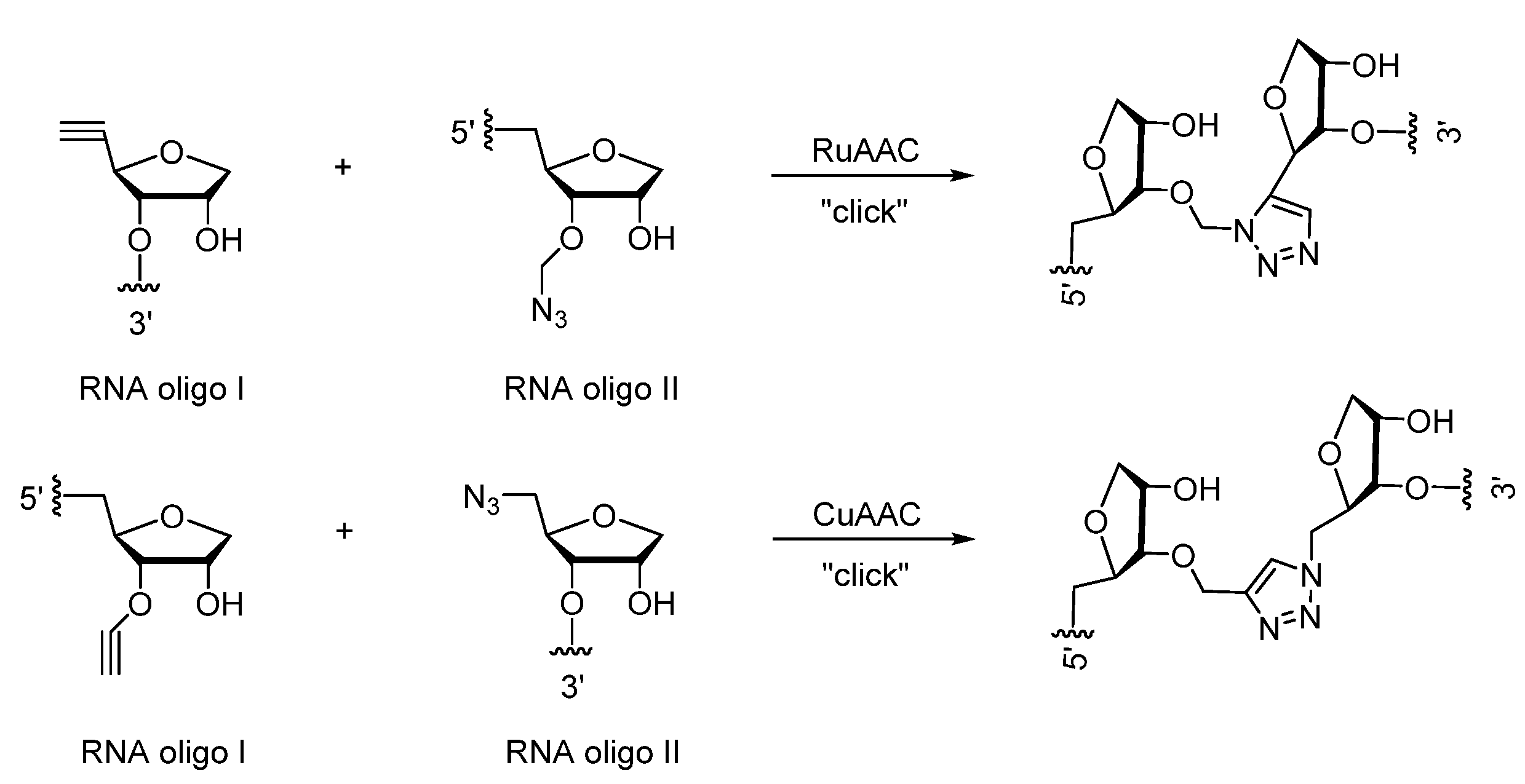
2. Triazole-Modified Oligonucleotides
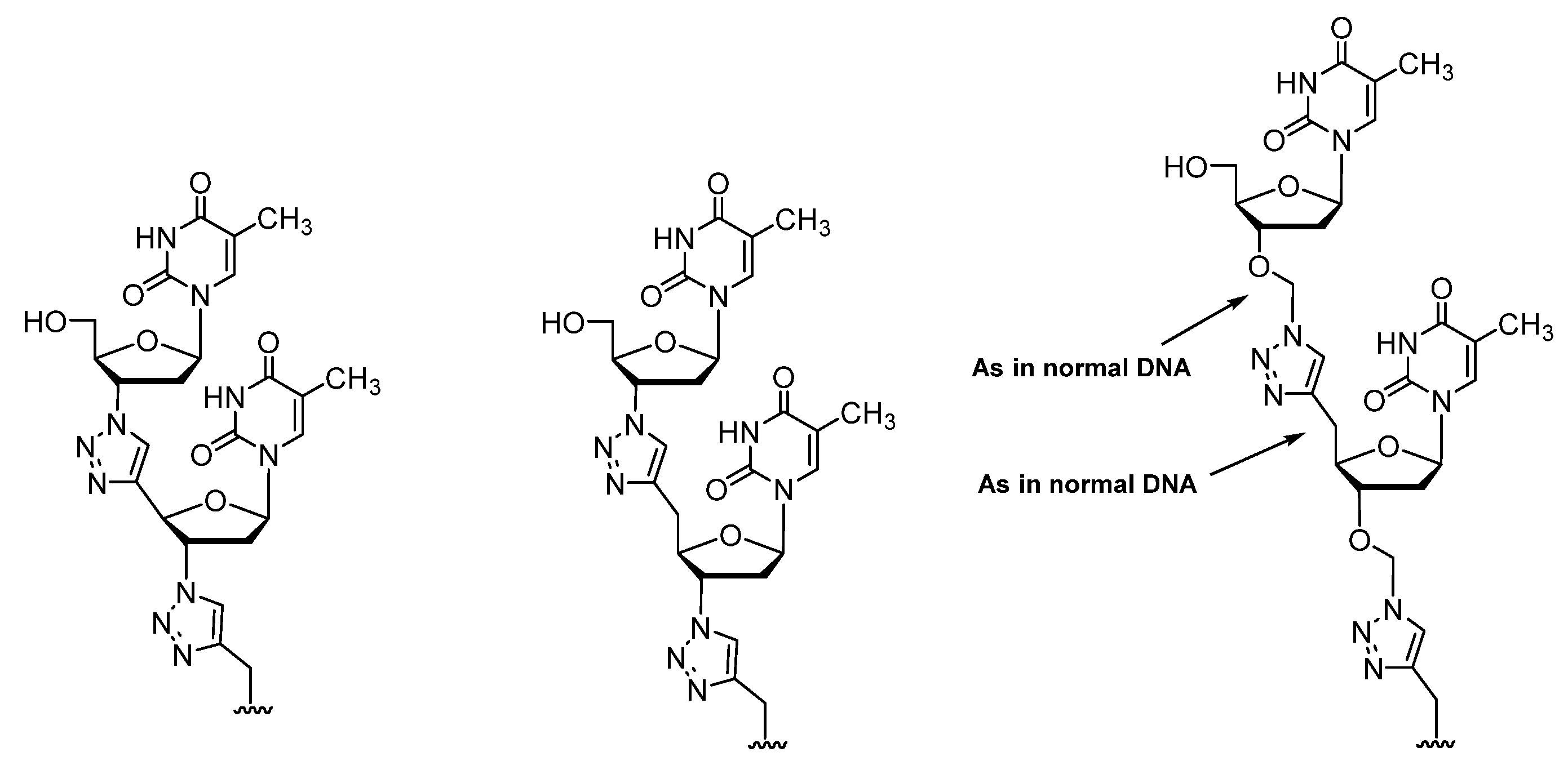
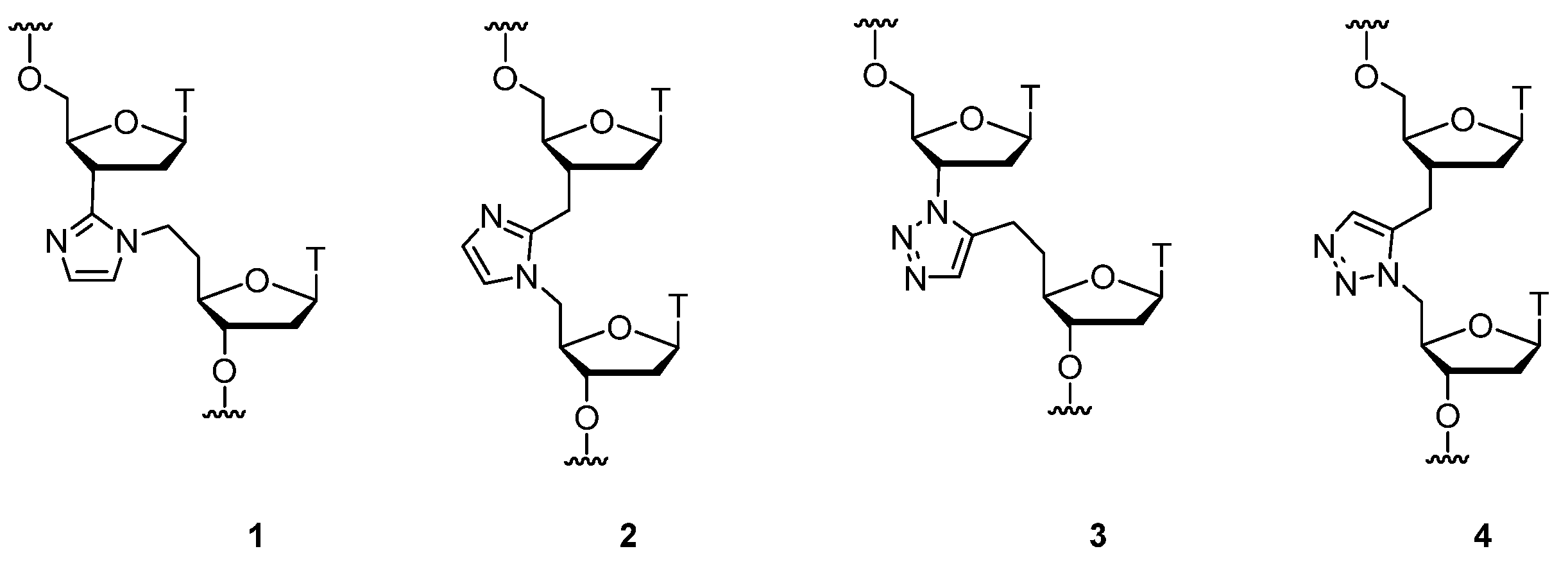
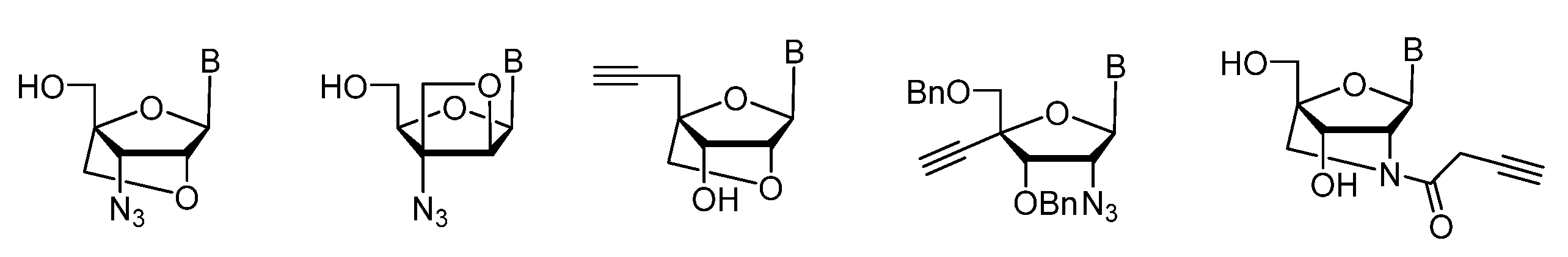
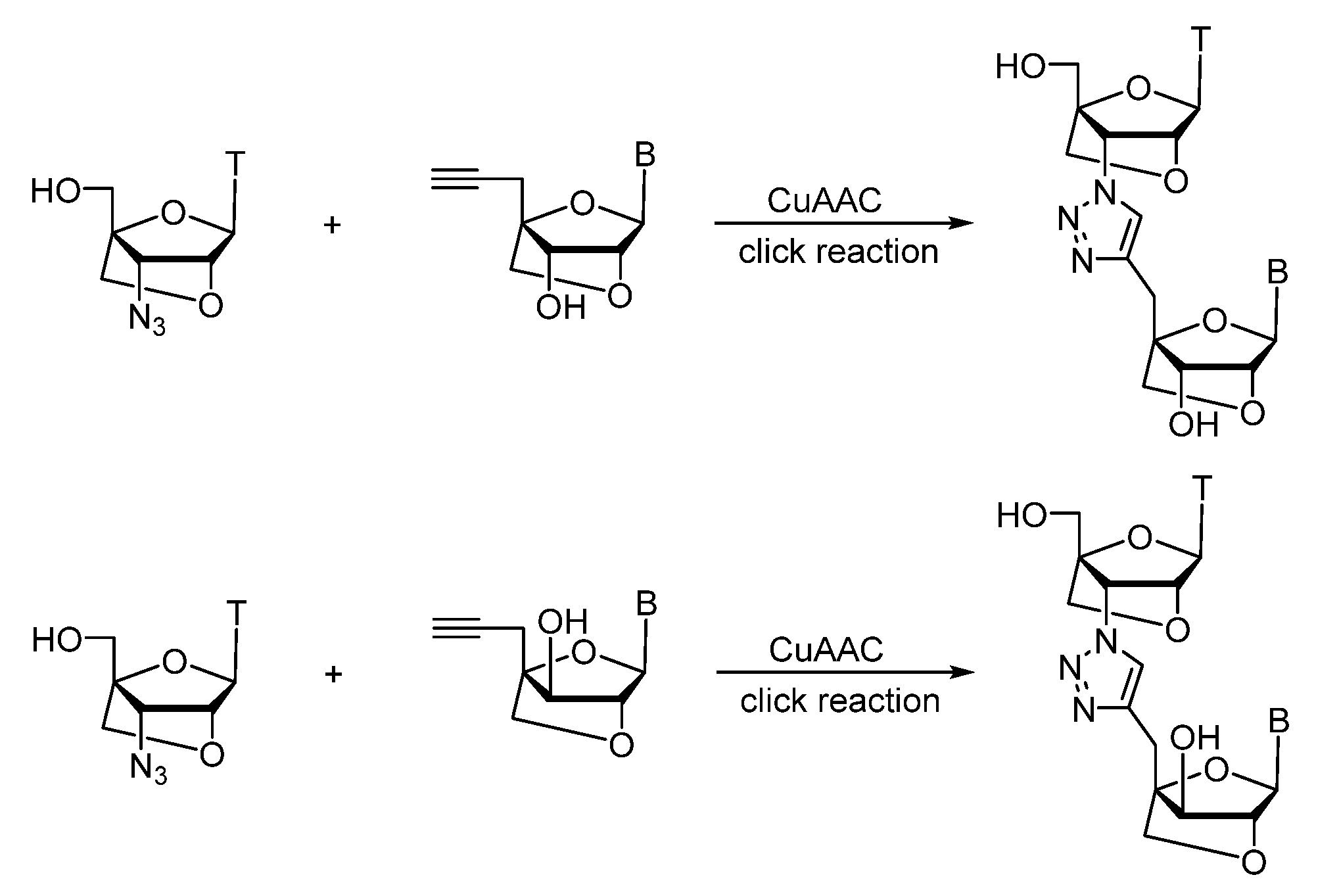
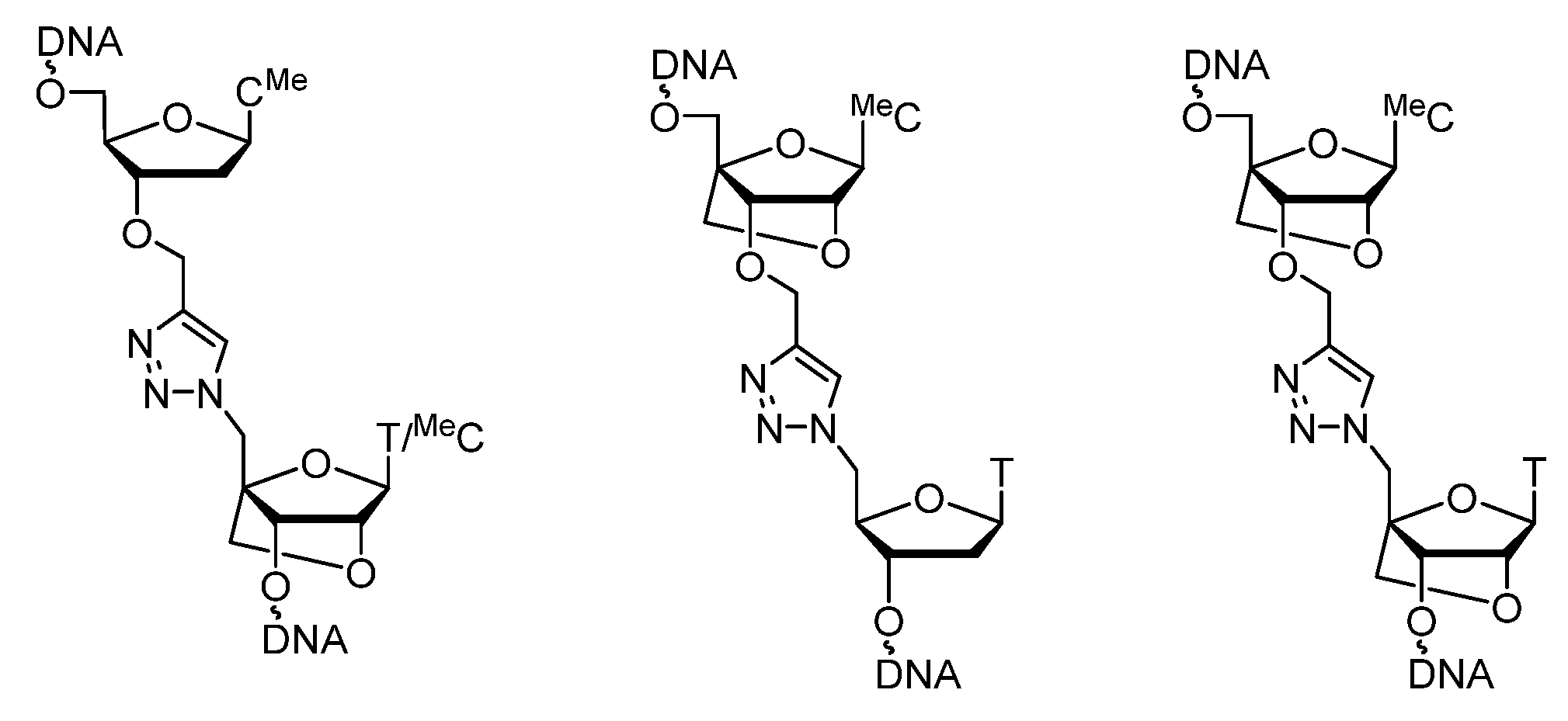
2.1. (TL)DNA Skeleton
2.2. (TL)RNA Skeleton
2.3. (TL)LNA and (TL)BNA Skeleton
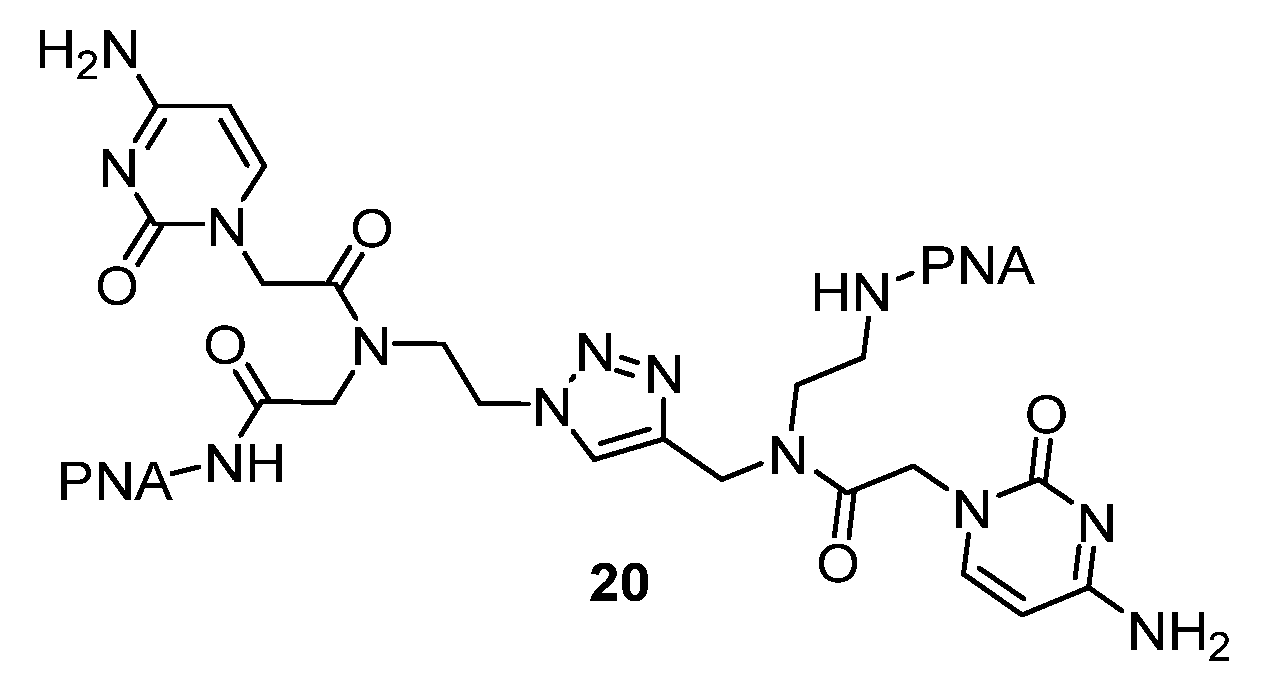
2.4. (TL)PNA Skeleton
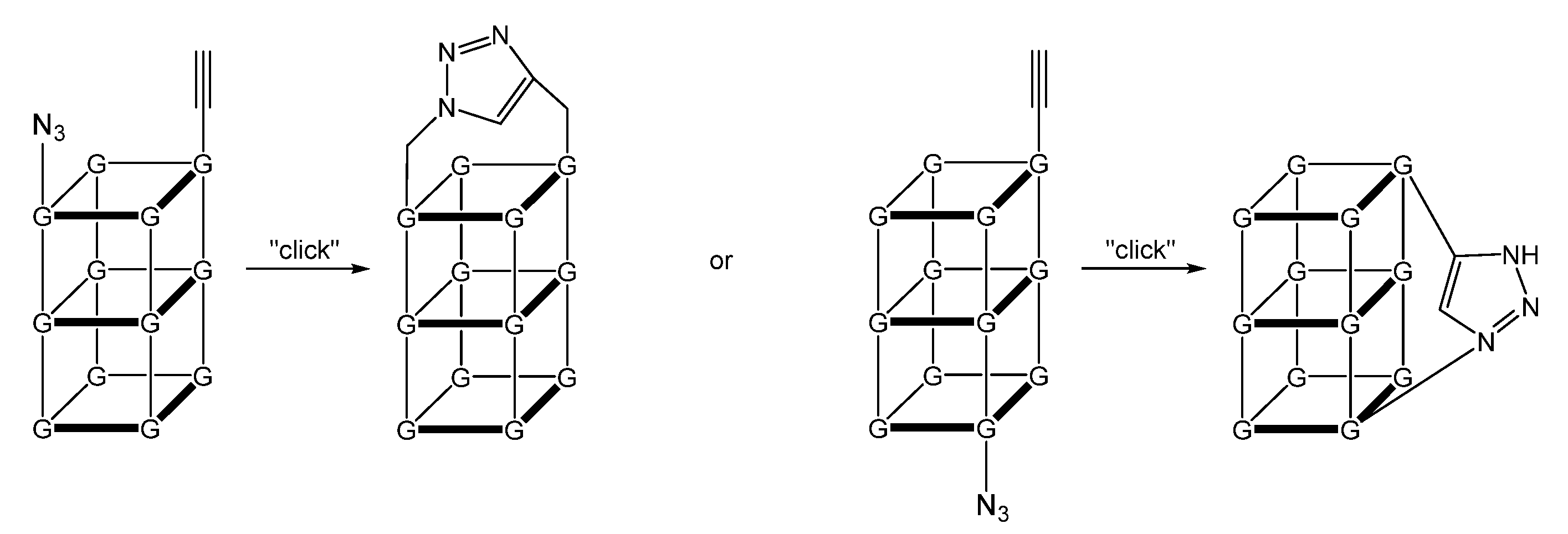
2.5. (TL)G-Quadruplexes
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| A | adenosine |
| Ac | acetyl protecting group |
| ADAR | adenosine deaminases |
| Ago2 | argonaute RISC catalytic component 2 |
| ASO | antisense oligonucleotide |
| asRNA | antisense ribonucleic acid |
| ATP | adenosine triphosphate |
| AZT | 3′-azido-3′-deoxythymidine |
| B | heterocyclic base |
| BCL2 | B-cell lymphoma 2 proteins |
| BNA | bridged nucleic acids |
| Bzl | benzoyl protecting group |
| C | cytidine |
| cat-ELCCA | catalytic enzyme-linked click chemistry assay |
| CD | circular dichroism spectroscopy |
| CeNA | cyclohexene nucleic acids |
| ceRNA | competing endogenous ribonucleic acid |
| circRNA | circular ribonucleic acid |
| cPNA | clickable peptide nucleic acids |
| CRISP-Cas9 | clustered regularly interspaced short palindromic repeats associated protein 9 |
| CuAAC | copper(I)-catalyzed alkyne-azide cycloaddition |
| CuI | copper(I) iodide |
| CuSO4 | copper(II) sulfate |
| DIPEA | N,N-diisopropylethylamine |
| DMF | dimethylformamide |
| DMTr | 4,4′ -dimethoxytrityl protecting group |
| DNA | 2′- deoxyribonucleic acid |
| dsDNA | double-stranded DNA |
| dU | 2′-deoxyuridine |
| dU*TP | alkyne-modified 2′-deoxyuridine triphosphate |
| eIF4E | eukaryotic translation initiation factor 4E |
| endo-siRNA | endogenous siRNA |
| EPR | electron paramagnetic resonance spectroscopy |
| eRNA | enhancer ribonucleic acid |
| EtOH | ethanol |
| FTR | functionality transfer reaction |
| G4 | G-quadruplex |
| gsRNA | germline small ribonucleic acid |
| HCV | hepatitis C virus |
| HIV | human immunodeficiency virus |
| iPr | isopropyl protecting group |
| lincRNA | intervening/intergenic ribonucleic acid |
| LNA | locked nucleic acids |
| lncRNA | long noncoding ribonucleic acid |
| Me | methyl |
| min | minute(s) |
| miRNA | micro ribonucleic acid |
| mRNA | messenger ribonucleic acid |
| MSNT | 1-(2-mesitylenesulfonyl)-3-nitro-1H-1,2,4-triazole or (2,4,6-trimethylphenyl)-(3-nitro-1,2,4-triazol-1-yl) sulfone |
| MW | microwave irradiation |
| NaAsc | sodium ascorbate |
| NAT | natural antisense transcript |
| ncRNA | non-coding ribonucleic acid |
| NMR | nuclear magnetic resonance spectroscopy |
| ODN | oligodeoxynucleotide |
| ON | oligonucleotide |
| PCR | polymerase chain reaction |
| piRNA | piwi-interacting ribonucleic acid |
| PNA | peptide nucleic acids |
| pRNA | promoter ribonucleic acid |
| rasiRNA | repeat associated siRNA |
| RCA | rolling circle amplification reaction |
| RNA | ribonucleic acid |
| RNAi | RNA interference |
| RNAi | RNA interference |
| RNaseP | ribonuclease P |
| rRNA | ribosomal ribonucleic acid |
| rt | room temperature |
| RuAAC | ruthenium(II)-catalyzed alkyne-azide cycloaddition |
| SAM | S-adenosyl-L-methionine |
| scaRNA | small Cajal body-specific ribonucleic acid |
| sdRNA | snoRNA-derived ribonucleic acid |
| SELEX | systematic evolution of ligands by exponential enrichment or in vitro selection, or in vitro evolution |
| sgRNA | single guide RNA |
| siRNA | small interfering ribonucleic acid |
| snoRNA | small nucleolar ribonucleic acid |
| snRNA | small nuclear ribonucleic acid |
| ssDNA | single-stranded DNA |
| stRNA | small temporal ribonucleic acid |
| T | thymidine |
| TBDMS | tert-butyldiphenylsilyl protecting group |
| TBS | tert-butyl(dimethyl)silyl protecting group |
| TBTA | tris(benzyltriazolylmethyl)amine |
| tcDNA | tricyclo-DNA |
| tiRNA | tRNA-derived stress-induced ribonucleic acid |
| (TL)BNA | triazole-modified BNA |
| (TL)DNA | triazole-modified DNA |
| (TL)LNA | triazole-modified LNA |
| (TL)PNA | triazole-modified PNA |
| (TL)quadruplexes | triazole-modified quadruplexes |
| (TL)RNA | triazole-modified RNA |
| Tm | melting temperature |
| TP | 1,2,3-triazolylphosphonate internucleotide linkage |
| tRNA | transfer ribonucleic acid |
| U | uridine |
| UTR | untranslated region of mRNA |
| UV | ultraviolet |
References
- Becker, S.; Schneider, C.; Crisp, A.; Carell, T. Non-canonical nucleosides and chemistry of the emergence of life. Nat. Commun. 2018, 9, 5174. [Google Scholar] [CrossRef] [PubMed]
- Lönnberg, H. Chemistry of Nucleic Acids; De Gruyter: Berlin, Germany, 2020. [Google Scholar]
- Duffy, K.; Arangundy-Franklin, S.; Holliger, P. Modified nucleic acids: Replication, evolution, and next-generation therapeutics. BMC Biol. 2020, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The Genetic Signatures of Noncoding RNAs. PLoS Genet. 2009, 5, 1–12. [Google Scholar] [CrossRef]
- Nie, P.; Bai, Y.; Mei, H. Synthetic Life with Alternative Nucleic Acids as Genetic Materials. Molecules 2020, 25, 3483. [Google Scholar] [CrossRef]
- Barciszewski, J.; Marquez, V.E.; Vasseur, J.-J.; Markiewicz, W.T. Chemical Biology of Nucleic Acids. ACS Chem. Biol. 2015, 10, 1358–1361. [Google Scholar] [CrossRef]
- Benner, S.A.; Karalkar, N.B.; Hoshika, S.; Laos, R.; Shaw, R.W.; Matsuura, M.; Fajardo, D.; Moussatche, P. Alternative Watson-Crick Synthetic Genetic Systems. Cold Spring Harb. Perspect. Biol. 2016, 8, a023770. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef]
- Biondi, E.; Benner, S.A. Artificially Expanded Genetic Information Systems for New Aptamer Technologies. Biomedicines 2018, 6, 53. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The Sequence of the Human Genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.W.; Bruford, E.A. Naming ‘junk’: Human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genom. 2011, 5, 90–98. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. The Role of MicroRNAs in Cancer. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Walter, N.G.; Engelke, D.R. Ribozymes: Catalytic RNAs that cut things, make things, and do odd and useful jobs. Biologist 2002, 49, 199–203. [Google Scholar]
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Kiss, T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004, 117, 5949–5951. [Google Scholar] [CrossRef]
- Turner, J.D.; Williamson, R.; Almefty, K.K.; Nakaji, P.; Porter, R.; Tse, V.; Kalani, M.Y. The many roles of microRNAs in brain tumor biology. Neurosurg. Focus 2010, 28, E3. [Google Scholar] [CrossRef]
- Szymanski, M.; Barciszewska, M.Z.; Erdmann, V.A.; Barciszewski, J. A new frontier for molecular medicine: Noncoding RNAs. Biochem. Biophys. Acta 2005, 1756, 65–75. [Google Scholar] [CrossRef]
- Espinosa, C.E.S.; Slack, F.J. The Role of MicroRNAs in Cancer. Yale J. Biol. Med. 2006, 79, 131–140. [Google Scholar]
- Staedel, C.; Tran, T.P.A.; Giraud, J.; Darfeuille, F.; Di Giorgio, A.; Tourasse, N.J.; Salin, F.; Uriac, P.; Duca, M. Modulation of oncogenic miRNA biogenesis using functionalized polyamines. Sci. Rep. 2018, 8, 1667. [Google Scholar] [CrossRef]
- Costales, M.G.; Aikawa, H.; Li, Y.; Childs-Disney, J.L.; Abegg, D.; Hoch, D.G.; Velagapudi, S.P.; Nakai, Y.; Khan, T.; Wang, K.W.; et al. Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 2406–2411. [Google Scholar] [CrossRef]
- Boriack-Sjodin, P.A.; Ribich, S.; Copeland, R.A. RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug. Discov. 2018, 17, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.C.; Kwok, W.K.; Chen, Z.; Ng, H.-K. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009, 117, 599–611. [Google Scholar] [CrossRef]
