Pursuing Orally Bioavailable Hepcidin Analogues via Cyclic N-Methylated Mini-Hepcidins
Abstract
1. Introduction
2. Experimental Section
2.1. General Methods
2.2. Peptide Synthesis
2.3. In Vitro Peptide Bioactivity Assay
2.4. Serum Stability Assay
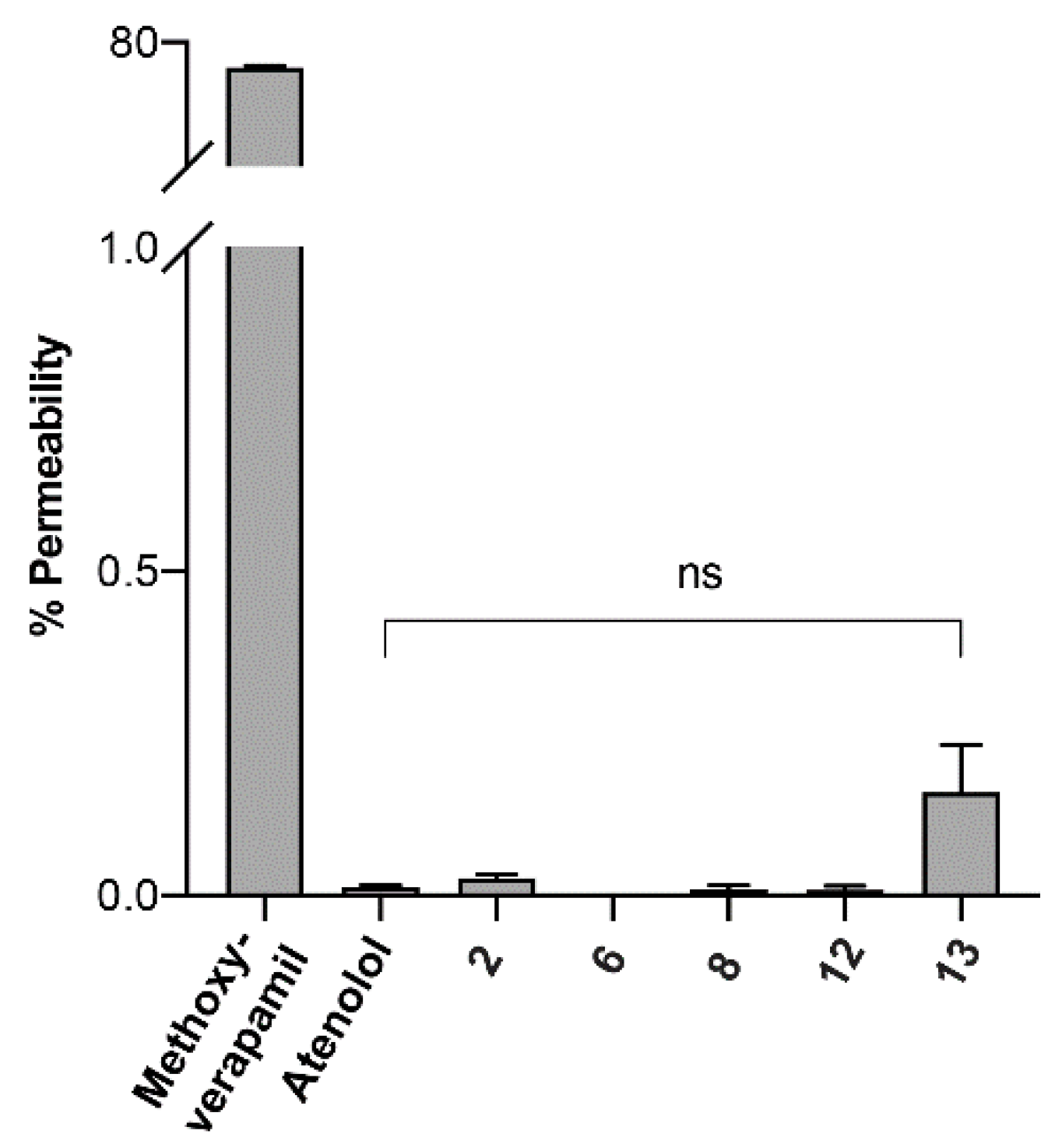
2.5. Parallel Artificial Membrane Permeability Assay (PAMPA)
3. Results
3.1. Peptide Design, Synthesis and Biological Activity
3.2. Structural Comparison of Acyclic and Cyclic N-Methylated Mini-Hepcidins
3.3. cHep9 and N-Methylated Mini-Hepcidins Show Improved Serum Stability
3.4. Cell Permeability of Rationally Designed N-Methylated Mini-Hepcidins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pietrangelo, A. Hepcidin in human iron disorders: Therapeutic implications. J. Hepatol. 2011, 54, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Pantopoulos, K.; Porwal, S.K.; Tartakoff, A.; Devireddy, L. Mechanisms of Mammalian Iron Homeostasis. Biochemistry 2012, 51, 5705–5724. [Google Scholar] [CrossRef] [PubMed]
- Aschemeyer, S.; Qiao, B.; Stefanova, D.; Valore, E.V.; Sek, A.C.; Ruwe, T.A.; Vieth, K.R.; Jung, G.; Casu, C.; Rivella, S.; et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood 2018, 131, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Vela, D. Hepcidin, an emerging and important player in brain iron homeostasis. J. Transl. Med. 2018, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tesfay, L.; Clausen, K.A.; Kim, J.W.; Hegde, P.; Wang, X.; Miller, L.D.; Deng, Z.; Blanchette, N.; Arvedson, T.; Miranti, C.K.; et al. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res. 2015, 75, 2254–2263. [Google Scholar] [CrossRef]
