1. Introduction
The structural homeostasis is challenging for sensory neurons, whose axons may extend up to more than one meter in humans and with an axonal volume reaching over a thousand times that of the cell body. In a typical mammalian cell, the gene, mRNA, translating machinery and protein destination are a few micrometers apart but the immense size of the axonal compartment challenges the capacity of the neuronal soma to supply and support the constant protein requirements of an entire nerve fiber. A fast microtubule-assisted transport machinery has been identified, conveying material at 20–40 cm/day. However, the capacity of the fast transport is orders of magnitude lower than bulk diffusion (at 1 mm/day), and the cargo seems to be mainly destined for the axon terminals, whereas intrinsic proteins of the axoplasm are not significantly represented [
1]. A body of evidence indicates that Schwann cells (SCs) support the axonal maintenance and regenerative responses by diverse mechanisms of cell–cell communication: SCs regulate a wide variety of axonal functions [
2], including passive functions associated with myelin formation with subsequent increase in the conduction velocity [
3], and more active roles such as enrichment of sodium channels at the nodes of Ranvier [
4], specification of the internodal distance [
5] as well as metabolic maintenance of the axonal compartment [
6]. The regulation of neuronal form and function by SCs has been found to be mediated by different forms of intercellular communication, including coupling via local currents in the periaxonal space, paracrine signaling (e.g., ATP, glutamate) and physical coupling via adhesion molecules and gap junctions [
7]. In addition to these classic mechanisms, recent findings suggest the occurrence of lateral molecular cargo transfer mediated by secreted extracellular vesicles (EVs) from SCs to axons [
8,
9,
10]. EVs constitute mainly two types of vesicles: exosomes and microvesicles, which are generated by all cell types and derive from multivesicular bodies or through bubbling of the plasma membrane, respectively [
11]. They contain, and are able to transport, proteins, lipids and genetic material such as DNA and RNA with variations in cargo composition depending on, e.g., the age, metabolic state and type of the donor cell [
12]. Therefore, in addition to export of cell waste [
13], EV-mediated intercellular signaling is an essential component of regulatory neuro-glial communication. In this regard, SC-derived exosomes were found to be internalized by peripheral axons, increasing in vitro neurite sprouting of sensory neurons as well as in vivo axonal regeneration by 50% following nerve crush injury [
9,
10]. These findings open a new dimension to the intercellular interaction in that the axonal cytoplasm may contain an incomplete translation machinery that is completed by molecules from the SC-derived EVs. The discrete and regular disposition of SCs along the axon may provide full coverage for axonal cargo delivery, suggesting SC-derived EVs participation in the specification of the phenotype of the underlying axon. The current knowledge of SC EVs transcription machinery is, however, very limited. A few recent studies have identified EV components as part of other examinations [
9,
10,
14,
15] but a systematic approach to the identification of small RNAs, which by definition are <200 nucleotide in length and usually non-coding RNA molecules, is lacking.
Previous studies have described how the p75 neurotrophin receptor (p75
NTR) is a key component of the Schwann cell–axon myelination program during development [
16] and, depending on binding to neurotrophins or pro-neurotrophins, may mediate cell survival and cytoskeletal remodeling or trigger cell death via p53 and c-Jun N-terminal kinase pathway activation (JNK), respectively [
17]. Interestingly, p75
NTR was found to be present in SC-derived exosomes, enabling them to lateral transfer to axons [
9]. This reveals unforeseen trafficking capabilities of the p75
NTR receptor and raises new questions regarding the role of p75
NTR for Schwann cell-EV content regulation and signaling. As for p75
NTR, the Vps10-domain receptor sortilin has also critical roles regarding neurotrophin signaling, as it was shown to be important for regulation of the anterograde axonal transport of Trk receptors and for positively modulating neurotrophin-induced sensory neuronal survival [
18]. Sortilin is abundantly expressed in the nervous system during development and adulthood [
19], with a perinuclear subcellular localization in intracellular vesicles and the trans-golgi network [
20]. When associated with pro-neurotrophins, sortilin forms a complex with p75
NTR to induce CNS neuronal apoptosis [
21]. In the peripheral nervous system (PNS), sortilin does not seem to be involved in neuron development [
18] nor to have a role in sensory neuron apoptosis triggered by a nerve injury [
22]. Interestingly, in human lung cancer cells, sortilin was found to be present in exosomes and was further closely linked to the exosomal release mechanism [
23]. Therefore, here, we identify small RNA cargos in secreted SC-derived EVs and disclose the paracrine function for p75
NTR and sortilin in glial EV production and composition. miRNA signatures identified here might have promising future applications as diagnostic or target tools for PNS-related disease states, which often show compromised neurotrophic signaling as a pathological hallmark.
2. Experimental Section
2.1. Sprague Dawley Rats
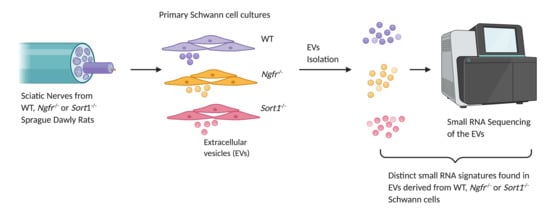
Pregnant wild-type (WT) Sprague Dawley rats were obtained from Janvier labs, while Ngfr−/− and Sort1−/− Sprague Dawley rats were purchased from Horizon Discovery and custom made, respectively, for breeding in house. Ngfr−/− is a deletion within exon 1, across the exon–intron boundary. We have determined that there were no alternative splicing fragments left, proving a clean Ngfr deletion. For Sort1−/−, a 5bp deletion in exon 9 was introduced, resulting in a stop codon, with subsequent completely destruction of the Vps10p-domain folding. No mRNA splice variants or protein fragments were detected by sequencing or Western blot analysis (data not shown).
Rats were housed under a 12 h light/12 h dark cycle in a pathogen-free environment, with water and food ad libitum.
The use of Sprague Dawley rats for obtaining primary Schwann cells was approved by the Danish Animal Experiments Inspectorate under the permission number 2017-15-0201-01192, (with approved date April 2017) and followed the Danish and European animal experimentation legislations (directive 2010/63/EU).