- Souckova, K.; Ivkovic, T.C.; Slaby, O. Non-coding RNA therapy in cancer. Precis. Med. Investig. Pract. Provid. 2020, 20, 211–220. [Google Scholar]
- Zhang, J.; Lau, M.W.; Ferré-D’Amaré, A.R. Ribozymes and Riboswitches: Modulation of RNA Function by Small Molecules. Biochemistry 2010, 49, 9123–9131. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a003533. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Edelman, E.R.; Artzi, N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: The jack-of-all-trades in cancer nanotheranostics? Adv. Drug Deliv. Rev. 2015, 81, 169–183. [Google Scholar] [CrossRef]
- Storz, G. An Expanding Universe of Noncoding RNAs. Science 2002, 296, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Baraniskin, A.; Hahn, S.A.; Mosel, F.; Bredemeier, M.; Wimberger, P.; Kimmig, R.; Kasimir-Bauer. S. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin. Chem. 2014, 60, 206–213. [Google Scholar] [CrossRef]
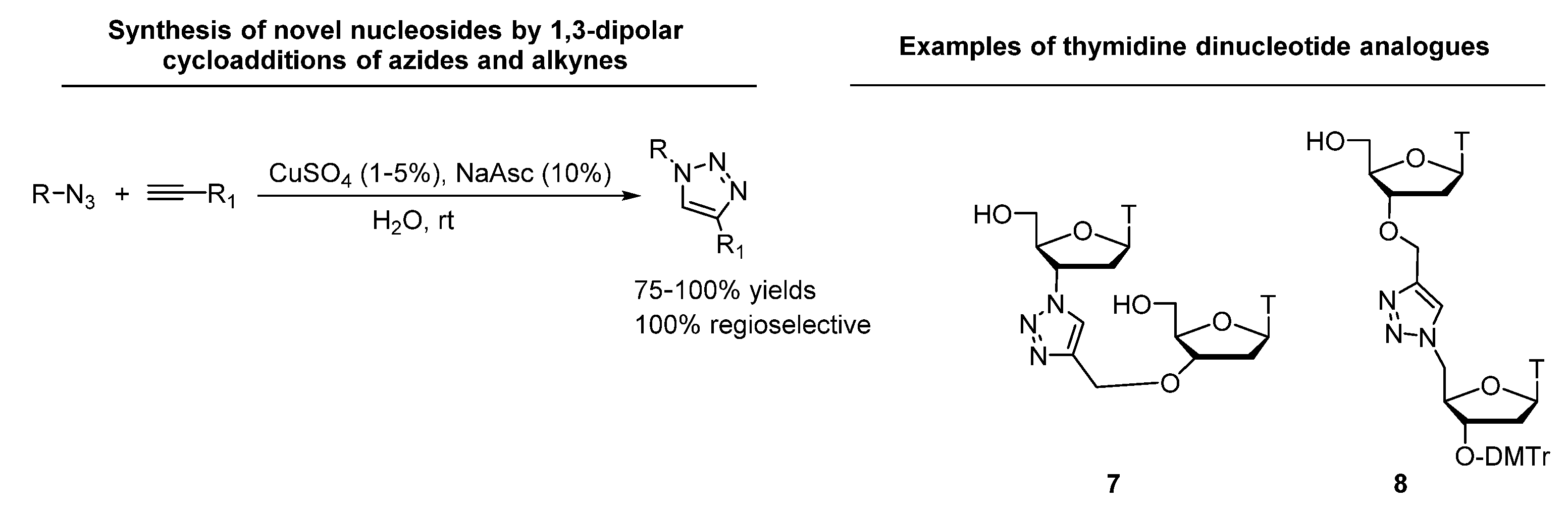
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1–3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Amblard, F.; Cho, J.H.; Schinazi, R.F. Cu(I)-Catalyzed Huisgen Azide−Alkyne 1,3-Dipolar Cycloaddition Reaction in Nucleoside, Nucleotide, and Oligonucleotide Chemistry. Chem. Rev. 2009, 109, 4207–4220. [Google Scholar] [CrossRef]
- Efthymiou, T.; Gong, W.; Desaulniers, J.-P. Chemical Architecture and Applications of Nucleic Acid Derivatives Containing 1,2,3-Triazole Functionalities Synthesized via Click Chemistry. Molecules 2012, 17, 12665–12703. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Mishra, B.B.; Mishra, K.B.; Mishra, N.; Singh, A.S.; Chen, X. Cu-Catalyzed Click Reaction in Carbohydrate Chemistry. Chem. Rev. 2016, 116, 3086–3240. [Google Scholar] [CrossRef]
- Zavgorodny, S.G.; Pechenov, A.E.; Shvets, V.I.; Miroshnikov, A.I. S,X-Acetals in Nucleoside Chemistry. III1. Synthesis of 2′-and 3′-O-Azidomethyl Derivatives of Ribonucleosides. Nucleos. Nucleot. Nucl. 2000, 19, 1977–1991. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, D.; Baranowski, D.; Ruszkowski, P.; Boryski, J. 3’-O- and 5’-O-Propargyl derivatives of 5-fluoro-2’-deoxyuridine: Synthesis, cytotoxic evaluation and conformational analysis. Nucleos. Nucleot. Nucl. 2016, 35, 178–194. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.; Rodríguez, H.; Albericio, F. CuAAC: An Efficient Click Chemistry Reaction on Solid Phase. ACS Comb. Sci. 2016, 18, 1–14. [Google Scholar] [CrossRef]
- Kropp, H.M.; Dürr, S.L.; Peter, C.; Diederichs, K.; Marx, A. Snapshots of a modified nucleotide moving through the confines of a DNA polymerase. Proc. Natl. Acad. Sci. USA 2018, 115, 9992–9997. [Google Scholar] [CrossRef]
- Lingala, S.; Nordstrøm, L.U.; Mallikaratchy, P.R. Synthesis of stable azide and alkyne functionalized phosphoramidite nucleosides. Tetrahedron Lett. 2019, 60, 211–213. [Google Scholar] [CrossRef]
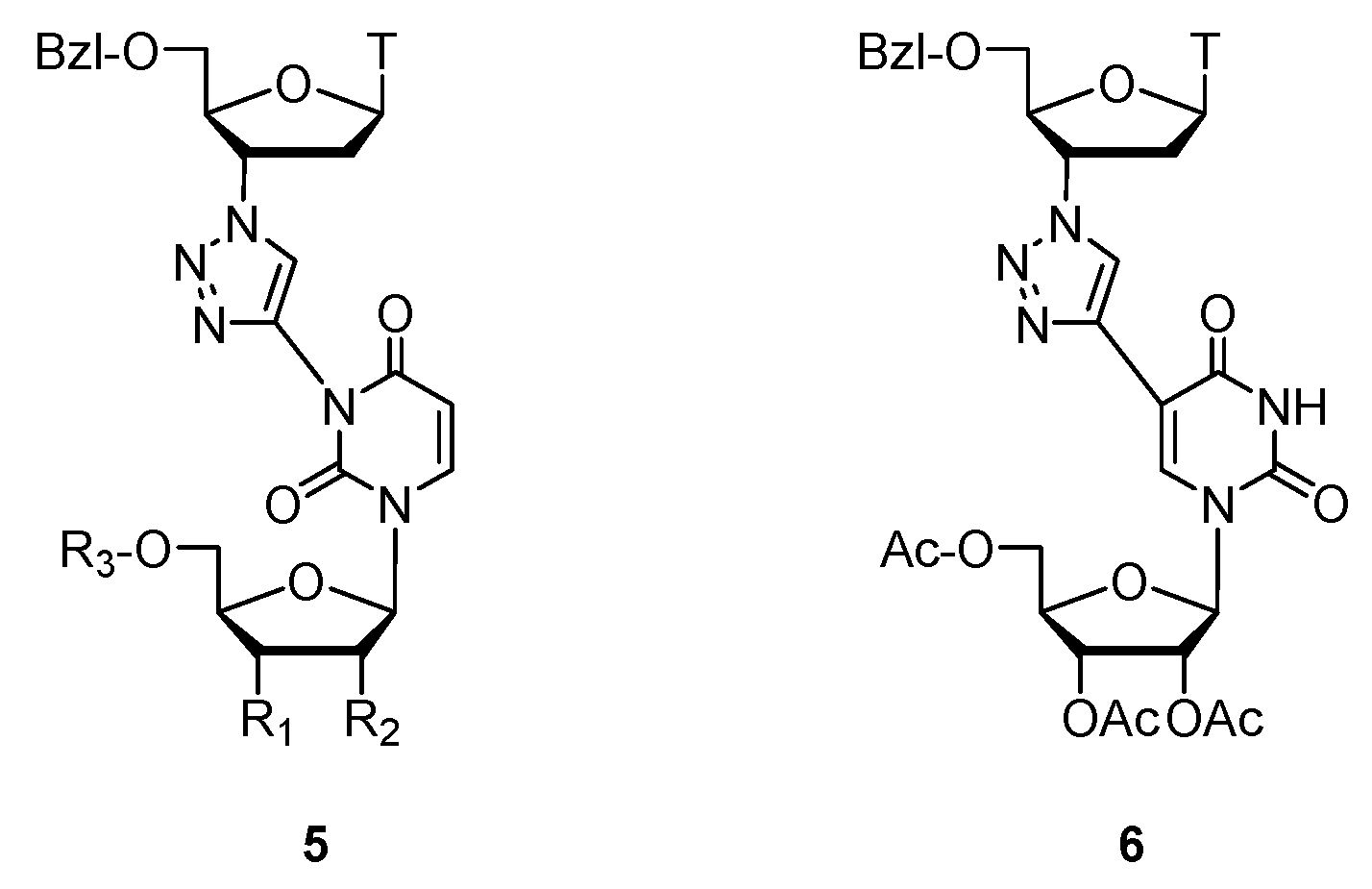
- Michalska, L.; Wawrzyniak, D.; Szymańska-Michalak, A.; Barciszewski, J.; Boryski, J.; Baraniak, D. Synthesis and biological assay of new 2′-deoxyuridine dimers containing a 1,2,3-triazole linker. Part, I. Nucleos. Nucleot. Nucl. 2019, 38, 218–235. [Google Scholar] [CrossRef]
- Baraniak, D.; Baranowski, D.; Ruszkowski, P.; Boryski, J. Nucleoside dimers analogues with 1,2,3-triazole linkage: Conjugation of floxuridine and thymidine provides novel tools for cancer treatment. Part II. Nucleos. Nucleot. Nucl. 2019, 38, 807–835. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, D.; Ruszkowski, P.; Baranowski, D.; Framski, G.; Boryski, J. Nucleoside dimers analogs containing floxuridine and thymidine with unnatural linker groups: Synthesis and cancer line studies. Part III. Nucleos. Nucleot. Nucl. 2019, 38, 980–1005. [Google Scholar] [CrossRef] [PubMed]
- Jawalekar, A.M.; Malik, S.; Verkade, J.M.M.; Gibson, B.; Barta, N.S.; Hodges, J.C.; Rowan, A.; van Delft, F.L. Oligonucleotide Tagging for Copper-Free Click Conjugation. Molecules 2013, 18, 7346–7363. [Google Scholar] [CrossRef]
- Sharma, V.K.; Sharma, R.K.; Singh, S.K. Antisense oligonucleotides: Modifications and clinical trials. Med. Chem. Commun. 2014, 5, 1454–1471. [Google Scholar] [CrossRef]
- Guo, F.; Li, Q.; Zhou, C. Synthesis and biological applications of fluoro-modified nucleic acids. Org. Biomol. Chem. 2017, 15, 9552–9565. [Google Scholar] [CrossRef]
- Chittepu, P.; Sirivolu, V.R.; Seela, F. Nucleosides and oligonucleotides containing 1,2,3-triazole residues with nucleobase tethers: Synthesis via the azide-alkyne ‘click’ reaction. Bioorg. Med. Chem. 2008, 16, 8427–8439. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xu, N.; Li, Z.; Zhang, S.; Wu, J.; Kim, D.H.; Marma, M.S.; Meng, Q.; Cao, H.; Li, X.; et al. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc. Natl. Acad. Sci. USA 2008, 105, 9145–9150. [Google Scholar] [CrossRef]
- Okholm, A.; Kjems, J.; Astakhova, K. Fluorescence detection of natural RNA using rationally designed “clickable” oligonucleotide probes. RSC Adv. 2014, 4, 45653–45656. [Google Scholar] [CrossRef]
- Horisawa, K. Specific and quantitative labeling of biomolecules using click chemistry. Front. Physiol. 2014, 5, 457. [Google Scholar] [CrossRef]
- Saito, Y.; Hudson, R.H.E. Base-modified fluorescent purine nucleosides and nucleotides for use in oligonucleotide probes. J. Photochem. Photobiol. C 2018, 36, 48–73. [Google Scholar] [CrossRef]
- Venkatesham, A.; Pillalamarri, S.R.; Wit, F.D.; Lescrinier, E.; Debyser, Z.; Van Aerschot, A. Propargylated Purine Deoxynucleosides: New Tools for Fluorescence Imaging Strategies. Molecules 2019, 24, 468. [Google Scholar] [CrossRef]
- Bag, S.S.; Das, S.K. Triazolyl C-nucleosides via the intermediacy of β-1′-ethynyl-2′-deoxyribose derived from a Nicholas reaction: Synthesis, photophysical properties and interaction with BSA. Tetrahedron 2019, 75, 3024–3037. [Google Scholar] [CrossRef]
- Klimkowski, P.; De Ornellas, S.; Singleton, D.; El-Sagheer, A.H.; Brown, T. Design of thiazole orange oligonucleotide probes for detection of DNA and RNA by fluorescence and duplex melting. Org. Biomol. Chem. 2019, 17, 5943–5950. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M. Tailored therapeutics based on 1,2,3-1H-triazoles: A mini review. Med. Chem. Commun. 2019, 10, 1302–1328. [Google Scholar] [CrossRef] [PubMed]
- Pujari, S.S.; Seela, F. Parallel Stranded DNA Stabilized with Internal Sugar Cross-Links: Synthesis and Click Ligation of Oligonucleotides Containing 2′-Propargylated Isoguanosine. J. Org. Chem. 2013, 78, 8545–8561. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Seela, F. Cross-Linked DNA: Site-Selective “Click” Ligation in Duplexes with Bis-Azides and Stability Changes Caused by Internal Cross-Links. Bioconjug. Chem. 2012, 23, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Tera, M.; Taji, Z.H.; Luedtke, N.W. Intercalation-enhanced “Click” Crosslinking of DNA. Angew. Chem. Int. Ed. 2018, 130, 15631–15635. [Google Scholar] [CrossRef]
- Best, M.D. Click Chemistry and Bioorthogonal Reactions: Unprecedented Selectivity in the Labeling of Biological Molecules. Biochemistry 2009, 48, 6571–6584. [Google Scholar] [CrossRef]
- Grammel, M.; Hang, H. Chemical reporters for biological discovery. Nat. Chem. Biol. 2013, 9, 475–484. [Google Scholar] [CrossRef]
- Ren, X.; Gerowska, M.; El-Sagheerac, A.H.; Brown, T. Enzymatic incorporation and fluorescent labelling of cyclooctyne-modified deoxyuridine triphosphates in DNA. Bioorg. Med. Chem. 2014, 22, 4384–4390. [Google Scholar] [CrossRef]