- Lakhal-Littleton, S.; Wolna, M.; Chung, Y.J.; Christian, H.C.; Heather, L.C.; Brescia, M.; Ball, V.; Diaz, R.; Santos, A.; Biggs, D.; et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. eLife 2016, 5, e19804. [Google Scholar] [CrossRef]
- Ramos, E.; Ruchala, P.; Goodnough, J.B.; Kautz, L.; Preza, G.C.; Nemeth, E.; Ganz, T. Minihepcidins prevent iron over-load in a hepcidin-deficient mouse model of severe hemochromatosis. Blood 2012, 120, 3829–3836. [Google Scholar] [CrossRef]
- Casu, C.; Oikonomidou, P.R.; Chen, H.; Nandi, V.; Ginzburg, Y.; Prasad, P.; Fleming, R.E.; Shah, Y.M.; Valore, E.V.; Nemeth, E.; et al. Minihepcidin peptides as disease modifiers in mice affected by be-ta-thalassemia and polycythemia vera. Blood 2016, 128, 265–276. [Google Scholar] [CrossRef]
- Valore, E.V.; Ganz, T. Posttranslational processing of hepcidin in human hepatocytes is mediated by the prohormone convert-ase furin. Blood Cells Mol. Dis. 2008, 40, 132–138. [Google Scholar] [CrossRef]
- Clark, R.J.; Tan, C.C.; Preza, G.C.; Nemeth, E.; Ganz, T.; Craik, D.J. Understanding the Structure/Activity Relationships of the Iron Regulatory Peptide Hepcidin. Chem. Biol. 2011, 18, 336–343. [Google Scholar] [CrossRef]
- Nemeth, E.; Preza, G.C.; Jung, C.L.; Kaplan, J.; Waring, A.J.; Ganz, T. The N-terminus of hepcidin is essential for its in-teraction with ferroportin: Structure-function study. Blood 2006, 107, 328–333. [Google Scholar] [CrossRef]