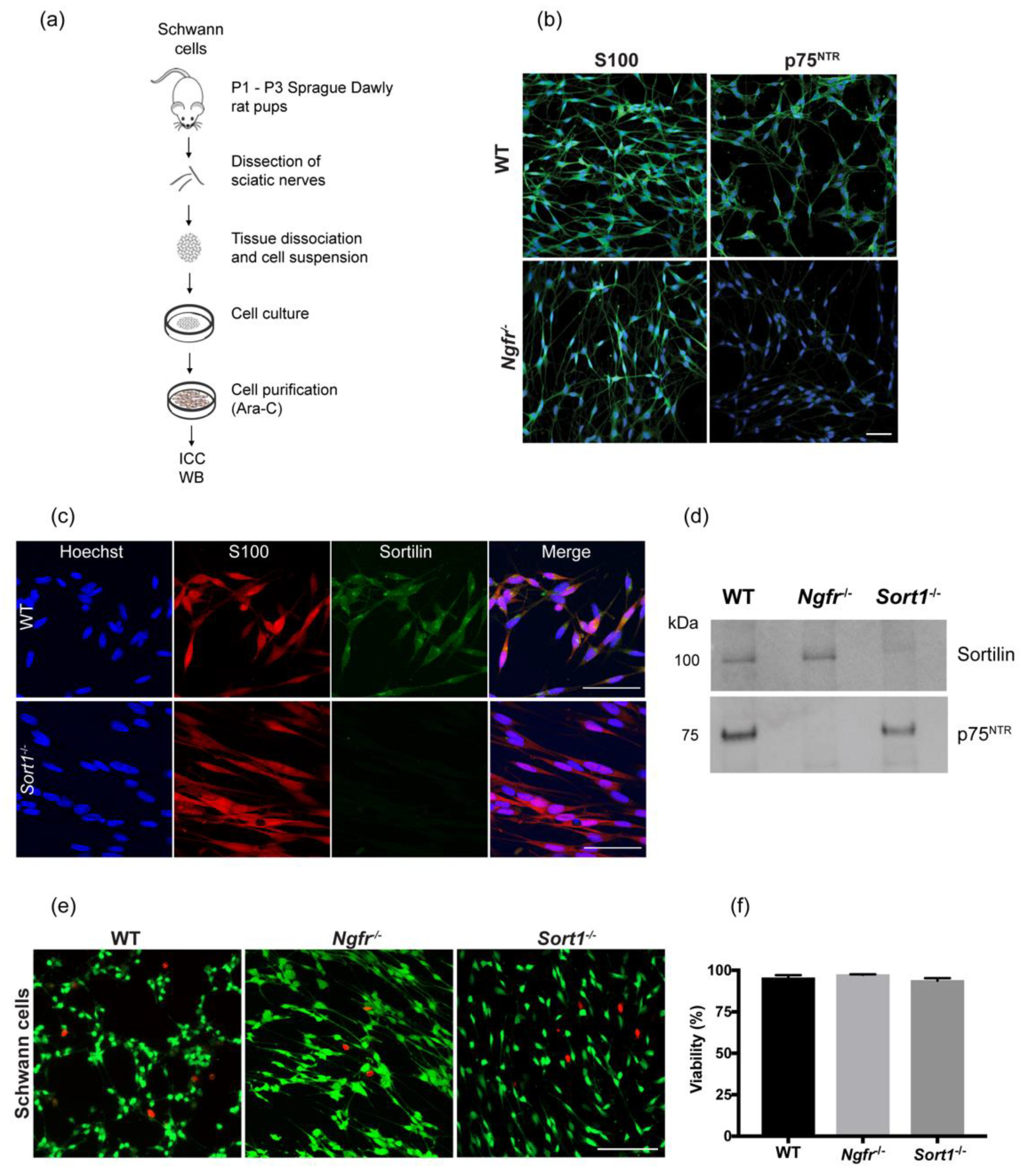
2.2. Primary Schwann Cell Culture
Primary Schwann cell cultures were prepared from neonatal (P1–P3) rat pups. In brief, dissected sciatic nerves were digested with 0.25% trypsin (Thermo Fisher Scientific, Waltham, MA, USA), and 0.1% collagenase I (Sigma, St. Louis, MO, USA) in L-15 media (Thermo Fisher), for 30 min at 37 °C. The anti-metabolic agent Cytosine-B-arabino furanoside hydrochloride (Ara-C; 10 μM; Sigma) was used for elimination of fibroblasts, in two cycles of 2–3 days. Purified primary Schwann cells were then expanded with growth media consisting of DMEM (Thermo Fisher) supplemented with 10% FBS (Thermo), 1% penicillin/streptomycin, 2.5 μM forskolin (Sigma) and 10 ng/mL recombinant human neuregulin-1-β1 EGF domain (R&D Systems, Minneapolis, MN, USA) at 37°C in a humidified incubator containing 5% CO2.
2.3. Immunocytochemistry
Primary Schwann cells were grown in 12 well-plate glass coverslips and fixed with 4% paraformaldehyde for 15 min. After 1 h blocking with 5% donkey serum and 1% bovine serum albumin (BSA) prepared in D-PBS, cells were incubated with rabbit anti-S100 (1:400, Dako, Agilent, Santa Clara, CA, USA), goat anti-sortilin (1:100, R&D Systems) or rabbit anti-p75NTR (1:500, Promega, Madison, WI, USA), diluted in blocking buffer, overnight at 4 °C. Secondary antibodies (Alexa Fluor donkey anti-goat 488 and Alexa Fluor donkey anti-rabbit 568; 1:500, Molecular Probes, Eugenes, OR, USA) were incubated for 2 h at room temperature (RT), together with Hoechst (1:10,000, Sigma) for nuclear staining. Sections were then mounted with Dako Fluorescent mounting medium (Dako) and images acquired with an LSM 780 confocal microscope (Carl Zeiss, Jena, Germany).
2.4. Live and Death Assay
The viability of WT, Ngfr−/− and Sort1−/− primary Schwann cells was analyzed using a combination of calcein-AM (4 μM) and ethidium homodimer-1 (4 μM) (Live/Death Viability/Cytotoxicity Kit, #L3224, Thermo). Primary Schwann cells were seeded into glass coverslips in a 12-well plate (500,000 cells/mL) in DMEM containing 10% FBS and 1% penicillin/streptomycin for 24 h and the assay performed accordingly to the manufacturer’s instructions. The staining was visualized under a fluorescent microscope, with n = 6 images per condition (in duplicates) acquired. The number of live (calcein-AM positive) and dead (ethidium homodimer-1 positive) cells were counted and the percentage of live cells (relative to total number of cells) calculated (n = 3 independent experiments).
Statistical analysis was accomplished using one-way ANOVA and Tukey’s multiple comparisons post hoc test, with Graph Pad Prism (version 8, San Diego, CA, USA). Quantitative data are reported as mean ± SEM.