- Seo, S.; Onizuka, K.; Nishioka, C.; Takahashi, E.; Tsuneda, S.; Abe, H.; Ito, Y. Phosphorylated 5-ethynyl-2′-deoxyuridine for advanced DNA labeling. Org. Biomol. Chem. 2015, 13, 4589–4595. [Google Scholar] [CrossRef]
- Sallustrau, A.; Bregant, S.; Chollet, C.; Audisio, D.; Taran, F. Scalable and practical synthesis of clickable Cu-chelating azides. Chem. Commun. 2017, 53, 7890–7893. [Google Scholar] [CrossRef] [PubMed]
- Godeau, G.; Staedel, C.; Barthélémy, P. Lipid-Conjugated Oligonucleotides via “Click Chemistry” Efficiently Inhibit Hepatitis C Virus Translation. J. Med. Chem. 2008, 51, 4374–4376. [Google Scholar] [CrossRef]
- Singh, Y.; Murat, P.; Defrancq, E. Recent developments in oligonucleotide conjugation. Chem. Soc. Rev. 2010, 39, 2054–2070. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zheng, L.; Zhang, P.; Li, J.; Zhang, Y. Development of Bioorthogonal Reactions and Their Applications in Bioconjugation. Molecules 2015, 20, 3190–3205. [Google Scholar] [CrossRef]
- Neumann, S.; Biewend, M.; Rana, S.; Binder, W.H. The CuAAC: Principles, Homogeneous and Heterogeneous Catalysts, and Novel Developments and Applications. Macromol. Rapid Commun. 2020, 41, 1900359. [Google Scholar] [CrossRef]
- Erbas-Cakmak, S.; Leigh, D.A.; McTernan, C.T.; Nussbaumer, A.L. Artificial Molecular Machines. Chem. Rev. 2015, 115, 10081–10206. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Rusling, D.A. Triplex-forming oligonucleotides: A third strand for DNA nanotechnology. Nucleic Acids Res. 2018, 46, 1021–1037. [Google Scholar] [CrossRef] [PubMed]
- Valsangkar, V.; Chandrasekaran, A.R.; Wang, R.; Haruehanroengra, P.; Levchenko, O.; Halvorsen, K.; Sheng, J. Click-based functionalization of a 2′-O-propargyl-modified branched DNA nanostructure. J. Mater. Chem. B 2017, 5, 2074–2077. [Google Scholar] [CrossRef]
- Takezawa, Y.; Shionoya, M. Supramolecular DNA Three-Way Junction Motifs with a Bridging Metal Center. Front. Chem. 2020, 7, 925. [Google Scholar] [CrossRef]
- Thirumurugan, P.; Matosiuk, D.; Jozwiak, K. Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 2013, 113, 4905–4979. [Google Scholar] [CrossRef]
- Garner, A.L. at-ELCCA: Catalyzing Drug Discovery Through Click Chemistry. Chem. Commun. 2018, 54, 6531–6539. [Google Scholar] [CrossRef] [PubMed]
- Corso, A.D.; Pignataro, L.; Belvisi, L.; Gennari, C. Innovative Linker Strategies for Tumor-Targeted Drug Conjugates. Chem. Eur. J. 2019, 25, 14740–14757. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Bernardes, G.J.L. Machine learning for target discovery in drug development. Curr. Opin. Chem. Biol. 2020, 56, 16–22. [Google Scholar] [CrossRef]
- Isobe, H.; Fujino, T. Triazole-linked analogues of DNA and RNA (TLDNA and TLRNA): Synthesis and functions. Chem. Rec. 2014, 14, 41–51. [Google Scholar] [CrossRef]
- Astakhova, K.; Ray, R.; Taskova, M.; Uhd, J.; Carstens, A.; Morris, K. “Clicking” Gene Therapeutics: A Successful Union of Chemistry and Biomedicine for New Solutions. Mol. Pharm. 2018, 15, 2892–2899. [Google Scholar] [CrossRef] [PubMed]
- Fairbanks, B.D.; Culver, H.R.; Mavila, S.; Bowman, C.N. Towards High-Efficiency Synthesis of Xenonucleic Acids. Trends Chem. 2019, 2, 43–56. [Google Scholar] [CrossRef]
- Fantoni, N.Z.; El-Sagheer, A.H.; Brown, T. A Hitchhiker’s Guide to Click-Chemistry with Nucleic Acids. Chem. Rev. 2021. [Google Scholar] [CrossRef]
- Brown, T.; El-Sagheer, A.H. Oligonucleotide ligation. U.S. Patent 8,846,883 B2, 30 September 2014. [Google Scholar]
- Howe, F.S.; Russell, A.; Lamstaes, A.R.; El-Sagheer, A.; Nair, A.; Brown, T.; Mellor, J. CRISPRi is not strand-specific at all loci and redefines the transcriptional landscape. eLife 2017, 6, e29878. [Google Scholar] [CrossRef]
- Taemaitree, L.; Shivalingam, A.; El-Sagheer, A.H.; Brown, T. An artificial triazole backbone linkage provides a split-and-click strategy to bioactive chemically modified CRISPR sgRNA. Nat. Commun. 2019, 10, 1610. [Google Scholar] [CrossRef]
- Filippova, J.; Matveeva, A.; Zhuravlev, E.; Stepanov, G. Guide RNA modification as a way to improve CRISPR/Cas9-based genome-editing systems. Biochimie 2019, 167, 49–60. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, Y.; Yin, H. Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing. Adv. Drug Deliv. Rev. 2021, 168, 246–258. [Google Scholar] [CrossRef]
- Tolle, F.; Brändle, G.M.; Matzner, D.; Mayer, G. A Versatile Approach Towards Nucleobase-Modified Aptamers. Angew. Chem. Int. Ed. 2015, 54, 10971–10974. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brändle, G.M.; Ewers, J.; Mayer, G. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Chen, T.; Sheng, K.; Liu, Z.; Zhang, Z.; Romesberg, F.E. Selection of Aptamers with Large Hydrophobic 2′-Substituents. J. Am. Chem. Soc. 2020, 142, 2125–2128. [Google Scholar] [CrossRef]
- Deiters, A.; Schultz, P.G. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorganic Med. Chem. Lett. 2005, 15, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Parrish, A.R.; Wang, L. Expanding the Genetic Code for Biological Studies. Chem. Biol. 2009, 16, 323–336. [Google Scholar] [CrossRef]
- Lu, K.; Duan, Q.-P.; Ma, L.; Zhao, D.-X. Chemical strategies for the synthesis of peptide-oligonucleotide conjugates. Bioconjug. Chem. 2010, 21, 187–202. [Google Scholar] [CrossRef]
- Ballikaya, S.; Lee, J.; Warnken, U.; Schnölzer, M.; Gebert, J.; Kopitz, J. De Novo proteome analysis of genetically modified tumor cells by a metabolic labeling/azide-alkyne cycloaddition approach. Mol. Cell. Proteom. 2014, 13, 3446–3456. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Monfregola, L.; Caruthers, M. Peptide-substituted oligonucleotide synthesis and non-toxic, passive cell delivery. Signal Transduct. Target. Ther. 2016, 1, e16019. [Google Scholar] [CrossRef] [PubMed]
- MacCulloch, T.; Buchberger, A.; Stephanopoulos, N. Emerging applications of peptide–oligonucleotide conjugates: Bioactive scaffolds, self-assembling systems, and hybrid nanomaterials. Org. Biomol. Chem. 2019, 17, 1668–1682. [Google Scholar] [CrossRef]
- Stephanopoulos, N. Peptide–Oligonucleotide Hybrid Molecules for Bioactive Nanomaterials. Bioconjug. Chem. 2019, 30, 1915–1922. [Google Scholar] [CrossRef]
- Liu, C.; Zou, Y.; Hu, H.; Jiang, Y.; Qin, L. pDobz/pDobb protected diaminodiacid as a novel building block for peptide disulfide-bond mimic synthesis. RSC Adv. 2019, 9, 5438–5444. [Google Scholar] [CrossRef]
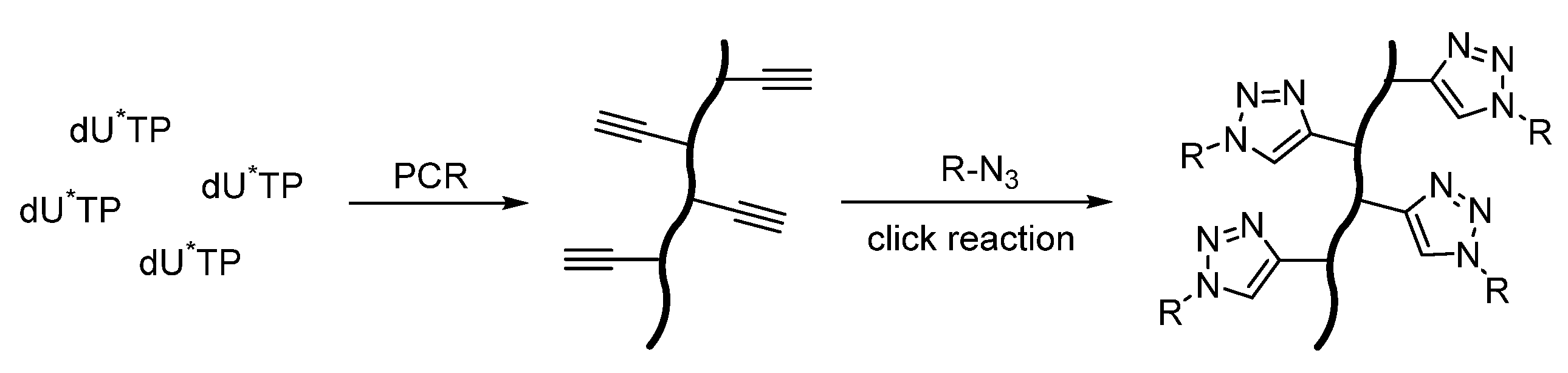
- Hocek, M. Enzymatic Synthesis of Base-Functionalized Nucleic Acids for Sensing, Cross-linking, and Modulation of Protein–DNA Binding and Transcription. Acc. Chem. Res. 2019, 52, 1730–1737. [Google Scholar] [CrossRef]
- Taskova, M.; Madsen, C.S.; Jensen, K.J.; Hansen, L.H.; Vester, B.; Astakhova, K. Antisense Oligonucleotides Internally Labeled with Peptides Show Improved Target Recognition and Stability to Enzymatic Degradation. Bioconjug. Chem. 2017, 28, 768–774. [Google Scholar] [CrossRef]
- Jain, N.; Smith, S.W.; Ghone, S.; Tomczuk, B. Current ADC Linker Chemistry. Pharm. Res. 2015, 32, 3526–3540. [Google Scholar] [CrossRef]
- Dovgan, I.; Koniev, O.; Kolodych, S.; Wagner, A. Antibody–Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjug. Chem. 2019, 30, 2483–2501. [Google Scholar] [CrossRef] [PubMed]
- Bernard, S.; Audisio, D.; Riomet, M.; Bregant, S.; Sallustrau, A.; Plougastel, L.; Decuypere, E.; Gabillet, S.; Kumar, R.A.; Elyian, J.; et al. Bioorthogonal Click and Release Reaction of Iminosydnones with Cycloalkynes. Angew. Chem. Int. Ed. 2017, 56, 15612–15616. [Google Scholar] [CrossRef]
- Hsu, N.-S.; Lee, C.-C.; Kuo, W.-C.; Chang, Y.-W.; Lo, S.-Y.; Wang, A.H.-J. Development of a Versatile and Modular Linker for Antibody–Drug Conjugates Based on Oligonucleotide Strand Pairing. Bioconjug. Chem. 2020, 31, 1804–1811. [Google Scholar] [CrossRef]