- Preza, G.C.; Ruchala, P.; Pinon, R.; Ramos, E.; Qiao, B.; Peralta, M.A.; Sharma, S.; Waring, A.; Ganz, T.; Nemeth, E. Minihepcidins are rationally designed small peptides that mimic hepcidin activity in mice and may be useful for the treatment of iron overload. J. Clin. Investig. 2011, 121, 4880–4888. [Google Scholar] [CrossRef] [PubMed]
- La Jolla Pharmaceutical Company. La Jolla Pharmaceutical Company Reports Positive Results from Phase 1 Study of LJPC-401; La Jolla Pharmaceutical Company: San Diego, CA, USA, 2016. [Google Scholar]
- Merganser Biotech Inc. Merganser Biotech Inc. Initiates First Clinical Trial; Business Wire: San Francisco, CA, USA, 2016. [Google Scholar]
- Protagonist Therapeutics. Protagonist Therapeutics Initiates Phase 1 Study with Novel Hepcidin Mimetic, PTG-300; PR Newswire: New York, NY, USA, 2017. [Google Scholar]
- Protagonist Therapeutics. Protagonist Therapeutics Initiates Phase 2 Trial of Novel Hepcidin Mimetic PTG-300 for the Treatment of Patients with Beta Tha-Lassemia; PR Newswire: New York, NY, USA, 2019. [Google Scholar]
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin Revisited, Disulfide Connectivity, Dynamics, and Structure. J. Biol. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef] [PubMed]
- Borel, J.F.; Feurer, C.; Gubler, H.U.; Stähelin, H. Biological effects of cyclosporin A: A new antilymphocytic agent. Agents Actions 1976, 6, 468–475. [Google Scholar] [CrossRef]