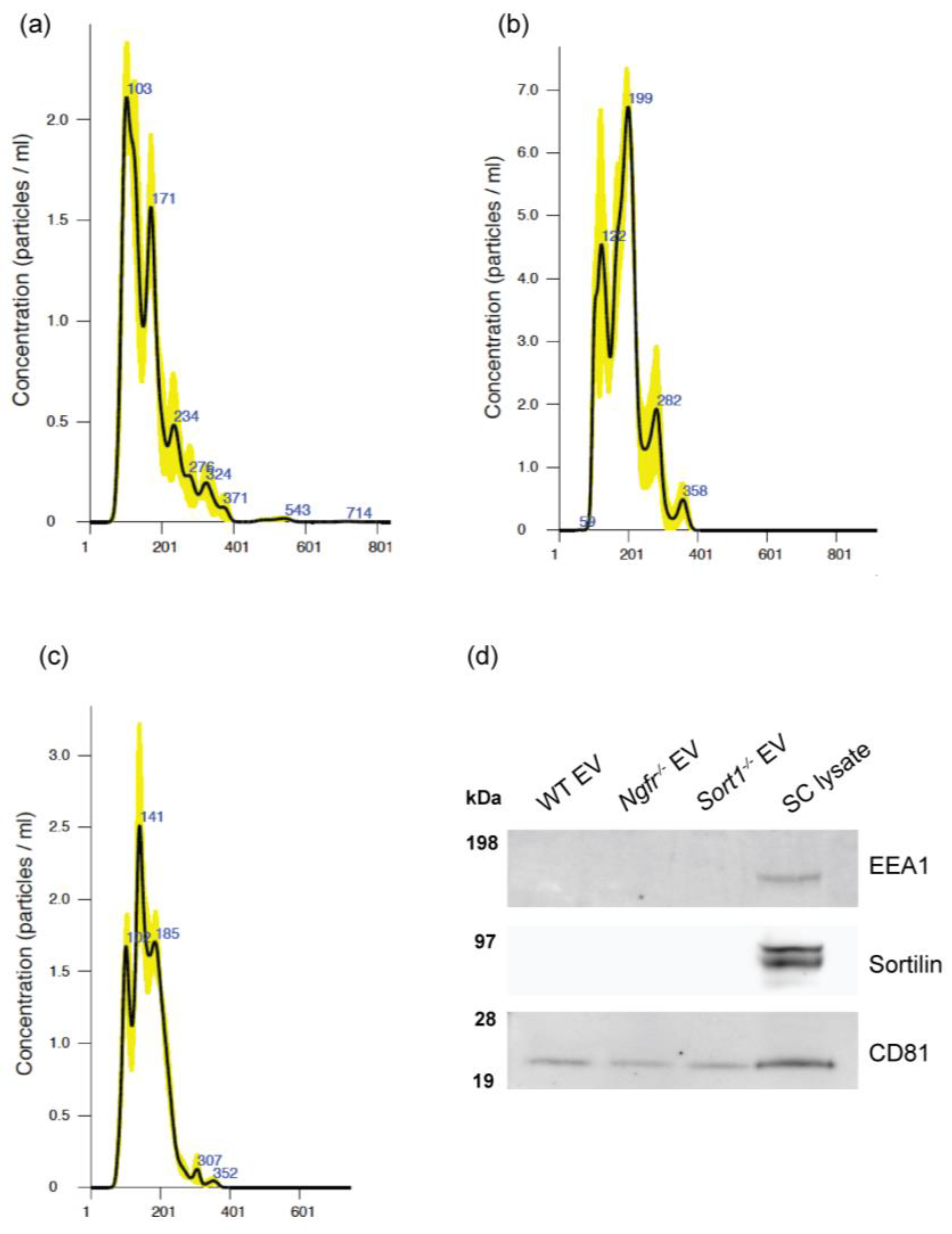
2.5. Isolation of EVs by Differential Ultracentrifugation
For EV isolation, purified primary Schwann cells were expanded in T175 flasks with growth media. When Schwann cells were about 80% confluent, media was replaced by an identical growth media but containing 10% of an exosome-depleted FBS (Thermo Fisher). After 48 h, media was collected for isolation of the extracellular vesicles (EVs). Two T175 flasks were used per condition and results represent n = 8–10 independent experiments. EVs were isolated from the supernatants of Schwann cells by differential ultracentrifugation. Large dead cells and cell debris were eliminated by successive centrifugations at 300× g for 10 min, 2000× g for 10 min and 10,000× g for 30 min, at 4 °C. Finally, supernatants were ultracentrifuged at 100,000× g for 120 min, at 4 °C, in a fixed angle rotor (Optima L-80-XP ultracentrifuge, 60 Ti rotor, Beckman-Coulter, Brea, CA, USA) to pellet the EVs. These were then resuspended in 250 μL PBS and immediately stored at −80 °C.
2.6. NanoSight Nanoparticle Tracking (NTA) Analysis
The Nanosight LM10 (Malvern Instruments, Malvern, UK) with a 405 nm laser was used to analyze the EVs. The resuspended EVs were diluted 1:100 in PBS and, subsequently, NTA measurements were performed in triplicates, with a 30 s video capture of each sample acquired using camera level set at 16 and detection threshold at 10. NTA software version 3.1 (Malvern Instruments) was used to analyze the data and extract the concentration and size of EVs in the samples.
2.7. Western Blotting
WT, Sort1−/− and Ngfr−/− primary Schwann cells and SC-derived EVs were lysed in standard ice-cold Tris-NaCI-EDTA lysis buffer (supplemented with protease and phosphatase inhibitors) and centrifuged at 10,000× g for 20 min at 4 °C. The supernatant was collected, and total protein concentration determined with the Bicinchoninic acid assay (Sigma). Fifty micrograms of total protein samples were separated on 4–12% Bis-Tris protein gels (Thermo Fisher) and electroblotted with nitrocellulose iBlot Gel Transfer Stacks (Invitrogen, Waltham, MA, USA) using the iBlot DryBlotting System (Invitrogen), according to manufactures guidelines. After 1 h blocking with tris buffer saline containing 2% skimmed milk and 2% Tween-20, membranes were incubated overnight at 4 °C with primary antibodies against Sortilin (1:250; R&D Systems) and p75NTR (1:500; Abcam, Cambridge, UK). For the Western blot with the EV pellets, 15 μg of total protein was used and primary antibodies consisted of CD81 (1:100; Santa Cruz Biotechnology, Dallas, TX, USA), as an exosome marker, EEA1 (1:1000; Cell Signaling, Danvers, MA, USA) as a marker for early endosomes and Sortilin (1:250; R&D Systems).
2.8. Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analysis of the EV Pellet
In-solution digest—The exosome sample was lyophilized and resuspended in 8 M urea in 0.1 M ammonium bicarbonate (Ambiec; pH 8.0). The sample was then reduced with 5 mM dithiothreitol (DTT) for 30 min and subsequently alkylated with 15 mM iodoacetamide in for another 30 min, while kept dark. The sample was diluted 5 times in 50 mM Ambic (pH 8.0) and digested overnight with 0.5 µg sequencing grade modified trypsin (Promega, Madison, WI, USA) at 37 °C. The digested sample was micro-purified using Octadecyl C18 Empore extractions disk (3M, St Paul, MN, USA) packed in P10 pipet tips. The purified peptides were suspended in 0.1% formic acid and stored at −20 °C until LC–MS/MS analysis.
LC–MS/MS analysis—The mass spectrometry analysis was performed on an Eksigent nanoLC 415 system (SCIEX, Framingham, MA, USA) connected to a TripleTOF 6600 mass spectrometer (SCIEX) equipped with a NanoSpray III source (SCIEX) and operated under Analyst TF 1.6.0 control (SCIEX). The sample was injected and trapped on a 2 cm in-house packed trap column (id = 100 μm) using RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH). Peptides were eluted from the trap column and separated on a 15 cm analytical column (id = 75 μm) pulled and packed in-house with RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH). Peptides were eluted from the column with a flow rate of 250 nL min−1, using a 30 min gradient going from 5% to 35% buffer B (acetonitrile with 0.1% formic acid) and sprayed directly into the mass spectrometer. The analysis relied on an information-dependent acquisition method collecting up to 25 MS/MS spectra in each 1.6 s cycle using an exclusion window of 6 s.
Data processing—Raw data obtained from the LC-MS/MS analysis were searched against the Swiss-prot and Trembl databases using the taxonomy rattus (2020_4; SwisProt: 8120 sequences; Trembl: 29,586 sequences) in Mascot 2.5.1 (Matrix science, London, UK). Trypsin was specified as digestion enzyme and allowing 1 miss cleavage. Carbamidomethyl modification of cysteines and oxidation of methionine were selected as fixed and variable modifications, respectively. Mass tolerance of the precursor and product ions was specified as 10 ppm and 0.2 Da using ESI-QUAD-TOF as the instrument setting. The significance threshold was set to 0.01 and the expected cut-off value to 0.005. The search result was imported to MS Data Miner version 1.3.0 (Sourceforge, San Diego, CA, USA) for further processing. Only proteins identified with at least two unique peptides with an ion score of 30 or above were reported as protein identifications (
Table S1).