- Wan, J.; Li, Y.; Jin, K.; Guo, J.; Xu, J.; Wang, C. Robust Strategy for Antibody–Polymer–Drug Conjugation: Significance of Conjugating Orientation and Linker Charge on Targeting Ability. ACS Appl. Mater. Interfaces 2020, 12, 23717–23725. [Google Scholar] [CrossRef]
- Dai, Z.; Zhang, X.-N.; Nasertorabi, F.; Cheng, Q.; Li, J.; Katz, B.B.; Smbatyan, G.; Pei, H.; Louie, S.G.; Lenz, H.-J.; et al. Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases. Sci. Adv. 2020, 6, eaba6752. [Google Scholar] [CrossRef]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9, 1432. [Google Scholar] [CrossRef]
- Bhuket, P.R.N.; Luckanagul, J.A.; Ratnatilaka, P.; Wang, Q. Chemical modification of enveloped viruses for biomedical applications. Integr. Biol. 2018, 10, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Micklefield, J. Backbone modification of nucleic acids: Synthesis, structure and therapeutic applications. Curr. Med. Chem. 2001, 8, 1157–1179. [Google Scholar] [CrossRef]
- Alvarez-Salas, L.M. Nucleic Acids as Therapeutic Agents. Curr. Top. Med. Chem. 2008, 8, 1379–1404. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, S.; Milam, V.T. Modified Nucleic Acids: Expanding the Capabilities of Functional Oligonucleotides. Molecules 2020, 25, 4659. [Google Scholar] [CrossRef]
- Opalinska, J.B.; Gewirtz, A.M. Nucleic-acid therapeutics: Basic principles and recent applications. Nat. Rev. Drug Discov. 2002, 1, 503–514. [Google Scholar] [CrossRef]
- Sharma, V.K.; Rungta, P.; Prasad, A.K. Nucleic acid therapeutics: Basic concepts and recent developments. RSC Adv. 2014, 4, 16618–16631. [Google Scholar] [CrossRef]
- Sharma, V.K.; Watts, J.K. Oligonucleotide therapeutics: Chemistry, delivery and clinical progres. Future Med. Chem. 2015, 7, 2221–2242. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.A.; Ellington, A.D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harb. Perspect. Biol. 2017, 9, a023812. [Google Scholar] [CrossRef] [PubMed]
- Kukwikila, M.; Gale, N.; El-Sagheer, A.H.; Brown, T.; Tavassoli, A. Assembly of a biocompatible triazole-linked gene by one-pot click-DNA ligation. Nat. Chem. 2017, 9, 1089–1098. [Google Scholar] [CrossRef]
- Heravi, M.M.; Tamimi, M.; Yahyavi, H.; Hosseinnejad, T. Huisgen’s Cycloaddition Reactions: A Full Perspective. Curr. Org. Chem. 2016, 20, 1591–1647. [Google Scholar] [CrossRef]
- Johansson, J.R.; Beke-Somfai, T.; Stålsmeden, A.S.; Kann, N. Ruthenium-Catalyzed Azide Alkyne Cycloaddition Reaction: Scope, Mechanism, and Applications. Chem. Rev. 2016, 116, 14726–14768. [Google Scholar] [CrossRef]
- Zhu, L.; Brassard, C.J.; Zhang, X.; Guha, P.M.; Clark, R.J. On the Mechanism of Copper(I)-Catalyzed Azide-Alkyne Cycloaddition. Chem. Rec. 2016, 16, 1501–1517. [Google Scholar] [CrossRef]
- Bonandi, L.; Christodoulou, M.S.; Fumagalli, G.; Perdicchia, D.; Rastelli, G.; Passarella, D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today 2017, 22, 1572–1581. [Google Scholar] [CrossRef]
- Eremeeva, E.; Herdewijn, P. Non canonical genetic material. Curr. Opin. Biotechnol. 2019, 57, 25–33. [Google Scholar] [CrossRef]
- Oliveira, B.L.; Guo, Z.; Bernardes, G.J.L. Inverse electron demand Diels–Alder reactions in chemical biology. Chem. Soc. Rev. 2017, 46, 4895–4950. [Google Scholar] [CrossRef]
- Quadrelli, P. Modern Applications of Cycloaddition Chemistry; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Uhlmann, E.; Peyman, A. Antisense Oligonucleotides: A New Therapeutic Principle. Chem. Rev. 1990, 90, 543–584. [Google Scholar] [CrossRef]
- Kurreck, J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003, 270, 1628–1644. [Google Scholar] [CrossRef]
- Dean, N.M.; Bennett, C.F. Antisense oligonucleotide-based therapeutics for cancer. Oncogene 2003, 22, 9087–9096. [Google Scholar] [CrossRef]
- Tamm, I.; Wagner, M. Antisense therapy in clinical oncology. Mol. Biotechnol. 2006, 33, 221–238. [Google Scholar] [CrossRef]
- Corey, D.R. Chemical modification: The key to clinical application of RNA interference? J. Clin. Investig. 2007, 117, 3615–3622. [Google Scholar] [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Gallas, A.; Alexander, C.; Davies, M.C.; Puri, S.; Allen, S. Chemistry and formulations for siRNA therapeutics. Chem. Soc. Rev. 2013, 42, 7983–7997. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther 2020, 5, 101. [Google Scholar] [CrossRef]
- Wan, W.B.; Seth, P.P. The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem. 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Freier, S.M.; Altmann, K.-H. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [Google Scholar] [CrossRef]
- Kasahara, Y.; Kuwahara, M. Artificial Specific Binders Directly Recovered from Chemically Modified Nucleic Acid Libraries. J. Nucleic Acids 2012, 2012, 156482. [Google Scholar] [CrossRef] [PubMed]
- von Matt, P.; Lochmann, T.; Altmann, K.-H. Replacement of the phosphodiester linkage in oligonucleotides by heterocycles: Synthesis of thymidine dinucleotide analogs with triazole-modified backbones. Bioorganic Med. Chem. Lett. 1997, 7, 1549–1552. [Google Scholar] [CrossRef]
- von Matt, T.; Altmann, K.-H. Replacement of the phosphodiester linkage in oligonucleotides by heterocycles: The effect of triazole-and imidazole-modified backbones on DNA/RNA duplex stability. Bioorganic Med. Chem. Lett. 1997, 7, 1553–1556. [Google Scholar] [CrossRef]
- Lazrek, H.B.; Engels, J.W.; Pfleiderer, W. Synthesis of Novel Branched Nucleoside Dimers Containing a 1,2,3-Triazolyl Linkage. Nucleos. Nucleot. 1998, 17, 1851–1856. [Google Scholar] [CrossRef]
- Zhou, L.; Amer, A.; Korn, M.; Burda, R.; Balzarini, J.; De Clercq, E.; Kern, E.R.; Torrence, P.F. Synthesis and antiviral activities of 1,2,3-triazole functionalized thymidines: 1,3-dipolar cycloadditionfor efficient regioselective diversity generation. Antivir. Chem. Chemother. 2005, 16, 375–383. [Google Scholar] [CrossRef]
- Nuzzi, A.; Massi, A.; Dondoni, A. Model Studies Toward the Synthesis of Thymidine Oligonucleotides with Triazole Internucleosidic Linkages Via Iterative Cu(I)-Promoted Azide–Alkyne Ligation Chemistry. QSAR Comb. Sci. 2007, 26, 1191–1199. [Google Scholar] [CrossRef]
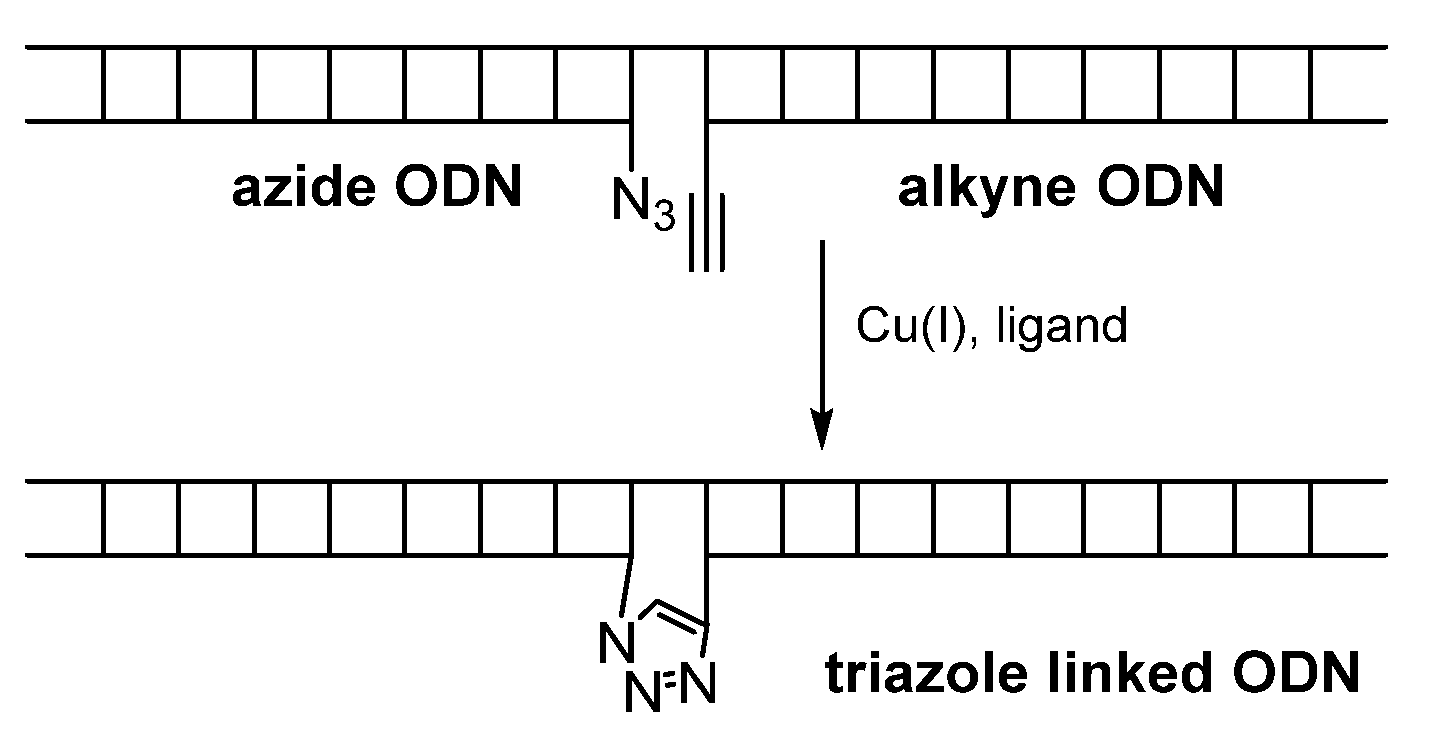
- Kumar, R.; El-Sagheer, A.; Tumpane, J.; Lincoln, P.; Wilhelmsson, L.M.; Brown, T. Template-Directed Oligonucleotide Strand Ligation, Covalent Intramolecular DNA Circularization and Catenation Using Click Chemistry. J. Am. Chem. Soc. 2007, 129, 6859–6864. [Google Scholar] [CrossRef] [PubMed]
- Sanzone, A.P.; El-Sagheer, A.H.; Brown, T.; Tavassoli, A. Assessing the biocompatibility of click-linked DNA in Escherichia coli. Nucleic Acids Res. 2012, 40, 10567–10575. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. A triazole linkage that mimics the DNA phosphodiester group in living systems. Q. Rev. Biophys. 2015, 48, 429–436. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Sanzone, A.P.; Gao, R.; Tavassoli, A.; Brown, T. Biocompatible artificial DNA linker that is read through by DNA polymerases and is functional in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 11338–11343. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Efficient RNA synthesis by in vitro transcription of a triazole-modified DNA template. Chem. Commun. 2011, 47, 12057–12058. [Google Scholar] [CrossRef] [PubMed]
- Routh, A.; Head, S.R.; Ordoukhanian, P.; Johnson, J.E. ClickSeq: Fragmentation-Free Next-Generation Sequencing via Click Ligation of Adaptors to Stochastically Terminated 3’-Azido cDNAs. J. Mol. Biol. 2015, 427, 2610–2616. [Google Scholar] [CrossRef] [PubMed]