- Naylor, M.R.; Bockus, A.T.; Blanco, M.-J.; Lokey, R.S. Cyclic peptide natural products chart the frontier of oral bioavailability in the pursuit of undruggable targets. Curr. Opin. Chem. Biol. 2017, 38, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.K.; Craik, D.J. Cyclic peptide oral bioavailability: Lessons from the past. Pept. Sci. 2016, 106, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Shepherd, N.E.; Xu, W.; Lucke, A.J.; Stoermer, M.J.; Fairlie, D.P. Orally Absorbed Cyclic Peptides. Chem. Rev. 2017, 117, 8094–8128. [Google Scholar] [CrossRef] [PubMed]
- Okumu, F.W.; Pauletti, G.M.; Vander Velde, D.G.; Siahaan, T.J.; Borchardt, R.T. Effect of restricted conformational flexibility on the permeation of model hexapeptides across Caco-2 cell monolayers. Pharm. Res. 1997, 14, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Pauletti, G.M.; Gangwar, S.; Knipp, G.T.; Nerurkar, M.M.; Okumu, F.W.; Tamura, K.; Siahaan, T.J.; Borchardt, R.T. Structural requirements for intestinal absorption of peptide drugs. J. Control. Release 1996, 41, 3–17. [Google Scholar] [CrossRef]
- Doedens, L.; Opperer, F.; Cai, M.; Beck, J.G.; Dedek, M.; Palmer, E.; Hruby, V.J.; Kessler, H. Multiple N-Methylation of MT-II Backbone Amide Bonds Leads to Melanocortin Receptor Subtype hMC1R Selectivity: Pharmacological and Conformational Studies. J. Am. Chem. Soc. 2010, 132, 8115–8128. [Google Scholar] [CrossRef]
- Biron, E.; Chatterjee, J.; Ovadia, O.; Langenegger, D.; Brueggen, J.; Hoyer, D.; Schmid, H.A.; Jelinek, R.; Gilon, C.; Hoffman, A.; et al. Improving Oral Bioavailability of Peptides by Multiple N-Methylation: Somatostatin Analogues. Angew. Chem. Int. Ed. 2008, 47, 2595–2599. [Google Scholar] [CrossRef]