2.9. Small RNA Library Preparation and Sequencing
Total RNA from the 250 μL purified EVs was isolated using miRNeasy Serum/Plasma Advanced kit (Qiagen, Hilden, Germany) following the manufacture’s protocol (n = 8–10 independent experiments, including different donors). The small RNA library was constructed using 5 μL of the eluted RNA following the manufacture’s protocol of QIAseq miRNA Library Kit (Qiagen). Briefly, 3′ and 5′ adapters were ligated to the RNA. Then the ligated RNAs were reverse transcribed to cDNA using the primer containing the UMI (unique molecule index). Finally, the sample index was introduced by amplification of the library and 22 PCR cycles were used. The quality of the library was checked on the Bioanalyzer using high sensitivity DNA analysis kit (Agilent, Santa Clara, CA, USA) and the quantity was measured using Kapa library quantification kit (Roche, Basel, Switzerland). The libraries were pooled and sequenced using single-end 75 bp sequencing on a Nextseq 500 sequencing machine (Illumina, San Diego, CA, USA).
2.10. Small RNA Data Analysis
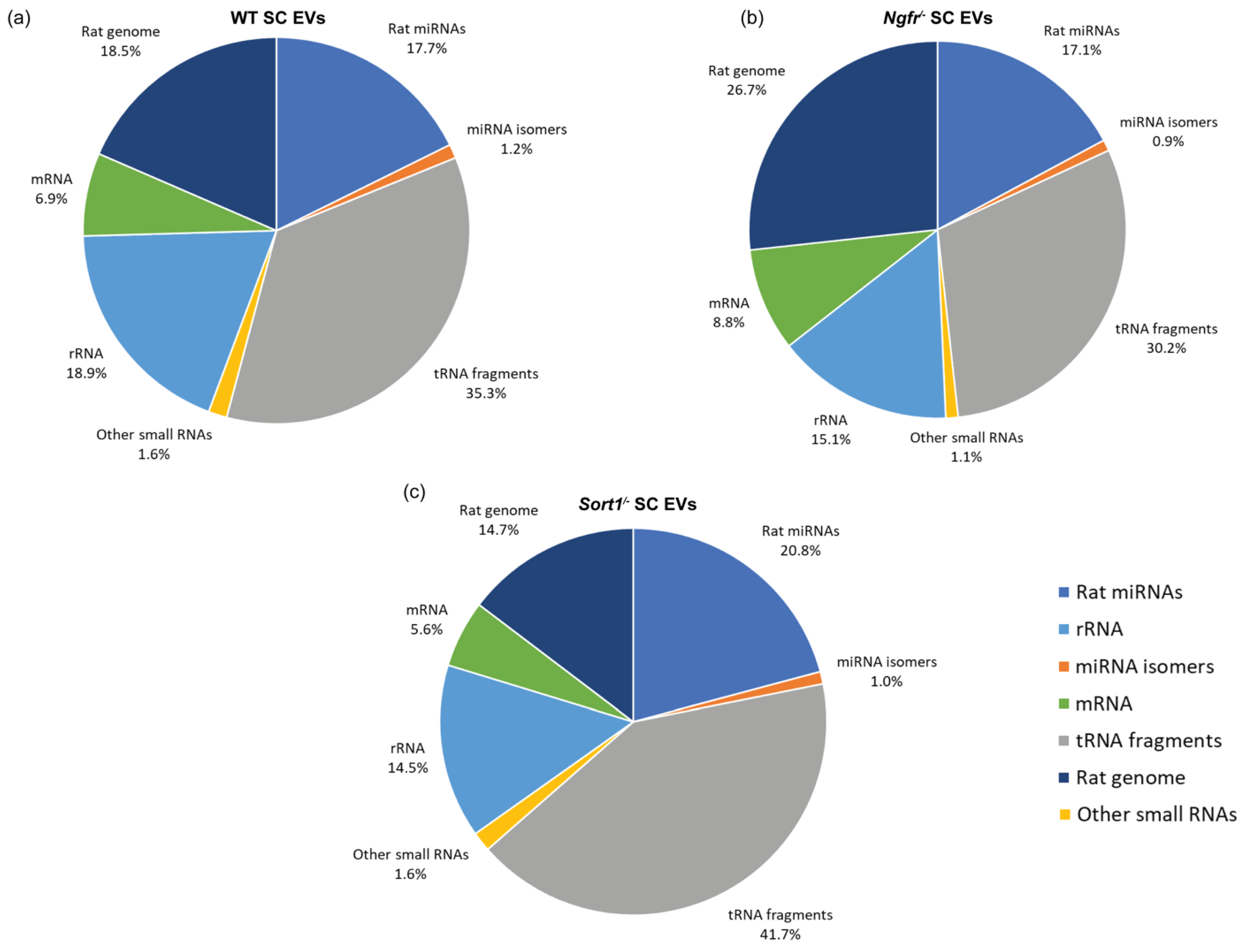
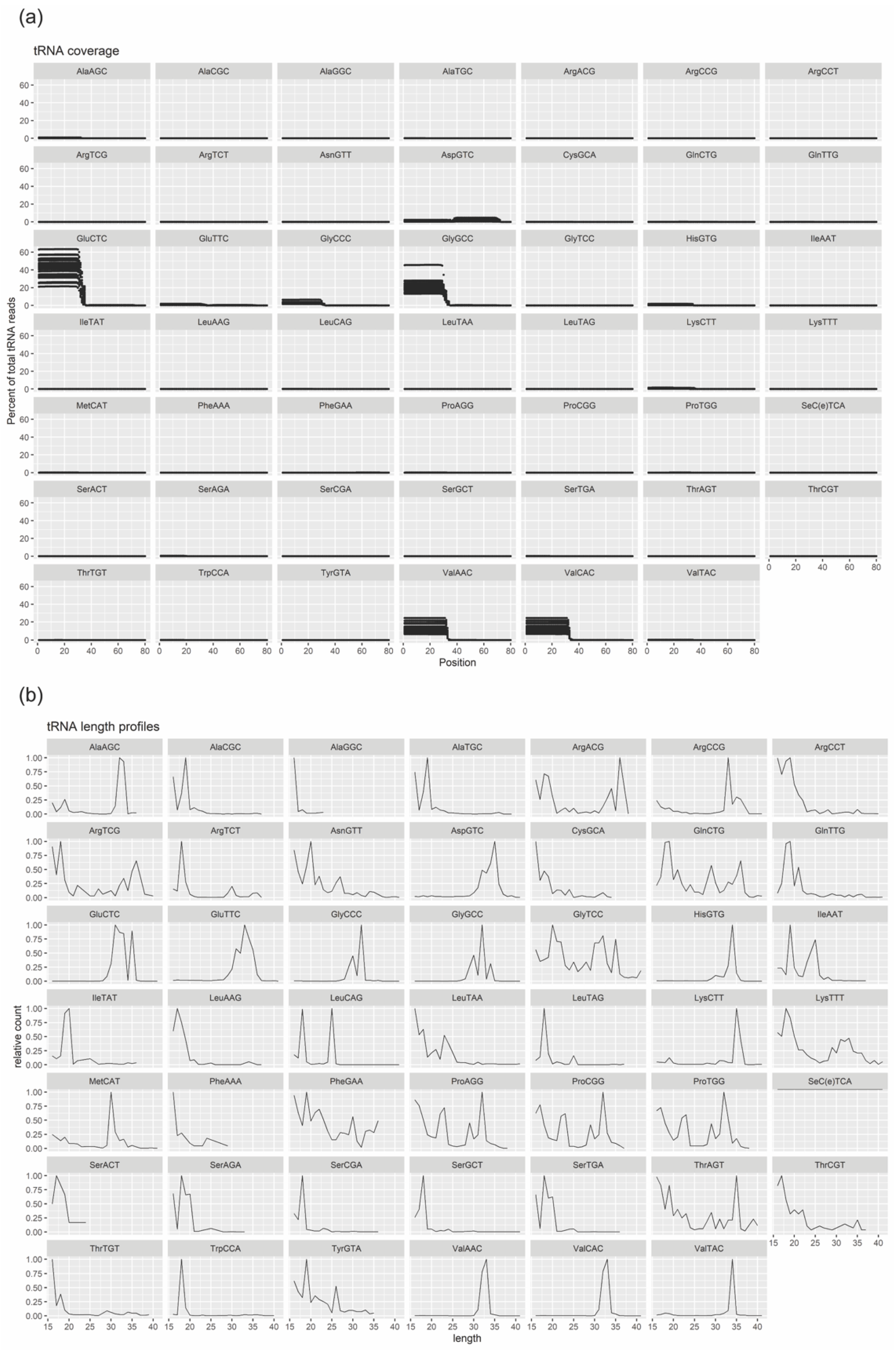
The raw data were quality filtered and trimmed by fastx_toolkit, and adaptor sequences were removed using Cutadapt. Quality control was performed using FastQC to ensure high quality data. Filtered reads were first mapped to rat tRNA sequences using Bowtie allowing 1 mismatch. Non-mapping reads were then mapped to miRNA sequences using Bowtie allowing 0 mismatches, though allowing the addition of A and T nucleotides at the 3′ end, since miRNAs often have untemplated A and U additions. Reads not mapping to miRNAs were mapped to other relevant transcriptomes (mRNA, rRNA and other small RNAs) and then to the rat (rn6) genome. The expression analysis was done both for miRNA and tRNA. The miRNA and tRNA mapping reads were deduplicated, meaning identical reads with identical UMIs were collapsed to a single read. Then the read counts of miRNA and tRNA were subjected to differential expression analysis using DESeq2 in R. The miRNAs and tRNAs were determined to be significantly changed if the adjusted p-values were below 0.05.