- Miura, F.; Fujino, T.; Kogashi, K.; Shibata, Y.; Miura, M.; Isobe, H.; Ito, T. Triazole linking for preparation of a next-generation sequencing library from single-stranded DNA. Nucleic Acids Res. 2018, 46, e95. [Google Scholar] [CrossRef]
- Osman, E.A.; Gadzikwa, T.; Gibbs, J.M. Quick Click: The DNA-Templated Ligation of 3’-O-Propargyl- and 5’-Azide-Modified Strands Is as Rapid as and More Selective than Ligase. ChemBioChem 2018, 19, 2081–2087. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Kumar, R.; Findlow, S.; Werner, J.M.; Lane, A.N.; Brown, T. A Very Stable Cyclic DNA Miniduplex with Just Two Base Pairs. ChemBioChem 2008, 9, 50–52. [Google Scholar] [CrossRef]
- Yang, H.; Seela, F. “Bis-Click” Ligation of DNA: Template-Controlled Assembly, Circularisation and Functionalisation with Bifunctional and Trifunctional Azides. Chem. Eur. J. 2017, 23, 3375–3385. [Google Scholar] [CrossRef]
- Xiong, H.; Seela, F. Stepwise “Click” Chemistry for the Template Independent Construction of a Broad Variety of Cross-Linked Oligonucleotides: Influence of Linker Length, Position, and Linking Number on DNA Duplex Stability. J. Org. Chem. 2011, 76, 5584–5597. [Google Scholar] [CrossRef]
- Lietard, J.; Meyer, A.; Vasseur, J.-J.; Morvan, F. New Strategies for Cyclization and Bicyclization of Oligonucleotides by Click Chemistry Assisted by Microwaves. J. Org. Chem. 2008, 73, 191–200. [Google Scholar] [CrossRef]
- Padgett, R.A.; Konarska, M.M.; Grabowski, P.J.; Hardy, S.F.; Sharp, P.A. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science 1984, 225, 898–903. [Google Scholar] [CrossRef]
- Nakane, M.; Ichikawa, S.; Matsuda, A. Triazole-Linked Dumbbell Oligodeoxynucleotides with NF-κB Binding Ability as Potential Decoy Molecules. J. Org. Chem. 2008, 73, 1842–1851. [Google Scholar] [CrossRef]
- Vasilyeva, S.V.; Levina, A.S.; Li-Zhulanov, N.S.; Shatskaya, N.V.; Baiborodin, S.I.; Repkova, M.N.; Zarytova, V.F.; Mazurkova, N.A.; Silnikov, V.N. SiO2 nanoparticles as platform for delivery of 3′-triazole analogues of AZT-triphosphate into cells. Bioorg. Med. Chem. 2015, 23, 2168–2175. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, V.; Oberleitner, B.; Sobotta, J.; Nickels, P.; Grossi, G.; Kempter, S.; Frischmuth, T.; Liedl, T.; Manetto, A. One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew. Chem. Int. Ed. 2015, 54, 7795–7798. [Google Scholar] [CrossRef]
- Wirges, C.T.; Gramlich, P.M.E.; Gutsmiedl, K.; Gierlich, J.; Burley, G.A.; Carell, T. Pronounced Effect of DNA Hybridization on Click Reaction Efficiency. QSAR Comb. Sci. 2007, 26, 1159–1164. [Google Scholar] [CrossRef]
- Gierlich, J.; Burley, G.A.; Gramlich, P.M.E.; Hammond, D.M.; Carell, T. Click Chemistry as a Reliable Method for the High-Density Postsynthetic Functionalization of Alkyne-Modified DNA. Org. Lett. 2006, 8, 3639–3642. [Google Scholar] [CrossRef]
- Burley, G.A.; Gierlich, J.; Mofid, M.R.; Nir, H.; Tal, S.; Eichen, Y.; Carell, T. Directed DNA Metallization. J. Am. Chem. Soc. 2006, 128, 1398–1399. [Google Scholar] [CrossRef]
- Gierlich, J.; Gutsmiedl, K.; Gramlich, P.M.; Schmidt, A.; Burley, G.A.; Carell, T. Synthesis of Highly Modified DNA by a Combination of PCR with Alkyne-Bearing Triphosphates and Click Chemistry. Chem. Eur. J. 2007, 13, 9486–9494. [Google Scholar] [CrossRef]
- Gramlich, P.M.E.; Wirges, C.T.; Gierlich, J.; Carell, T. Synthesis of Modified DNA by PCR with Alkyne-Bearing Purines Followed by a Click Reaction. Org. Lett. 2008, 10, 249–251. [Google Scholar] [CrossRef]
- Su, M.; Kirchner, A.; Stazzoni, S.; Müller, M.; Wagner, M.; Schröder, A.; Carell, T. 5-Formylcytosine Could Be a Semipermanent Base in Specific Genome Sites. Angew. Chem. Int. Ed. 2016, 55, 11797–11800. [Google Scholar] [CrossRef] [PubMed]
- Gramlich, P.M.E.; Wirges, C.T.; Manetto, A.; Carell, T. Postsynthetic DNA modification through the copper-catalyzed azide-alkyne cycloaddition reaction. Angew. Chem. Int. Ed. 2008, 47, 8350–8358. [Google Scholar] [CrossRef]
- O’Mahony, G.; Ehrman, E.; Grøtli, M. Synthesis of adenosine-based fluorosides containing a novel heterocyclic ring system. Tetrahedron Lett. 2005, 46, 6745–6748. [Google Scholar] [CrossRef]
- Mathew, S.C.; By, Y.; Berthault, A.; Virolleaud, M.-A.; Carrega, L.; Chouraqui, G.; Commeiras, L.; Condo, J.; Attolini, M.; Gaudel-Siri, A.; et al. Expeditious synthesis and biological evaluation of new C-6 1,2,3-triazole adenosine derivatives A1 receptorantagonists or agonists. Org. Biomol. Chem. 2010, 8, 3874–3881. [Google Scholar] [CrossRef] [PubMed]
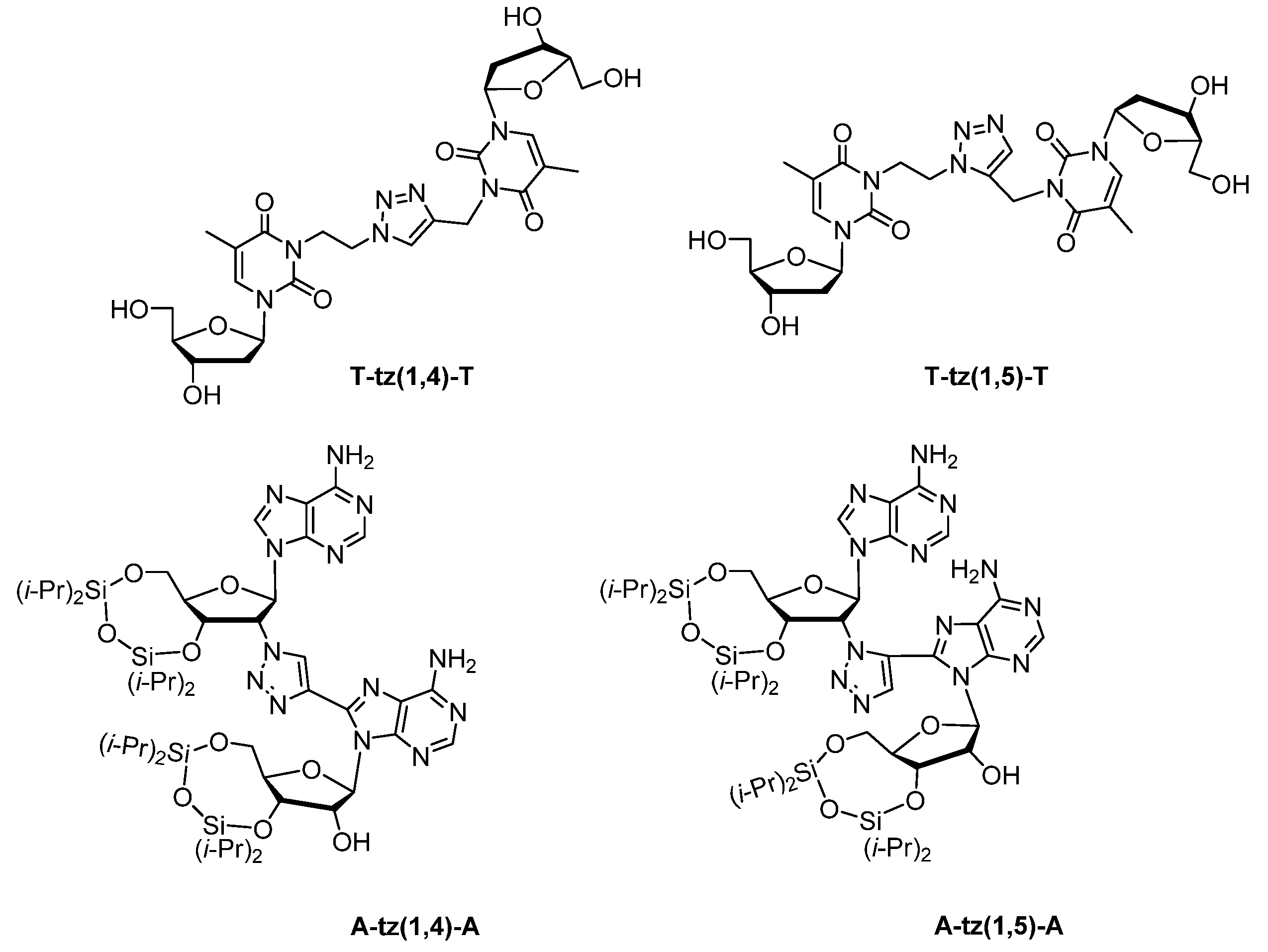
- Baker, Y.R.; Traoré, D.; Wanat, P.; Tyburn, A.; El-Sagheer, A.H.; Brown, T. Searching for the ideal triazole: Investigating the 1,5-triazole as a charge neutral DNA backbone mimic. Tetrahedron 2020, 76, 130914. [Google Scholar] [CrossRef]
- Kumar, A.S. Design and synthesis of double-headed nucleosides by using click chemistry approach. Chem. Data Collect. 2020, 28, 100468. [Google Scholar]
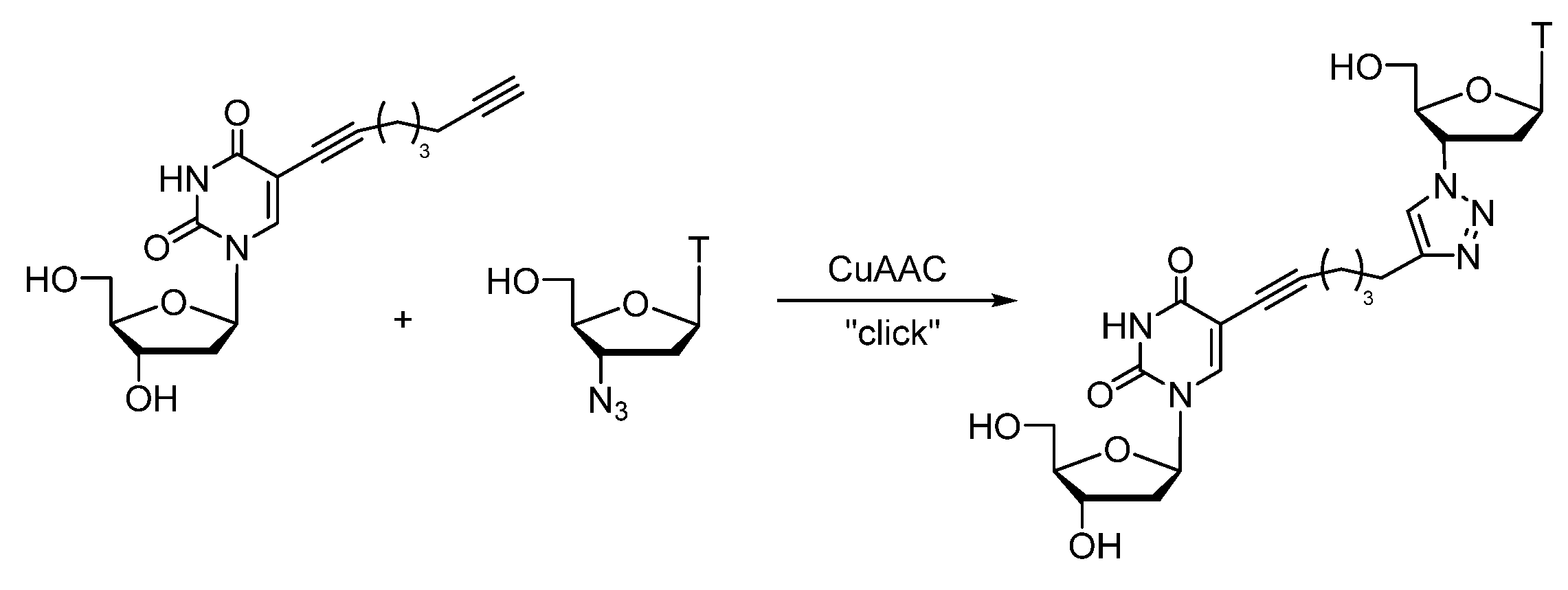
- Seela, F.; Sirivolu, V.R. DNA Containing Side Chains with Terminal Triple Bonds: Base-Pair Stability and Functionalization of Alkynylated Pyrimidines and 7-Deazapurines. Chem. Biodivers. 2006, 3, 509–514. [Google Scholar] [CrossRef]
- Seela, F.; Ming, X. Oligonucleotides Containing 7-Deaza-2′-deoxyinosine as Universal Nucleoside: Synthesis of 7-Halogenated and 7-Alkynylated Derivatives, Ambiguous Base Pairing, and Dye Functionalization by the Alkyne–Azide ‘Click’ Reaction. Helv. Chim. Acta 2008, 91, 1181–1200. [Google Scholar] [CrossRef]
- Seela, F.; Sirivolu, V.R. Nucleosides and Oligonucleotides with Diynyl Side Chains: Base Pairing and Functionalization of 2′-Deoxyuridine Derivatives by the Copper(I)-Catalyzed Alkyne–Azide ‘Click’ Cycloaddition. Helv. Chim. Acta 2007, 90, 535–552. [Google Scholar] [CrossRef]
- Sirivou, V.R.; Chittepu, P.; Seela, F. DNA with Branched Internal Side Chains: Synthesis of 5-Tripropargylamine-dU and Conjugation by an Azide-Alkyne Double Click Reaction. ChemBioChem 2008, 9, 2305–2316. [Google Scholar] [CrossRef]
- Seela, F.; Sirivolu, V.R. Pyrrolo-dColigonucleotides bearing alkynyl side chains with terminal triple bonds: Synthesis, base pairing and fluorescent dye conjugates prepared by the azide–alkyne “click” reaction. Org. Biomol. Chem. 2008, 6, 1674–1687. [Google Scholar] [CrossRef]
- Seela, F.; Sirivolu, V.R.; Chittepu, P. Modification of DNA with Octadiynyl Side Chains: Synthesis, Base Pairing, and Formation of Fluorescent Coumarin Dye Conjugates of Four Nucleobases by the Alkyne−Azide “Click” Reaction. Bioconjugate Chem. 2008, 19, 211–224. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click chemistry with DNA. Chem. Soc. Rev. 2010, 39, 1388–1405. [Google Scholar] [CrossRef]
- Berndl, S.; Herzig, N.; Kele, P.; Lachmann, D.; Li, X.; Wolfbeis, O.S.; Wagenknecht, H.-A. Comparison of a Nucleosidic vs. Non-Nucleosidic Postsynthetic “Click” Modification of DNA with Base-Labile Fluorescent Probes. Bioconjug. Chem. 2009, 20, 558–564. [Google Scholar] [CrossRef]
- Géci, I.; Filichev, V.V.; Pedersen, E.B. Stabilization of Parallel Triplexes by Twisted Intercalating Nucleic Acids (TINAs) Incorporating 1,2,3-Triazole Units and Prepared by Microwave-Accelerated Click Chemistry. Chem. Eur. J. 2007, 13, 6379–6386. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.; Neto, V.; Bouazza, A.H.; Zerrouki, R.; Granet, R.; Krausz, P.; Champavier, Y. Microwave-assisted synthesis of a triazole-linked 3′–5′ dithymidine using click chemistry. Tetrahedron Lett. 2008, 49, 1004–1007. [Google Scholar] [CrossRef]