- Wang, C.K.; Northfield, S.E.; Colless, B.; Chaousis, S.; Hamernig, I.; Lohman, R.-J.; Nielsen, D.S.; Schroeder, C.I.; Liras, S.; Price, D.A.; et al. Rational design and synthesis of an orally bioavailable peptide guided by NMR amide temperature coefficients. Proc. Natl. Acad. Sci. USA 2014, 111, 17504–17509. [Google Scholar] [CrossRef]
- Skinner, S.P.; Fogh, R.H.; Boucher, W.; Ragan, T.J.; Mureddu, L.G.; Vuister, G.W. CcpNmr AnalysisAssign: A flexible platform for integrated NMR analysis. J. Biomol. NMR 2016, 66, 111–124. [Google Scholar] [CrossRef]
- Shelton, P.T.; Jensen, K.J. Linkers, Resins, and General Procedures for Solid-Phase Peptide Synthesis. In Advanced Structural Safety Studies; Springer Nature: Cham, Switzerland, 2013; Volume 1047, pp. 23–41. [Google Scholar]
- Qiao, B.; Sugianto, P.; Fung, E.; Del-Castillo-Rueda, A.; Moran-Jimenez, M.-J.; Ganz, T.; Nemeth, E. Hepcidin-Induced Endocytosis of Ferroportin Is Dependent on Ferroportin Ubiquitination. Cell Metab. 2012, 15, 918–924. [Google Scholar] [CrossRef]
- Cierpicki, T.; Otlewski, J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 2001, 21, 249–261. [Google Scholar] [CrossRef]
- Roodbeen, R.; Pedersen, S.L.; Hosseini, M.; Jensen, K.J. Microwave Heating in the Solid-Phase Synthesis of N-Methylated Peptides: When Is Room Temperature Better? Eur. J. Org. Chem. 2012, 2012, 7106–7111. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Piriou, F.; Lintner, K.; Fermandjian, S.; Fromageot, P.; Khosla, M.C.; Smeby, R.R.; Bumpus, F.M. Amino acid side chain conformation in angiotensin II and analogs: Correlated results of circular dichroism and 1H nuclear magnetic resonance. Proc. Natl. Acad. Sci. USA 1980, 77, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Peslova, G.; Petrak, J.; Kuzelova, K.; Hrdy, I.; Halada, P.; Kuchel, P.W.; Soe-Lin, S.; Ponka, P.; Sutak, R.; Becker, E.; et al. Hepcidin, the hormone of iron metabolism, is bound specifically to alpha-2-macroglobulin in blood. Blood 2009, 113, 6225–6236. [Google Scholar] [CrossRef]
- Huang, M.L.-H.; Austin, C.J.D.; Sari, M.-A.; Rahmanto, Y.S.; Ponka, P.; Vyoral, D.; Richardson, D.R. Hepcidin Bound to α2-Macroglobulin Reduces Ferroportin-1 Expression and Enhances Its Activity at Reducing Serum Iron Levels. J. Biol. Chem. 2013, 288, 25450–25465. [Google Scholar] [CrossRef]
- Fasano, A. Modulation of Intestinal Permeability: An Innovative Method of Oral Drug Delivery for the Treatment of Inherited and Acquired Human Diseases. Mol. Genet. Metab. 1998, 64, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, J.; Gilon, C.; Hoffman, A.; Kessler, H. N-Methylation of Peptides: A New Perspective in Medicinal Chemistry. Acc. Chem. Res. 2008, 41, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Conradi, R.A.; Hilgers, A.R.; Ho, N.F.H.; Burton, P.S. The Influence of Peptide Structure on Transport Across Caco-2 Cells. II. Peptide Bond Modification Which Results in Improved Permeability. Pharm. Res. 1992, 9, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Barker, P.L.; Bullens, S.; Bunting, S.; Burdick, D.J.; Chan, K.S.; Deisher, T.; Eigenbrot, C.; Gadek, T.R.; Gantzos, R.; Lipari, M.T.; et al. Cyclic RGD peptide analogues as antiplatelet antithrombotics. J. Med. Chem. 1992, 35, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Parquet, N.; Reigneau, O.; Humbert, H.; Guignard, M.; Ribaud, P.; Socié, G.; Devergie, A.; Espérou, H.; Gluckman, E. New oral formulation of cyclosporin A (Neoral) pharmacokinetics in allogeneic bone marrow transplant recipients. Bone Marrow Transplant. 2000, 25, 965–968. [Google Scholar] [CrossRef]
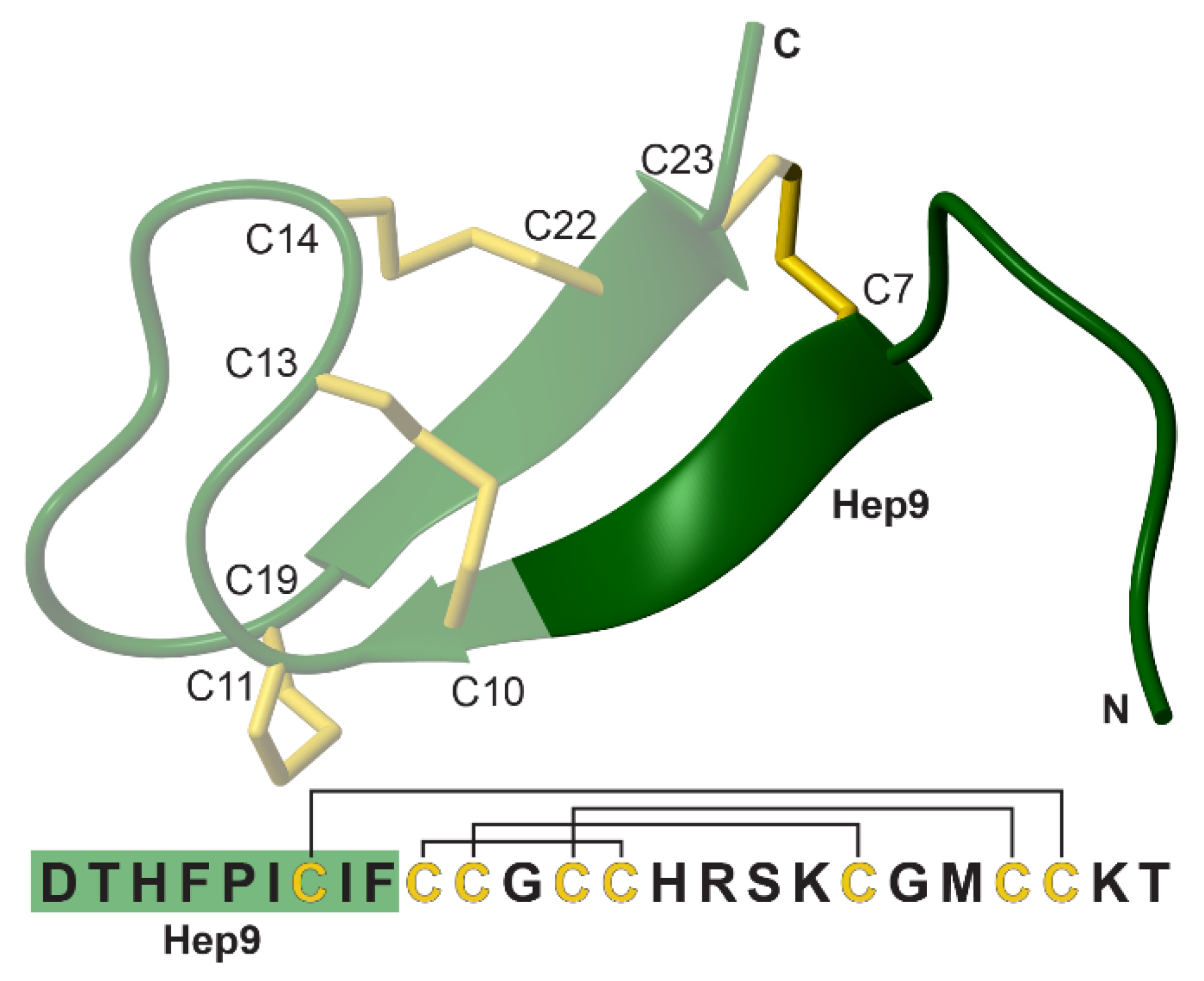
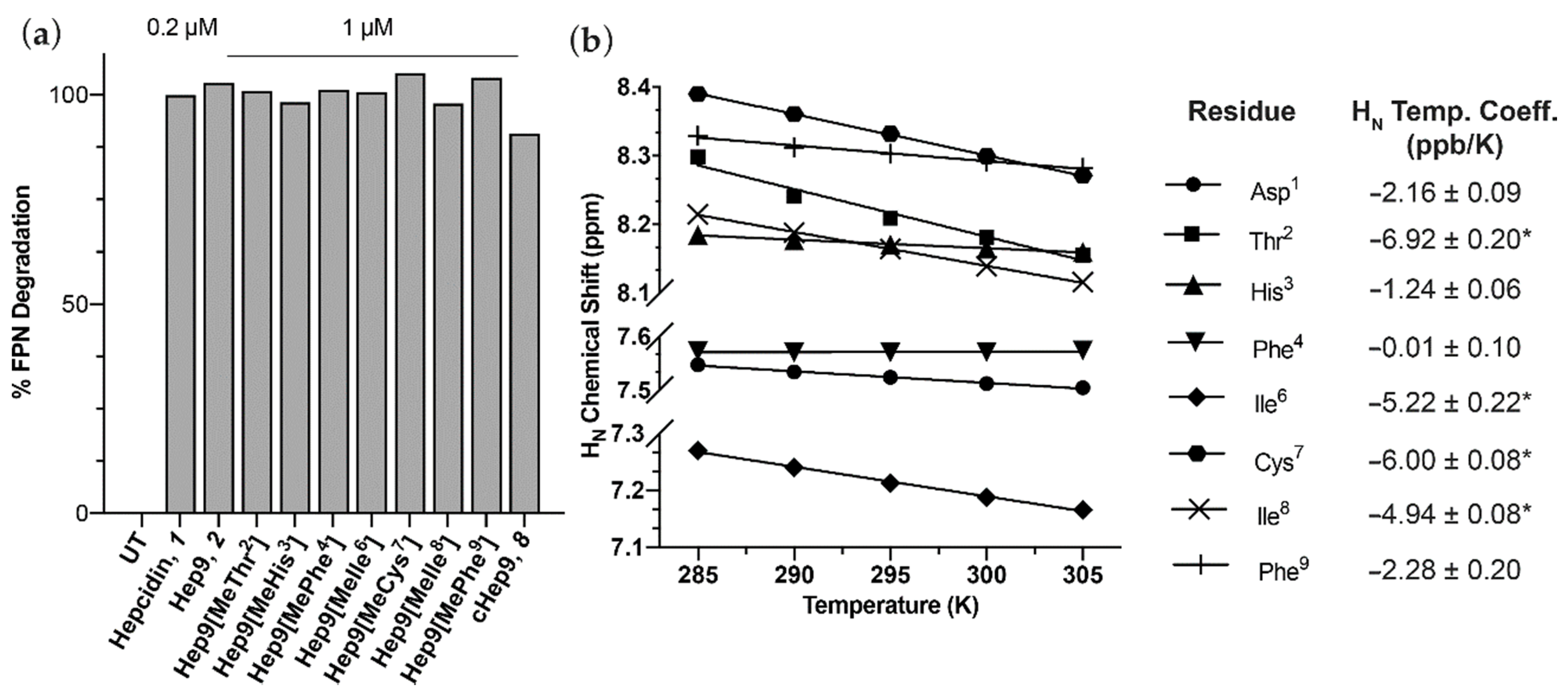
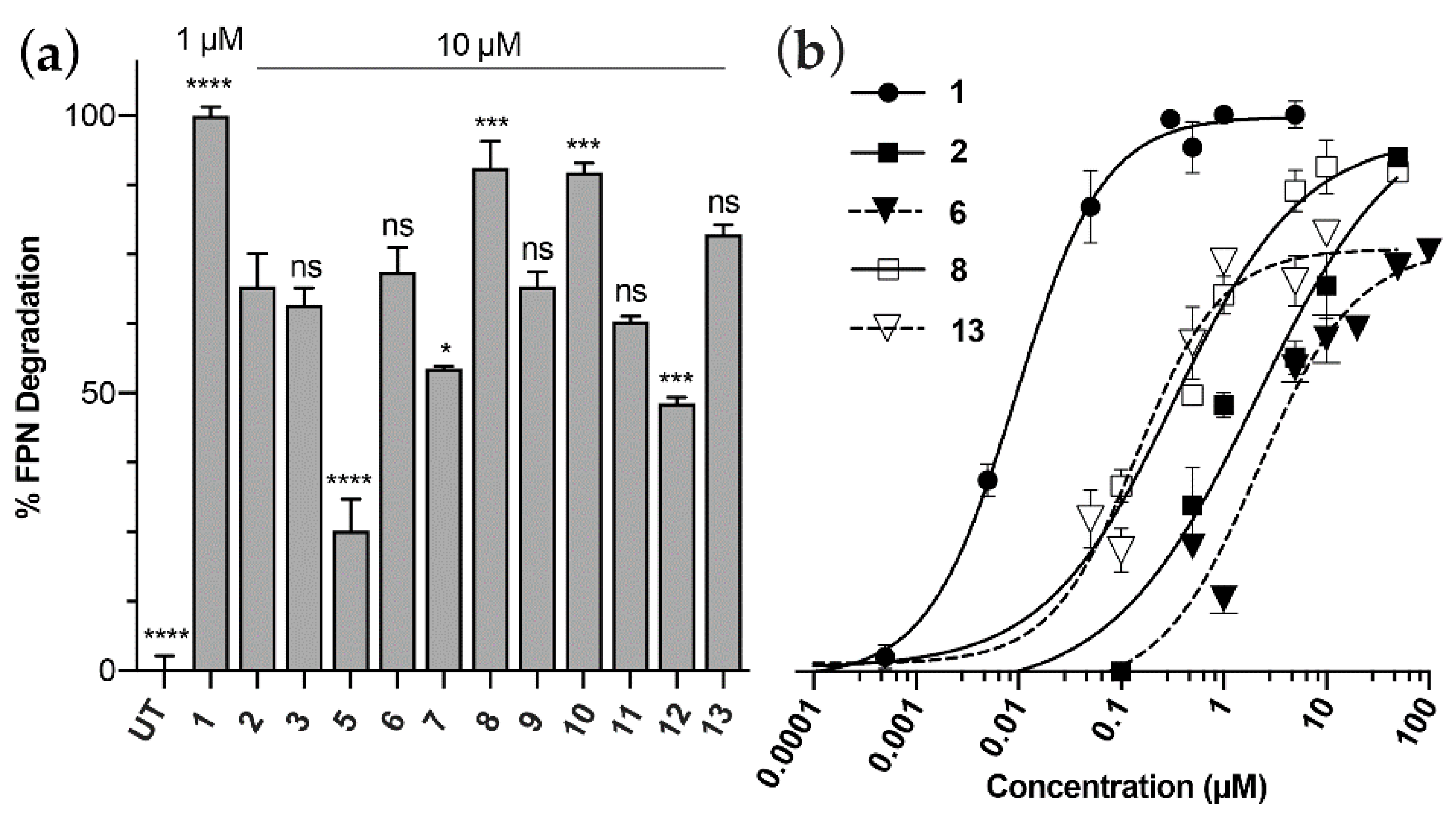
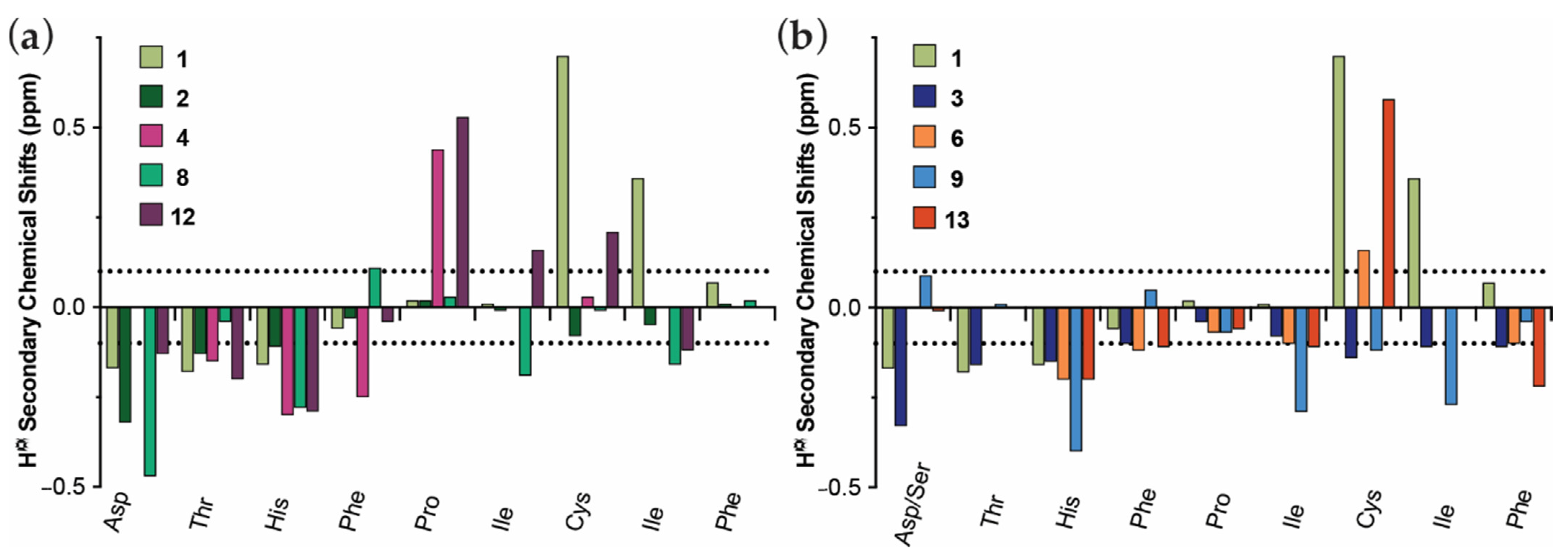
| # | Peptide | Sequence 1 | FPN Degradation % ± SEM 2 | pEC50 ± SEM |
|---|---|---|---|---|
| 1 | Hepcidin | DTHFPICIFCCGCCHRSKCGMCCKT | 100 ± 2.6 | 8.028 ± 0.081 |
| 2 | Hep9 | DTHFPICIF | 69 ± 10 | 5.676 ± 0.296 |
| 3 | Hep9[Ser1] | STHFPICIF | 66 ± 5.3 | 6.487 ± 0.124 |
| 4 | Hep9[MeIle6, MeIle8] | DTHFP[N-Me I]C[N-Me I]F | Not active | - |
| 5 | Hep9[Ser1, MeThr2, MeIle6] | S[N-Me T]HFP[N-Me I]CIF | 25 ± 9.9 | 4.39 ± 0.982 |
| 6 | Hep9[Ser1, MeThr2, MeIle8] | S[N-Me T]HFPIC[N-Me I]F | 72 ± 7.7 | 5.663 ± 0.117 |
| 7 | Hep9[Ser1, MeThr2, MeIle6, MeIle8] | S[N-Me T]HFP[N-Me I]C[N-Me I]F | 54 ± 0.8 | 5.723 ± 0.117 |
| 8 | cHep9 | c(DTHFPICIF) | 91 ± 8.2 | 6.487 ± 0.124 |
| 9 | cHep9[Ser1] | c(STHFPICIF) | 69 ± 4.4 | 6.575 ± 0.096 |