4. Discussion
Peripheral nerve roots, trunks and terminals are connected with SCs, the principal glial cells of the PNS. They derive from the neural crest cells and in their mature state SCs can be divided into myelinating SCs, which insulate axons with concentric layers of a lipid-rich myelin sheath, nonmyelinating SCs, that ensheath numerous small caliber axons in organized structures classified as Remak bundles, and terminal or perisynaptic SCs, located at the neuromuscular junction [
27,
28]. Recently, new SC subtypes have been described, such as the nociceptive SCs in the skin [
29] and the repair SCs, originating in the distal segment of an injured nerve to promote repair [
30]. Something they all have in common is their critical role in maintaining axonal and neuronal homeostasis under normal physiological conditions by bidirectional SC-axon communication and by interacting with the extracellular matrix and other cell types [
31]. This intercell communication is crucial for SC-mediated myelination [
32], survival [
33], as well as for directing their migration patterns [
34,
35] or activating dedifferentiation after a PNS injury [
36]; but also for axonal support and PNS plasticity [
10]. Several are the proposed signaling transmitting mechanisms, however, in the last decade, axo-glial communication through the release of exosomes has gained considerable insights. In vitro released SC-derived exosomes were shown to be internalized by sensory neurons, stimulating axonal regeneration both in vitro and in vivo [