- Lucas, R.; Zerrouki, R.; Granet, R.; Krausz, P.; Champavier, Y. A rapid efficient microwave-assisted synthesis of a 3′,5′-pentathymidine by copper(I)-catalyzed [3 + 2] cycloaddition. Tetrahedron 2008, 64, 5467–5471. [Google Scholar] [CrossRef]
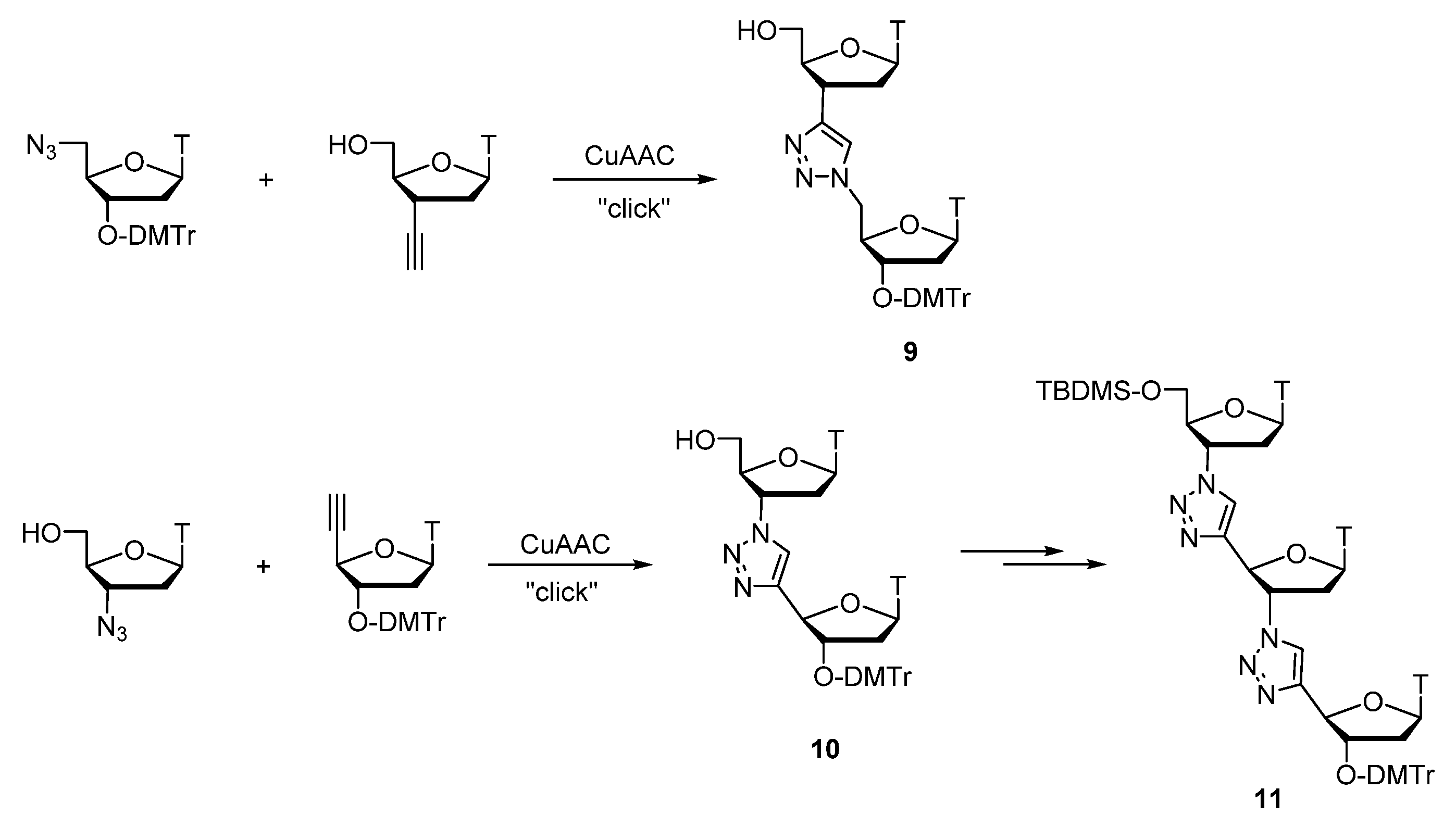
- Isobe, H.; Fujino, T.; Yamazaki, N.; Guillot-Nieckowski, M.; Nakamura, E. Triazole-Linked Analogue of Deoxyribonucleic Acid (TLDNA): Design, Synthesis, and Double-Strand Formation with Natural DNA. Org. Lett. 2008, 10, 3729–3732. [Google Scholar] [CrossRef] [PubMed]
- Varizhuk, A.; Chizhov, A.; Smirnov, I.; Kaluzhny, D.; Florentiev, V. Triazole-Linked Oligonucleotides with Mixed-Base Sequences: Synthesis and Hybridization Properties. Eur. J. Org. Chem. 2012, 11, 2173–2179. [Google Scholar] [CrossRef]
- Fujino, T.; Tsunaka, N.; Guillot-Nieckowski, M.; Nakanishi, W.; Iwamoto, T.; Nakamura, E.; Isobe, H. Synthesis and structures of deoxyribonucleoside analogues for triazole-linked DNA (TLDNA). Tetrahedron Lett. 2010, 51, 2036–2038. [Google Scholar] [CrossRef]
- Chandrasekhar, S.; Srihari, P.; Nagesh, C.; Kiranmai, N.; Nagesh, N.; Idris, M.M. Synthesis of Readily Accessible Triazole-Linked Dimer Deoxynucleoside Phosphoramidite for Solid-Phase Oligonucleotide Synthesis. Synthesis 2010, 21, 3710–3714. [Google Scholar] [CrossRef]
- Varizhuk, A.; Chizhov, A.; Florentiev, V. Synthesis and hybridization data of oligonucleotide analogs with triazole internucleotide linkages, potential antiviral and antitumor agents. Bioorg. Chem. 2011, 39, 127–131. [Google Scholar] [CrossRef]
- Fujino, T.; Yamazaki, N.; Isobe, H. Convergent synthesis of oligomers of triazole-linked DNA analogue (TLDNA) in solution phase. Tetrahedron Lett. 2009, 50, 4101–4103. [Google Scholar] [CrossRef]
- Varizhuk, A.M.; Kaluzhny, D.N.; Novikov, R.A.; Chizhov, A.O.; Smirnov, I.P.; Chuvilin, A.N.; Tatarinova, O.N.; Fisunov, G.Y.; Pozmogova, G.E.; Florentiev, V.L. Synthesis of Triazole-Linked Oligonucleotides with High Affinity to DNA Complements and an Analysis of Their Compatibility with Biosystems. J. Org. Chem. 2013, 78, 5964–5969. [Google Scholar] [CrossRef]
- Madhuri, V.; Kumar, V.A. Design and Synthesis of Dephosphono DNA Analogues Containing 1,2,3-Triazole Linker and Their UV-Melting Studies with DNA/RNA. Nucleos. Nucleot. Nucl. 2012, 31, 97–111. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.; Brown, T. Synthesis and Polymerase Chain Reaction Amplification of DNA Strands Containing an Unnatural Triazole Linkage. J. Am. Chem. Soc. 2009, 131, 3958–3964. [Google Scholar] [CrossRef]
- Kočalka, P.; El-Sagheer, A.H.; Brown, T. Rapid and Efficient DNA Strand Cross-Linking by Click Chemistry. ChemBioChem 2008, 9, 1280–1285. [Google Scholar] [CrossRef]
- Dallmann, A.; El-Sagheer, A.H.; Dehmel, L.; Mügge, C.; Griesinger, C.; Ernsting, N.P.; Brown, T. Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained. Chem. Eur. J. 2011, 17, 14714–14717. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Combined nucleobase and backbone modifications enhance DNA duplex stability and preserve biocompatibility. Chem. Sci. 2014, 5, 253–259. [Google Scholar] [CrossRef]
- Shivalingam, A.; Tyburn, A.E.S.; El-Sagheer, A.H.; Brown, T. Molecular Requirements of High-Fidelity Replication-Competent DNA Backbones for Orthogonal Chemical Ligation. J. Am. Chem. Soc. 2017, 139, 1575–1583. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click Nucleic Acid Ligation: Applications in Biology and Nanotechnology. Acc. Chem. Res. 2012, 45, 1258–1267. [Google Scholar] [CrossRef]
- Birts, C.N.; Sanzone, A.P.; El-Sagheer, A.H.; Blaydes, J.P.; Brown, T.; Tavassoli, A. Transcription of click-linked DNA in human cells. Angew. Chem. Int. Ed. 2014, 53, 2362–2365. [Google Scholar] [CrossRef]
- Chen, J.; Baker, Y.R.; Brown, A.; El-Sagheer, A.H.; Brown, T. Enzyme-free synthesis of cyclic single-stranded DNA constructs containing a single triazole, amide or phosphoramidate backbone linkage and their use as templates for rolling circle amplification and nanoflower formation. Chem. Sci. 2018, 9, 8110–8120. [Google Scholar] [CrossRef] [PubMed]
- Krishna, H.; Caruthers, M.H. Alkynyl Phosphonate DNA: A Versatile “Click” able Backbone for DNA-Based Biological Applications. J. Am. Chem. Soc. 2012, 134, 11618–11631. [Google Scholar] [CrossRef]
- Guo, P.; Haque, F.; Hallahan, B.; Reif, R.; Li, H. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 2012, 22, 226–245. [Google Scholar] [CrossRef]
- Habibian, M.; Harikrishna, S.; Fakhoury, J.; Barton, M.; Ageely, E.A.; Cencic, R.; Fakih, H.H.; Katolik, A.; Takahashi, M.; Rossi, J.; et al. Effect of 2′-5′/3′-5′ phosphodiester linkage heterogeneity on RNA interference. Nucleic Acids Res. 2020, 48, 4643–4657. [Google Scholar] [CrossRef]
- Kumar, V.A.; Ganesh, K.N. Structure-Editing of Nucleic Acids for Selective Targeting of RNA. Curr. Top. Med. Chem. 2007, 7, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Sosson, M.; Pfeffer, D.; Richert, C. Enzyme-free ligation of dimers and trimers to RNA primers. Nucleic Acids Res. 2019, 47, 3836–3845. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, C. Dynamic RNA Modifications in Posttranscriptional Regulation. Mol. Cell 2014, 56, 5–12. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, B.S.; He, C. Nucleic Acid Modifications in Regulation of Gene Expression. Cell Chem. Biol. 2016, 23, 74–85. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef]
- Lee, Y.J.; Moon, T.S. Design rules of synthetic non-coding RNAs in bacteria. Methods 2018, 143, 58–69. [Google Scholar] [CrossRef]
- Khawar, M.B.; Mehmood, R.; Abbasi, M.H.; Sheikh, N. Multifactorial role of long non-coding RNAs (LncRNAs) in hematopoiesis. Front. Biosci. 2018, 10, 119–126. [Google Scholar]
- Yu, A.-M.; Jian, C.; Yu, A.H.; Tu, M.-J. RNA therapy: Are we using the right molecules? Pharmacol. Ther. 2019, 196, 91–104. [Google Scholar] [CrossRef]
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.-J.; Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.I.; Bukhat, S.; Rasul, S.; Manzoor, H.; Manzoor, M. RNA therapeutics: Identification of novel targets leading to drug discovery. J. Cell. Biochem. 2020, 121, 898–929. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-M.; Choi, Y.H.; Tu, M.-J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef] [PubMed]
- Costales, M.G.; Childs-Disney, J.L.; Haniff, H.S.; Disney, M.D. How We Think about Targeting RNA with Small Molecules. J. Med. Chem. 2020, 63, 8880–8900. [Google Scholar] [CrossRef] [PubMed]
- Akman, H.B.; Bensan, A.E.E. Noncoding RNAs and cancer. Turk. J. Biol. 2014, 38, 817–828. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, H.; Chan, M.T.V.; Wu, W.K.K. HULC: An oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 2017, 21, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Selvam, C.; Mutisya, D.; Prakash, S.; Ranganna, K.; Thilagavathi, R. Therapeutic potential of chemically modified siRNA: Recent trends. Chem. Biol. Drug Des. 2017, 90, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Circular RNAs as Therapeutic Agents and Targets. Front. Physiol. 2018, 9, 1262. [Google Scholar] [CrossRef]
- Seok, H.; Lee, H.; Jang, E.-S.; Chi, S.W. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell. Mol. Life Sci. 2018, 75, 797–814. [Google Scholar] [CrossRef]
- Onizuka, K.; Shibata, A.; Taniguchi, Y.; Sasaki, S. Pin-point chemical modification of RNA with diverse molecules through the functionality transfer reaction and the copper-catalyzed azide–alkyne cycloaddition reaction. Chem. Commun. 2011, 47, 5004–5006. [Google Scholar] [CrossRef]