| 10 | cHep9-Gly4 | c(DTHFPICIFGGGG) | 90 ± 2.8 | 6.800 ± 0.072 |
| 11 | cHep9[MePhe4, MePhe9] | c(DTH[N-Me F]PICI[N-Me F]) | 63 ± 1.6 | - |
| 12 | cHep9[MeIle6, MeIle8] | c(DTHFP[N-Me I]C[N-Me I]F) | 48 ± 1.9 | 5.537 ± 0.162 |
| 13 | cHep9[Ser1, MeThr2, MeIle8] | c(S[N-Me T]HFPIC[N-Me I]F) | 79 ± 3.0 | 6.853 ± 0.129 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goncalves Monteiro, D.; van Dijk, J.W.A.; Aliyanto, R.; Fung, E.; Nemeth, E.; Ganz, T.; Rosengren, J.; Clark, R.J. Pursuing Orally Bioavailable Hepcidin Analogues via Cyclic N-Methylated Mini-Hepcidins. Biomedicines 2021, 9, 164. https://doi.org/10.3390/biomedicines9020164
Goncalves Monteiro D, van Dijk JWA, Aliyanto R, Fung E, Nemeth E, Ganz T, Rosengren J, Clark RJ. Pursuing Orally Bioavailable Hepcidin Analogues via Cyclic N-Methylated Mini-Hepcidins. Biomedicines. 2021; 9(2):164. https://doi.org/10.3390/biomedicines9020164
Chicago/Turabian StyleGoncalves Monteiro, Daniela, Johannes W. A. van Dijk, Randy Aliyanto, Eileen Fung, Elizabeta Nemeth, Tomas Ganz, Johan Rosengren, and Richard J. Clark. 2021. "Pursuing Orally Bioavailable Hepcidin Analogues via Cyclic N-Methylated Mini-Hepcidins" Biomedicines 9, no. 2: 164. https://doi.org/10.3390/biomedicines9020164
APA StyleGoncalves Monteiro, D., van Dijk, J. W. A., Aliyanto, R., Fung, E., Nemeth, E., Ganz, T., Rosengren, J., & Clark, R. J. (2021). Pursuing Orally Bioavailable Hepcidin Analogues via Cyclic N-Methylated Mini-Hepcidins. Biomedicines, 9(2), 164. https://doi.org/10.3390/biomedicines9020164