9], which opened a new dimension in our understanding of glia-to-axon transfer of molecular signals. Nevertheless, up to date, little is known regarding SC exosome cargo. Very recently, the proteomic profile of rat primary SC-derived exosomes was characterized [
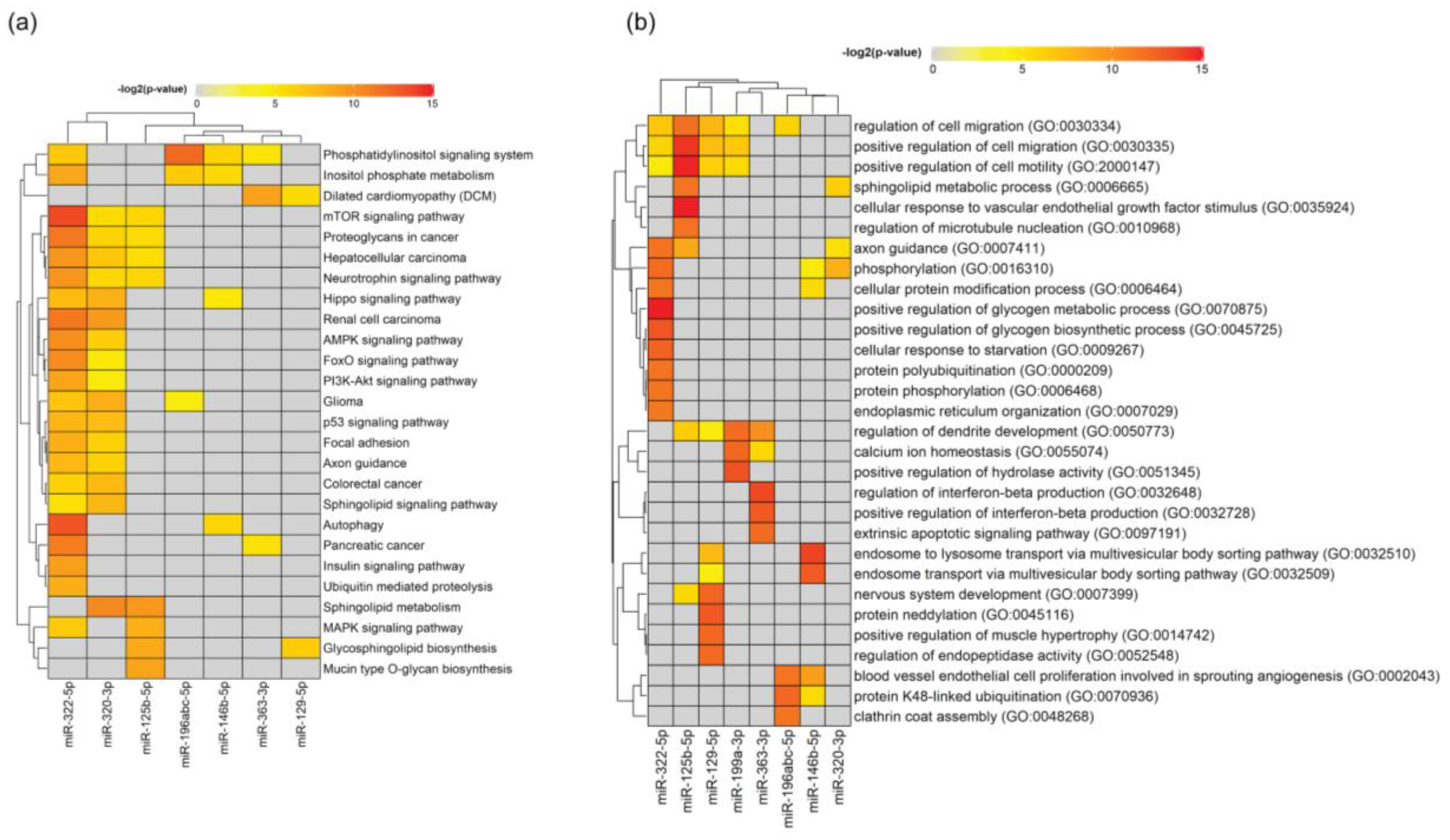
37]. Similar to those findings, with regard to enriched KEGG pathways within the exosome proteins, also in the present study we found that the regulation of actin cytoskeleton, axon guidance, PI3K-Akt and neurotrophin signaling pathways to be amongst the most enriched pathways regulated by the identified miRNA in SC-derived EVs. Hence, this suggests that miRNA present among the SC EV cargo may regulate neurorestorative genes, proteins and pathways in the receiving cell, opening a new avenue for potential therapeutic applications in regions of the nervous system with poor regenerative capabilities, such as the CNS [
38]. When compared with a recent study [
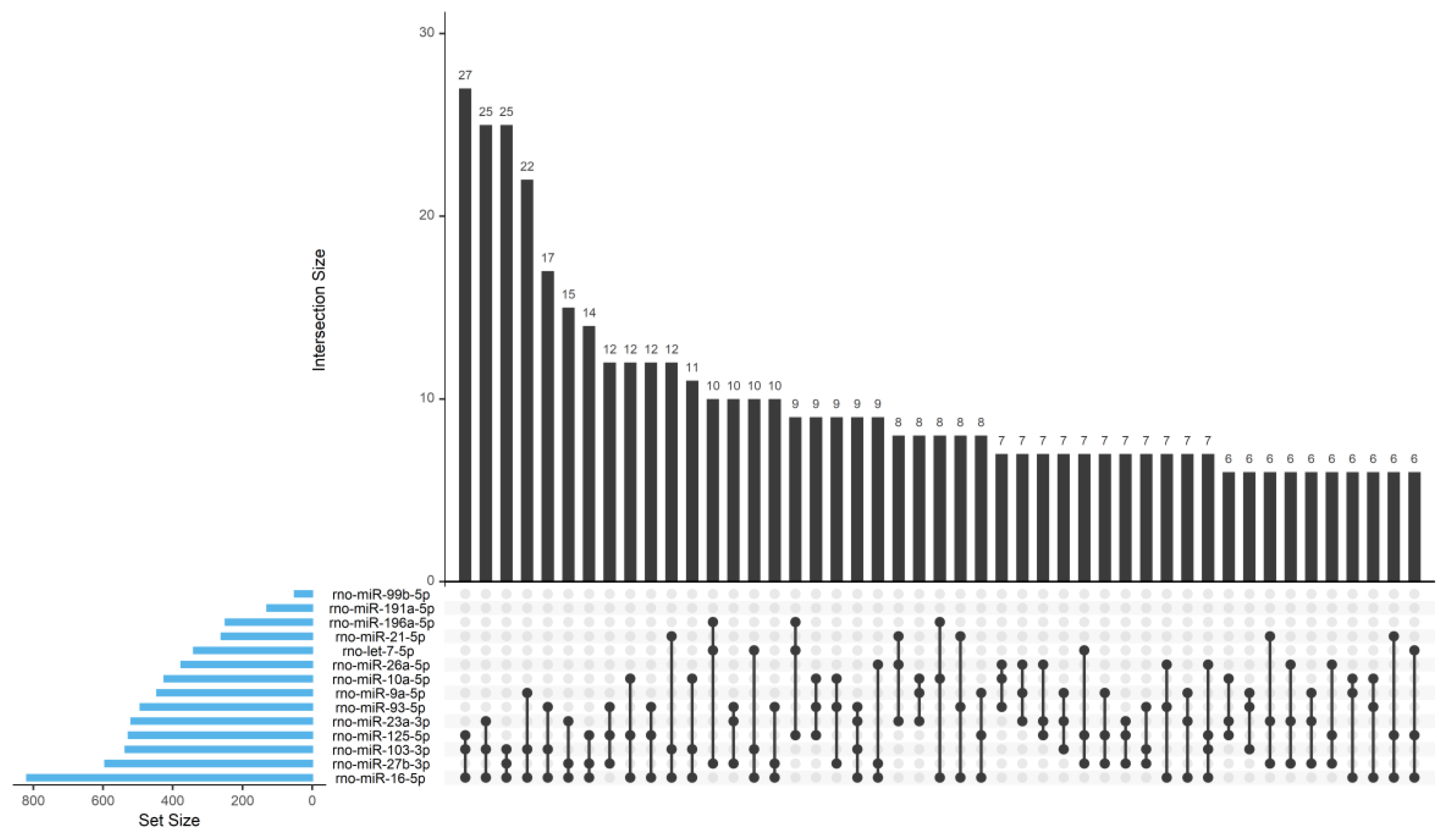
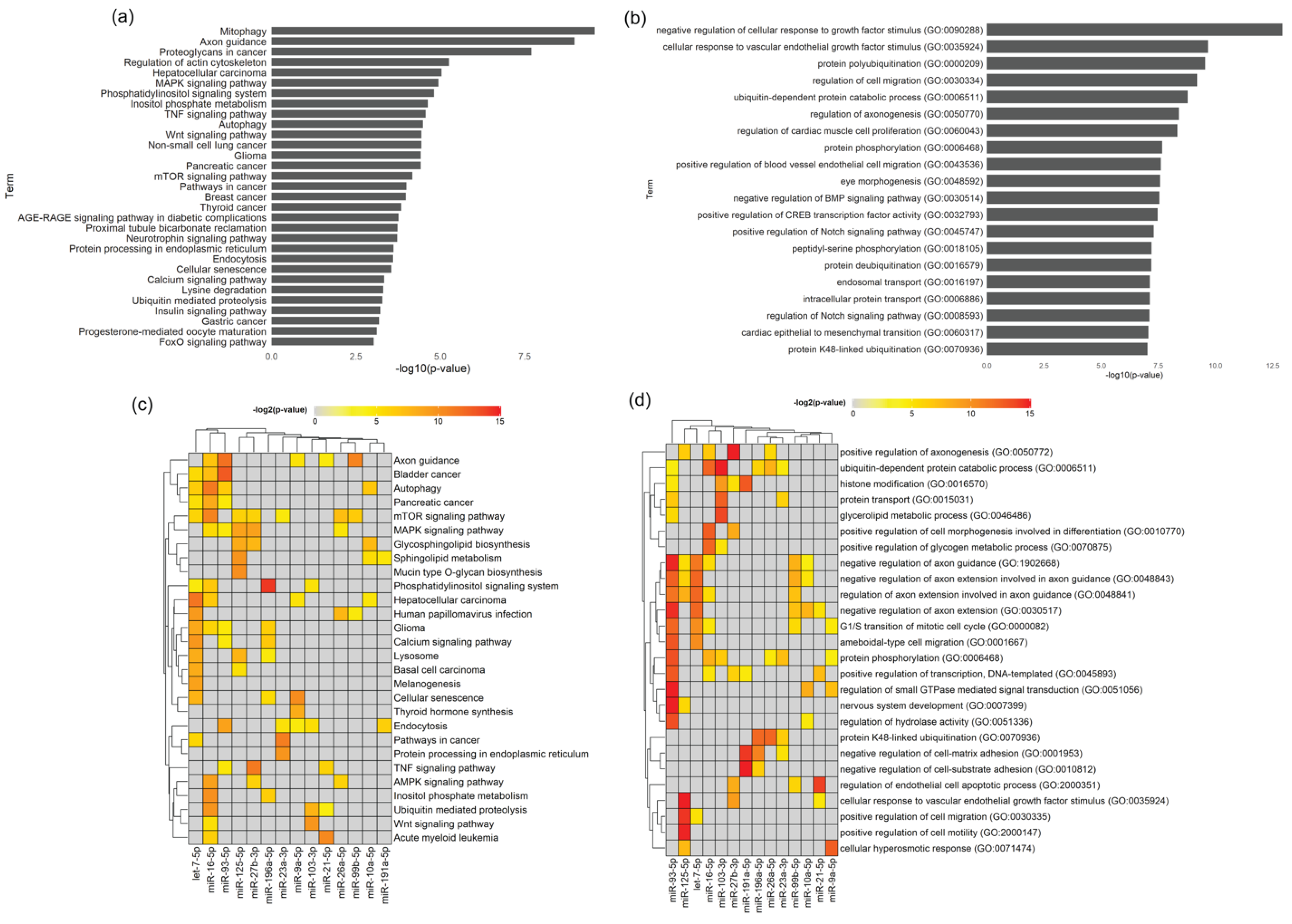
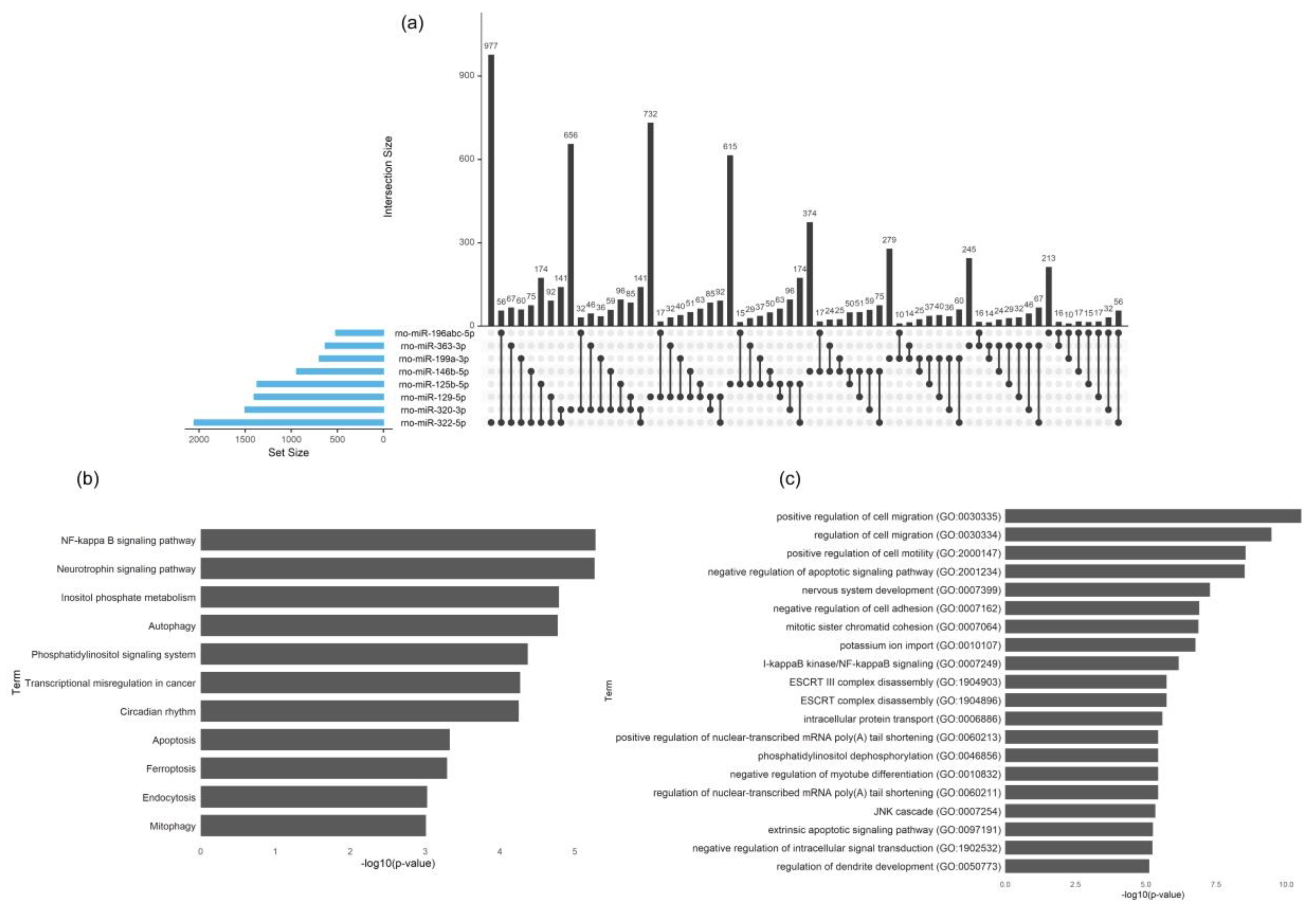
39], a lower percentage of miRNA and a higher percentage of tRNA in SC-derived EVs were here identified, which might be related with different culture methodologies, environmental factors or distinctive parameters/cut-offs involved in the analysis. The identified miRNA cargo of the Schwann-cell-derived EVs is involved in the basic biological pathways like endocytosis, mitophagy, autophagy and regulation of cytoskeleton, indicating that EVs can reprogram the phenotype and regulate the function of the recipient cells in the peripheral nerve. This EV-mediated crosstalk may be critical to maintain the homeostasis of the glial–axon syncytium.