- Pujari, S.S.; Leonard, P.; Seela, F. Oligonucleotides with “Clickable” Sugar Residues: Synthesis, Duplex Stability, and Terminal versus Central Interstrand Cross-Linking of 2′-O-Propargylated 2-Aminoadenosine with a Bifunctional Azide. J. Org. Chem. 2014, 79, 4423–4437. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, K.; Qu, D.; Chi, W.; Xiong, W.; Huang, Y.; Wen, J.; Feng, S.; Zhang, B. One pot conjugation of small molecules to RNA using click chemistry. Tetrahedron Lett. 2012, 53, 6747–6750. [Google Scholar] [CrossRef]
- Peel, B.J.; Hagen, G.; Krishnamurthy, K.; Desaulniers, J.-P. Conjugation and Evaluation of Small Hydrophobic Molecules to Triazole-Linked siRNAs. ACS Med. Chem. Lett. 2015, 6, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Costales, M.G.; Rzuczek, S.G.; Disney, M.D. Comparison of small molecules and oligonucleotides that target a toxic, non-coding RNA. Bioorganic Med. Chem. Lett. 2016, 26, 2605–2609. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Beal, P.A. Synthesis and evaluation of an alkyne-modified ATP analog for enzymatic incorporation into RNA. Bioorganic Med. Chem. Lett. 2016, 26, 1799–1802. [Google Scholar] [CrossRef]
- Atdjian, C.; Coelho, D.; Iannazzo, L.; Ethève-Quelquejeu, M.; Braud, E. Synthesis of Triazole-Linked SAM-Adenosine Conjugates: Functionalization of Adenosine at N-1 or N-6 Position without Protecting Groups. Molecules 2020, 25, 3241. [Google Scholar] [CrossRef]
- Sau, S.P.; Hrdlicka, P.J. C2′-Pyrene-Functionalized Triazole-Linked DNA: Universal DNA/RNA Hybridization Probes. J. Org. Chem. 2012, 77, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Velema, W.A.; Kool, E.T. The chemistry and applications of RNA 2′-OH acylation. Nat. Rev. Chem. 2020, 4, 22–37. [Google Scholar] [CrossRef]
- Haugland, M.M.; El-Sagheer, A.; Porter, R.J.; Peña, J.; Brown, T.; Anderson, E.A.; Lovett, J.E. 2′-Alkynylnucleotides: A Sequence- and Spin Label-Flexible Strategy for EPR Spectroscopy in DNA. J. Am. Chem. Soc. 2016, 138, 9069–9072. [Google Scholar] [CrossRef]
- Lorenz, D.A.; Garner, A.L. A click chemistry-based microRNA maturation assay optimized for high-throughput screening. Chem. Commun. 2016, 52, 8267–8270. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 2010, 107, 15329–15334. [Google Scholar] [CrossRef]
- Aigner, A.; Hartl, M.; Fauster, K.; Steger, J.; Bister, K.; Micura, R. Chemical Synthesis of Site-Specifically 2′-Azido-Modified RNA and Potential Applications for Bioconjugation and RNA Interference. ChemBioChem 2011, 12, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Fauster, K.; Hartl, M.; Santner, T.; Aigner, M.; Kreutz, C.; Bister, K.; Ennifar, E.; Micura, R. 2′-Azido RNA, a Versatile Tool for Chemical Biology: Synthesis, X-ray Structure, siRNA Applications, Click Labeling. ACS Chem. Biol. 2012, 7, 581–589. [Google Scholar] [CrossRef]
- Staudinger, H.; Meyer, J. Über neue organische Phosphorverbindungen III. Phosphinmethylenderivate und Phosphinimine. Helv. Chim. Acta 1919, 2, 635–646. [Google Scholar] [CrossRef]
- Micura, R. Cyclic Oligoribonucleotides (RNA) by Solid-Phase Synthesis. Chem. Eur. J. 1999, 5, 2077–2082. [Google Scholar] [CrossRef]
- Mutisya, D.; Selvam, C.; Kennedy, S.D.; Rozners, E. Synthesis and properties of triazole-linked RNA. Bioorganic Med. Chem. Lett. 2011, 21, 3420–3422. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Endo, K.; Yamazaki, N.; Isobe, H. Synthesis of Triazole-linked Analogues of RNA (TLRNA). Chem. Lett. 2012, 41, 403–405. [Google Scholar] [CrossRef]
- Fujino, T.; Suzuki, T.; Ooi, T.; Ikemoto, K.; Isobe, H. Duplex-forming Oligonucleotide of Triazole-linked RNA. Chem. Asian J. 2019, 14, 3380–3385. [Google Scholar] [CrossRef]
- Paredes, E.; Das, S.R. Click chemistry for rapid labeling and ligation of RNA. ChemBioChem 2011, 12, 125–131. [Google Scholar] [CrossRef]
- Muthmann, N.; Hartstock, K.; Rentmeister, A. Chemo-enzymatic treatment of RNA to facilitate analyses. WIREs RNA 2020, 11, e1561. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, C.M.; Beal, P.A. Nucleoside analogs in the study of the epitranscriptome. Methods 2019, 156, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Meter, E.N.V.; Onyango, J.A.; Teske, K.A. A review of currently identified small molecule modulators of microRNA function. Eur. J. Med. Chem. 2020, 188, 112008. [Google Scholar] [CrossRef]
- Desaulniers, J.-P.; Hagen, G.; Anderson, J.; McKim, C.; Roberts, B. Effective gene-silencing of siRNAs that contain functionalized spacer linkages within the central region. RSC Adv. 2017, 7, 3450–3454. [Google Scholar] [CrossRef]
- Efthymiou, T.C.; Huynh, V.; Oentoro, J.; Peel, B.; Desaulniers, J.-P. Efficient synthesis and cell-based silencing activity of siRNAS that contain triazole backbone linkages. Bioorganic Med. Chem. Lett. 2012, 22, 1722–1726. [Google Scholar] [CrossRef]
- Hagen, G.; Peel, B.J.; Samis, J.; Desaulniers, J.-P. Synthesis and in vitro assessment of chemically modified siRNAs targeting BCL2 that contain 2′-ribose and triazole-linked backbone modifications. Med. Chem. Commun. 2015, 6, 1210–1215. [Google Scholar] [CrossRef]
- Piecyk, M.; Jankowska-Anyszka, M. Chemical conjugation of an mRNA cap analogue with a cell-penetrating peptide as a potential membrane permeable translation inhibitor. Tetrahedron Lett. 2014, 55, 606–609. [Google Scholar] [CrossRef]
- Shanmugasundaram, M.; Charles, I.; Kore, A.R. Design, synthesis and biological evaluation of dinucleotide mRNA cap analog containing propargyl moiety. Bioorg. Med. Chem. 2016, 24, 1204–1208. [Google Scholar] [CrossRef]
- Gillingham, D.; Shahid, R. Catalysts for RNA and DNA modification. Curr. Opin. Chem. Biol. 2015, 25, 110–114. [Google Scholar] [CrossRef]
- Warminski, M.; Sikorski, P.J.; Kowalska, J.; Jemielity, J. Applications of Phosphate Modification and Labeling to Study (m)RNA Caps. Top. Curr. Chem. 2017, 375, 16. [Google Scholar] [CrossRef]
- Mamot, A.; Sikorski, P.J.; Warminski, M.; Kowalska, J.; Jemielity, J. Azido-Functionalized 5’ Cap Analogues for the Preparation of Translationally Active mRNAs Suitable for Fluorescent Labeling in Living Cells. Angew. Chem. Int. Ed. 2017, 56, 15628–15632. [Google Scholar] [CrossRef]
- Walczak, S.; Nowicka, A.; Kubacka, D.; Fac, K.; Wanat, P.; Mroczek, S.; Kowalska, J.; Jemielity, J. A novel route for preparing 5′ cap mimics and capped RNAs: Phosphate-modified cap analogues obtained via click chemistry. Chem. Sci. 2017, 8, 260–267. [Google Scholar] [CrossRef]
- A Nobel Prize for genetic scissors. Nat. Mater. 2021, 20, 1. [CrossRef]
- Fernholm, A. Genetic Scissors: A Tool for Rewriting the Code of Life; The Royal Swedish Academy of Sciences: Stockholm, Sweden, 2020. [Google Scholar]
- Hagedorn, P.H.; Persson, R.; Funder, E.D.; Albæk, N.; Diemer, S.L.; Hansen, D.J.; Møller, M.R.; Papargyri, N.; Christiansen, H.; Hansen, B.R.; et al. Locked nucleic acid: Modality, diversity, and drug discovery. Drug Discov. Today 2018, 23, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Braasch, D.A.; Corey, D.R. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001, 8, 1–7. [Google Scholar] [CrossRef]
- Campbell, M.A.; Wengel, J. Locked vs. unlocked nucleic acids (LNA vs.UNA): Contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011, 40, 5680–5689. [Google Scholar] [CrossRef]
- Imanishi, T.; Obika, S. BNAs: Novel nucleic acid analogs with a bridged sugar moiety. Chem. Commun. 2002, 1653–1659. [Google Scholar] [CrossRef]
- Mitsuoka, Y.; Yamamoto, T.; Kugimiya, A.; Waki, R.; Wada, F.; Tahara, S.; Sawamura, M.; Noda, M.; Fujimura, Y.; Kato, Y.; et al. Triazole- and Tetrazole-Bridged Nucleic Acids: Synthesis, Duplex Stability, Nuclease Resistance, and in Vitro and in Vivo Antisense Potency. J. Org. Chem. 2017, 82, 12–24. [Google Scholar] [CrossRef]