Importantly, the axon–SC interlaced union during development and throughout life is crucial for the homeostasis of the PNS but also upon disease. Axon guidance, one of the main regulated pathways by miRNA present in SC-derived EVs found in this study, is involved in the formation of neuronal networks at the growth cone and is directed by several families of proteins including semaphorins, ephrins, slits, netrins and their corresponding receptors [
40]. Recent reports implicated this pathway and some of these proteins as being involved in the pathogenesis of diabetic retinopathy and nephropathy [
41,
42], as well as associated with circulating miRNA profiles found in plasma of hyperglycemic patients with type I diabetes [
43]. Due to the important documented role of SCs for the development of diabetic neuropathy [
25,
44], it might be worth exploring if axon guidance related molecules are also involved in the development of this main diabetes complication. In fact, it was recently demonstrated that exosomes derived from high glucose-stimulated primary SCs suppress axonal growth in vitro and promote the development of diabetic peripheral neuropathy in the db/db mouse model [
14]. In parallel with this, a positive therapeutic effect of exosomes derived from healthy SCs on the db/db type 2 diabetic peripheral neuropathy mouse line was documented [
15], but with no assessment of exosome cargo. Therefore, results from the present study with disclosure of SC EV small RNA contents can help us gain a deeper understanding of potential therapeutic mechanisms for nerve regeneration and peripheral neuropathies.