- Obika, S.; Rahman, S.M.A.; Fujisaka, A.; Kaawada, Y.; Baba, T.; Imanishi, T. Bridged Nucleic Acids: Development, Synthesis and Properties. Heterocycles 2010, 81, 1347–1392. [Google Scholar] [CrossRef]
- Hari, Y. Bridged Nucleosides as Building Blocks of Oligonucleotides: Synthesis and Properties. Heterocycles 2020, 100, 681–717. [Google Scholar] [CrossRef]
- Bryld, T.; Sørensen, M.H.; Nielsen, P.; Koch, T.; Nielsen, C.; Wengel, J. Synthesis and antiviral evaluation of novel conformationally locked nucleosides and masked 5′-phosphate derivatives thereof. J. Chem. Soc. Perkin Trans. 1 2002, 14, 1655–1662. [Google Scholar] [CrossRef]
- Sharma, V.K.; Rungta, P.; Maikhuri, V.K.; Prasad, A.K. An astute synthesis of locked nucleic acid monomers. Sustain. Chem. Process. 2015, 3, 2. [Google Scholar] [CrossRef][Green Version]
- Sharma, V.K.; Singh, S.K.; Krishnamurthy, P.M.; Alterman, J.F.; Haraszti, R.A.; Khvorova, A.; Prasad, A.K.; Watts, J.K. Synthesis and biological properties of triazole-linked locked nucleic acid. Chem. Commun. 2017, 53, 8906–8909. [Google Scholar] [CrossRef]
- Yamashita, S.; Nishida, K.; Osawa, T.; Nakanishi, A.; Ito, J.; Hari, Y. Synthesis of Oligonucleotides Containing 2′-N-alkylaminocarbonyl-2′-amino-LNA (2′-urea-LNA) Moieties Using Post-Synthetic Modification Strategy. Molecules 2020, 25, 346. [Google Scholar] [CrossRef]
- Singh, S.K.; Sharma, V.K.; Bohra, K.; Olsen, C.E.; Prasad, A.K. Synthesis of Triazole-linked LNA-based Non-ionic Nucleoside Dimers Using Cu(I)- Catalyzed ‘Click’ Reaction. Curr. Org. Synth. 2014, 11, 757–766. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, S.K.; Sharma, V.K.; Mangla, P.; Olsen, C.E.; Prasad, A.K. Design and Synthesis of Triazole-Linked xylo-Nucleoside Dimers. Nucleos. Nucleot. Nucl. 2015, 34, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; El-Sagheer, A.H.; Truong, L.; Brown, T. Locked nucleic acid (LNA) enhances binding affinity of triazole-linked DNA towards RNA. Chem. Commun. 2017, 53, 8910–8913. [Google Scholar] [CrossRef]
- Kumar, P.; Truong, L.; Baker, Y.R.; El-Sagheer, A.H.; Brown, T. ; Synthesis, Affinity for Complementary RNA and DNA, and Enzymatic Stability of Triazole-Linked Locked Nucleic Acids (t-LNAs). ACS Omega 2018, 3, 6976–6987. [Google Scholar] [CrossRef]
- Larsen, H.J.; Bentin, T.; Nielsen, P.E. Antisense properties of peptide nucleic acid. Biochim. Biophys. Acta 1999, 1489, 159–166. [Google Scholar] [CrossRef]
- Soomets, U.; Hällbrink, M.; Langel, Ü. Antisense properties of peptide nucleic acid. Front. Biosci. 1999, 4, 782–786. [Google Scholar] [CrossRef]
- Četojević-Simin, D.; Jakimov, D.; Mrđanović, J.; Bogdanović, V.; Kojić, V.; Andrijević, L.; Bogdanović, G. Peptide nucleic acid: Sequence specific recognition in cancer diagnostics and gene therapy. Arch. Oncol. 2001, 9, 33–37. [Google Scholar]
- Piacenti, V.; Langella, E.; Autiero, I.; Nolan, J.C.; Piskareva, O.; Adamo, M.F.A.; Saviano, M.; Moccia, M. A combined experimental and computational study on peptide nucleic acid (PNA) analogues of tumor suppressive miRNA-34a. Bioorg. Chem. 2019, 91, 103165. [Google Scholar] [CrossRef]
- Holub, J.M.; Kirshenbaum, K. Tricks with clicks: Modification of peptidomimetic oligomers via copper-catalyzed azide-alkyne [3 + 2] cycloaddition. Chem. Soc. Rev. 2010, 39, 1325–1337. [Google Scholar] [CrossRef]
- Amant, A.H.S.; Engbers, C.; Hudson, R.H.E. A solid-phase CuAAC strategy for the synthesis of PNA containing nucleobase surrogates. Artif. Dna Pna Xna 2013, 4, 4–10. [Google Scholar] [CrossRef][Green Version]
- Chouikhi, D.; Barluenga, S.; Winssinger, N. Clickable peptide nucleic acids (cPNA) with tunable affinity. Chem. Commun. 2010, 46, 5476–5478. [Google Scholar] [CrossRef]
- Thomson, S.A.; Josey, J.A.; Cadilla, R.; Gaul, M.D.; Hassman, C.F.; Luzzio, M.J.; Pipe, A.J.; Reed, K.L.; Ricca, D.J.; Wiethe, R.W.; et al. Fmoc mediated synthesis of Peptide Nucleic Acids. Tetrahedron 1995, 51, 6179–6194. [Google Scholar] [CrossRef]
- Efthymiou, T.C.; Desaulniers, J.-P. Synthesis and properties of oligonucleotides that contain a triazole-linked nucleic acid dimer. J. Heterocyclic Chem. 2011, 48, 533–539. [Google Scholar] [CrossRef]
- Vergnaud, J.; Faugerasa, P.-A.; Chaleix, V.; Champavier, Y.; Zerrouki, R. Design of a new oligotriazole peptide nucleic acid analogue (oT-PNA). Tetrahedron Lett. 2011, 52, 6185–6189. [Google Scholar] [CrossRef]
- Marin, V.L.; Armitage, B.A. RNA Guanine Quadruplex Invasion by Complementary and Homologous PNA Probes. J. Am. Chem. Soc. 2005, 127, 8032–8033. [Google Scholar] [CrossRef]
- Gudanis, D.; Kaniowski, D.; Kulik, K.; Baranowski, D.; Gdaniec, Z.; Nawrot, B. Formation of an RNA Quadruplex-Duplex Hybrid in Living Cells between mRNA of the Epidermal Growth Factor Receptor (EGFR) and a G-Rich Antisense Oligoribonucleotide. Cells 2020, 9, 2375. [Google Scholar] [CrossRef]
- Solier, S.; Müller, S.; Rodriguez, R. Whole-genome mapping of small-molecule targets for cancer medicine. Curr. Opin. Chem. Biol. 2020, 56, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. 2003, 42, 2946–2974. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Xu, Y.; Suzuki, Y.; Komiyama, M. Click chemistry for the identification of G-quadruplex structures: Discovery of a DNA-RNA G-quadruplex. Angew. Chem. Int. Ed. 2009, 48, 3281–3284. [Google Scholar] [CrossRef]
- Saha, P.; Panda, D.; Dash, J. The application of click chemistry for targeting quadruplex nucleic acids. Chem. Commun. 2019, 55, 731–750. [Google Scholar] [CrossRef]
- Varizhuk, A.M.; Tsvetkov, V.B.; Tatarinova, O.N.; Kaluzhny, D.N.; Florentiev, V.L.; Timofeev, E.N.; Shchyolkina, A.K.; Borisova, O.F.; Smirnov, I.P.; Grokhovsky, S.L.; et al. Synthesis, characterization and in vitro activity of thrombin-binding DNA aptamers with triazole internucleotide linkages. Eur. J. Med. Chem. 2013, 67, 90–97. [Google Scholar] [CrossRef]
- Xu, Y.; Ishizuka, T.; Yang, J.; Ito, K.; Katada, H.; Komiyama, M.; Hayashi, T. Oligonucleotide Models of Telomeric DNA and RNA Form a Hybrid G-quadruplex Structure as a Potential Component of Telomeres. J. Biol. Chem. 2012, 287, 41787–41796. [Google Scholar] [CrossRef]
- Xu, Y.; Suzuki, Y.; Ishizuka, T.; Xiao, C.-D.; Liu, X.; Hayashi, T. Finding a human telomere DNA–RNA hybrid G-quadruplex formed by human telomeric 6-mer RNA and 16-mer DNA using click chemistry: A protective structure for telomere end. Bioorg. Med. Chem. 2014, 22, 4419–4421. [Google Scholar] [CrossRef]
- Ishizuka, T.; Xu, Y. Click Chemistry Takes a Snapshot of DNA-RNA Hybrid G-Quadruplex in Living Cells. J. Nat. Sci. 2016, 2, e237. [Google Scholar]
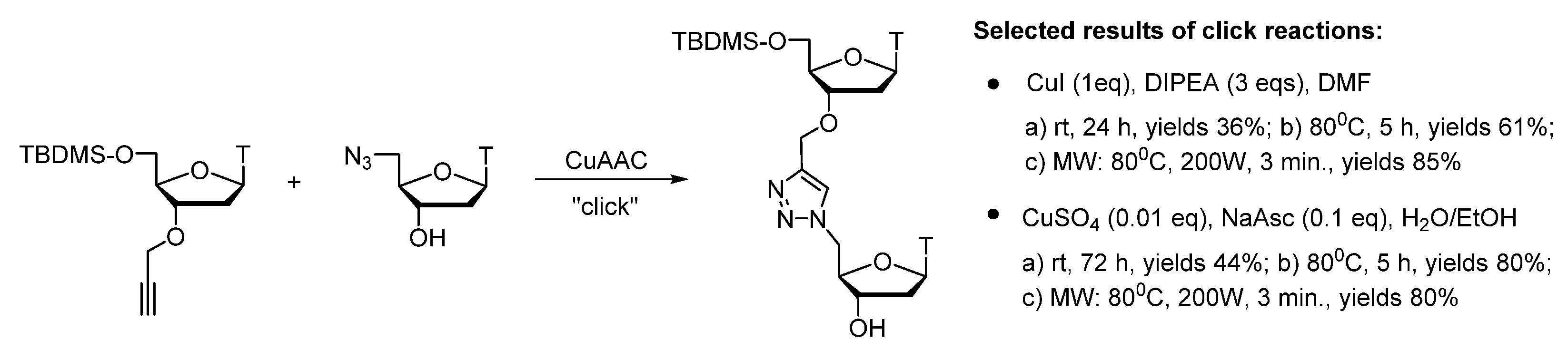

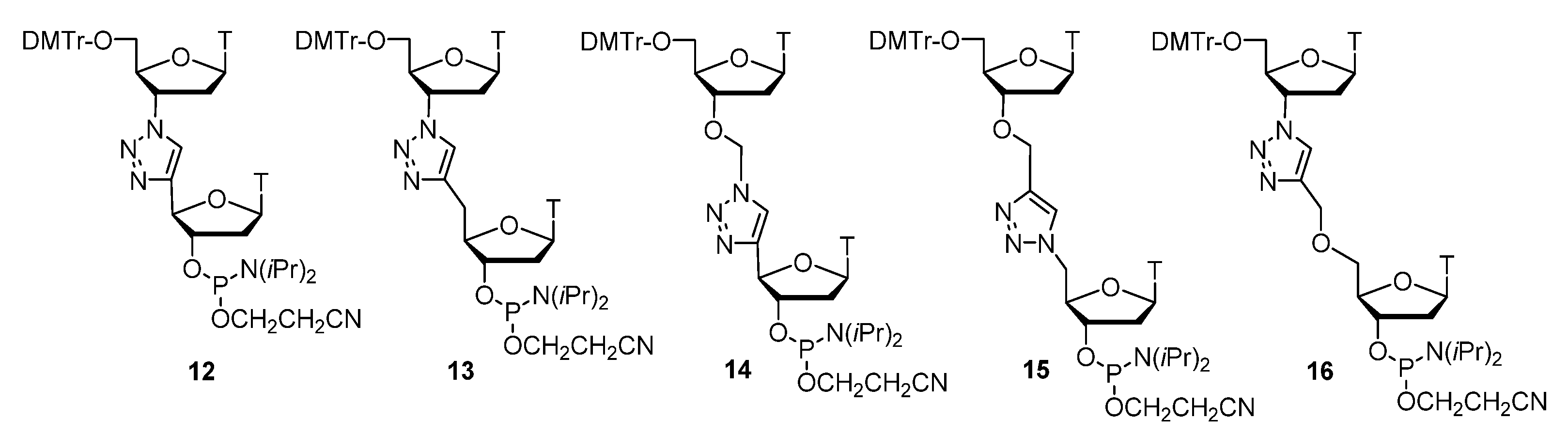
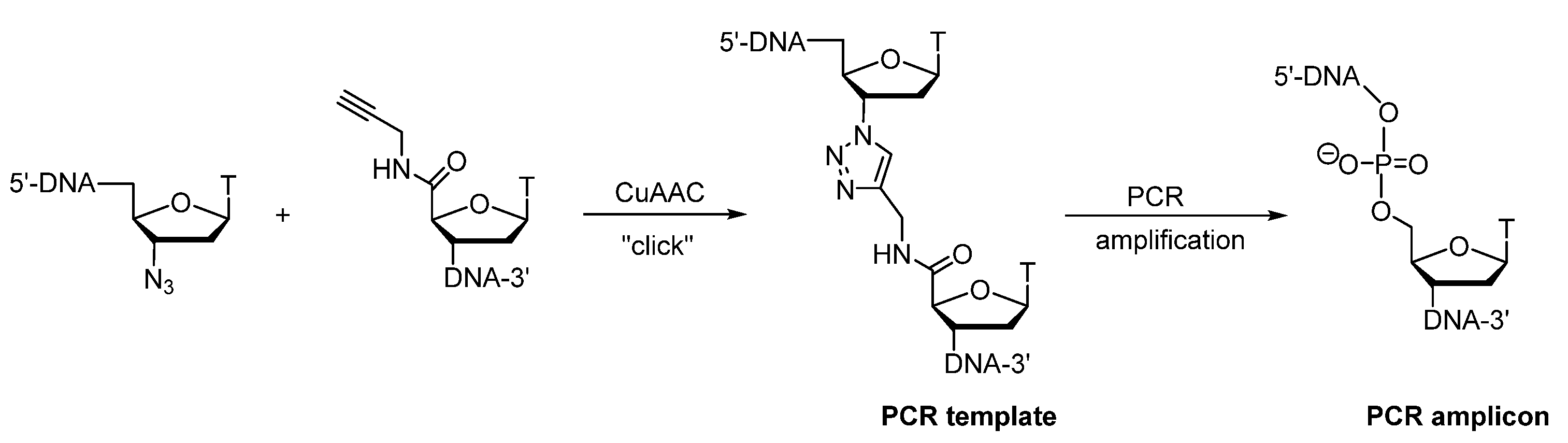
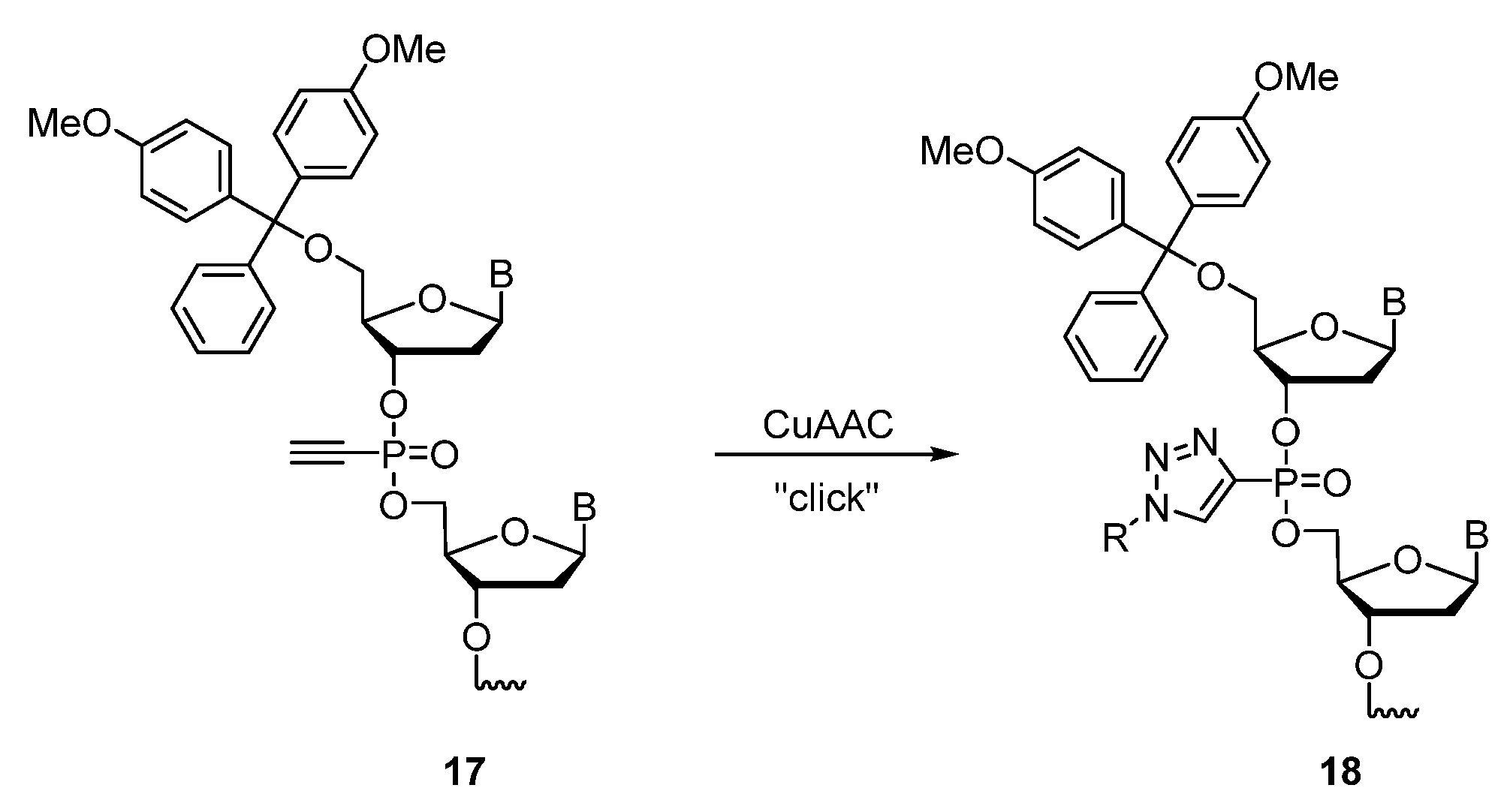


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baraniak, D.; Boryski, J. Triazole-Modified Nucleic Acids for the Application in Bioorganic and Medicinal Chemistry. Biomedicines 2021, 9, 628. https://doi.org/10.3390/biomedicines9060628
Baraniak D, Boryski J. Triazole-Modified Nucleic Acids for the Application in Bioorganic and Medicinal Chemistry. Biomedicines. 2021; 9(6):628. https://doi.org/10.3390/biomedicines9060628
Chicago/Turabian StyleBaraniak, Dagmara, and Jerzy Boryski. 2021. "Triazole-Modified Nucleic Acids for the Application in Bioorganic and Medicinal Chemistry" Biomedicines 9, no. 6: 628. https://doi.org/10.3390/biomedicines9060628
APA StyleBaraniak, D., & Boryski, J. (2021). Triazole-Modified Nucleic Acids for the Application in Bioorganic and Medicinal Chemistry. Biomedicines, 9(6), 628. https://doi.org/10.3390/biomedicines9060628