In the neurotrophin signaling pathway, we found that 16 out of 121 assigned genes are predicted to be targeted by more than three regulated miRNAs. Neurotrophins play multiple roles in neural development, neurodegeneration, inflammation or neuropathic pain [
45,
46] with well-known impact upon the pathogenesis of peripheral neuropathies, such as diabetic neuropathy [
47] or Charcot-Marie Tooth disease [
48]. Neurotrophins can bind and activate two different classes of receptors, p75
NTR and the Trk family of tyrosine kinase receptors [
49], with different signaling outcomes attained, involving cascades mediated by MAP kinase, PI-3K or Jun. In the PNS, the pleiotropic receptor p75
NTR is expressed by both SCs and sensory neurons facilitating cell death or survival, stimulating or restraining axonal growth and accelerating or weakening proliferation, depending on the cellular context and on binding to its co-receptors, such as sortilin [
18,
50]. Previously, it was found that p75
NTR was present as cargo in SC-secreted extracellular vesicles, being thus further suggested as a potential marker for SC-derived exosomes [
9]. Clinically, both p75
NTR and sortilin have been implicated in the pathophysiology of some degenerative diseases, including Alzheimer’s disease [
51,
52]. In the PNS, the role of p75
NTR is still somewhat unresolved as it was recently shown that conditional ablation of this receptor in SCs does not impact remyelination after peripheral nerve crush injury [
53]. Sortilin is also expressed by SCs in the periphery, but its involvement in nerve regeneration or PNS disease is yet to be determined. Therefore, we aimed at exploring the impact of deleting p75
NTR expression or its co-receptor sortilin on the release and small RNA content of primary SC-derived EVs. SCs lacking p75
NTR or sortilin were still able to produce and release EVs and, in contrast with what was observed for exosomes deriving from human lung carcinoma cells [
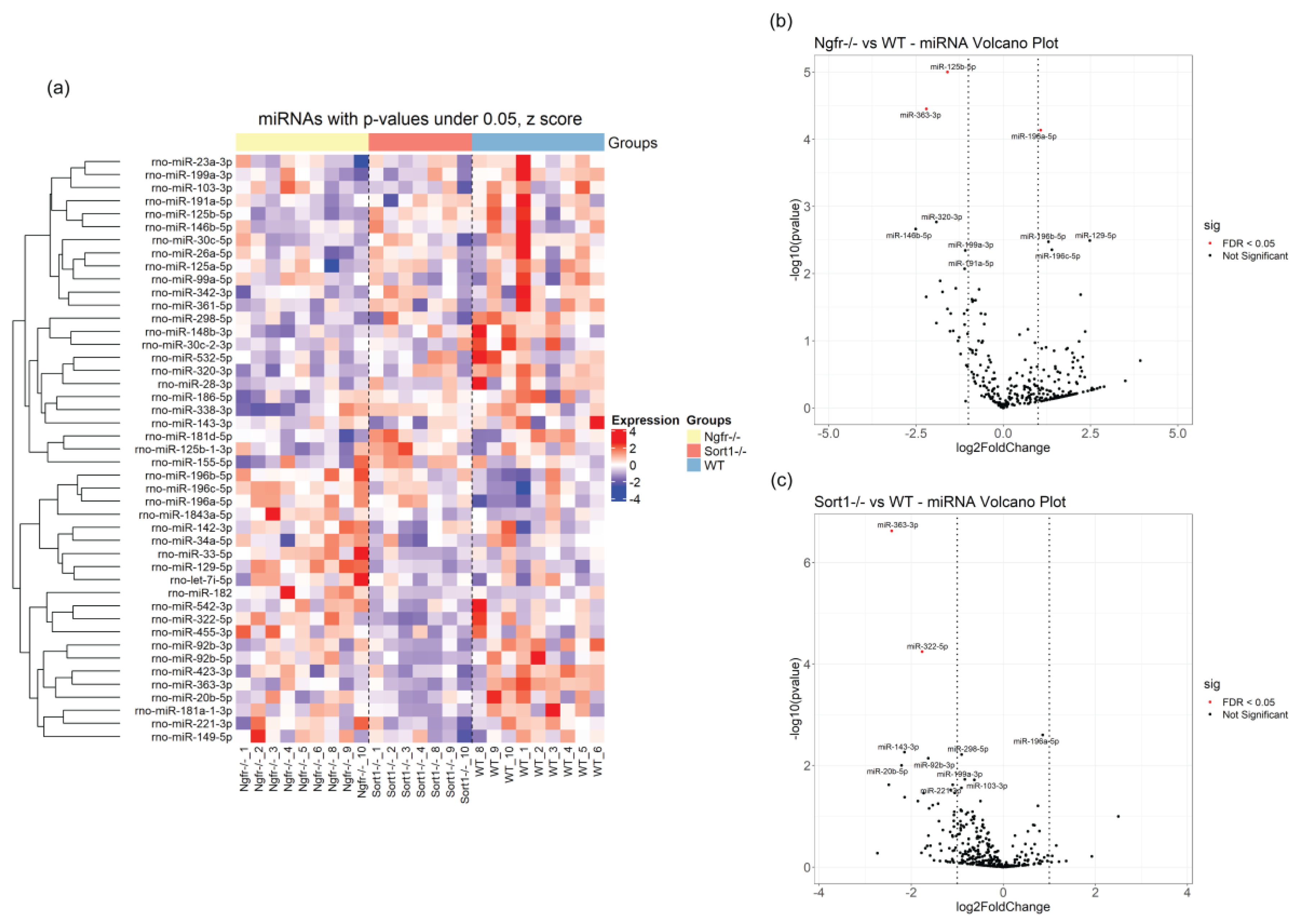
23], SC-derived EVs do not seem to carry sortilin—a finding that highlights the specificity of EV cargo from cell to cell. Overall, few EV miRNAs were differentially expressed but with potential of targeting a dozen of pathways and hundreds or thousands of genes. Most of the enriched miRNAs found in EVs from the transgenic primary SCs had gene targets in the NF-kB and PI3K signaling pathways, suggesting that these EVs may incorporate more information to increase immune regulation and survival mechanisms in recipient cells. miR-125b-5p, found down-regulated in EVs from
Ngfr−/− SCs, was previously associated with IL-1β induced inflammation [
54], which is very interesting due to the known relationship between IL-1 and p75
NTR [
55,
56]. Several studies highlighted different functions of sortilin in the immune system [
57,
58], but this receptor has also been linked to type 2 diabetes and obesity, cancer and cardiovascular pathologies. In this regard, miR-363-3p, that was here identified as downregulated in both transgenic derived SC EVs, has been linked to insulin resistance and post-stroke depressive behavior [
59,
60]. This, together with the reduced levels of miR-322-5p that was previously found important for targeting IGF-1 signaling [
61], suggests a potential involvement of SC expression of sortilin in diabetes or diabetic complications. It was very recently demonstrated that SC p75
NTR deficiency amplifies diabetic neuropathy disease by over-activating immune-related pathways and an increase in lysosomal stress [
62]. One hypothesis is that miR-196a and/or miR-125b-5p, here found enriched in
Ngfr−/− SC EVs, could potentially play a role in the observed diabetic phenotype in mice lacking SC p75
NTR expression as they were recently associated with type 1 diabetes [
43] and in the regulation of body fat distribution [
63].
The most abundant small RNA in SC-derived EVs in our study was tRNA-derived small RNAs, which is consistent to other studies using other cell types [
64,
65]. We found that tRFs derived from PheAAA, GlnCTG and GlnTTG were differentially expressed in the EVs derived from
Ngfr−/− SCs compared to WT EVs, while the sortilin knockdown did not result in any significant change of expression for tRNA fragments. It has been shown that tRFs can function as miRNAs by binding Argonaute proteins and repressing gene expression [
66]. Some studies demonstrated the association between the abnormal expression of tRFs and neurological disorders, such as Parkinson’s disease [
67], ischemia-reperfusion injury [
68] and cerebellar neurodegeneration [
69]. However, further investigation is needed to understand the function of the small RNAs derived from tRNAs in EVs secreted from SCs and the association with glial p75
NTR.
The role of EVs for cell-to-cell communication has gained substantial attention in recent years. They seem to be crucial factors for glia-to-axon transfer of proteins and RNA, regulating this functional syncytium in health and disease. miRNA signatures show promise as future biomarkers for several disease states due to their cell type-specific expression patterns. Therefore, the present preliminary work contributes especially to the knowledge of SC miRNA expression patterns to be used in future research targeting physiology in health and disease. Further complementary functional studies will be necessary to understand specific biological mechanisms related with particular miRNA targets in the context of PNS homeostasis.