Evolutionary Trajectory of Plasmodium falciparum: From Autonomous Phototroph to Dedicated Parasite
Abstract
1. Introduction
2. Genomic and Metabolic Adaptations
3. Molecular Mechanisms of Host Interaction
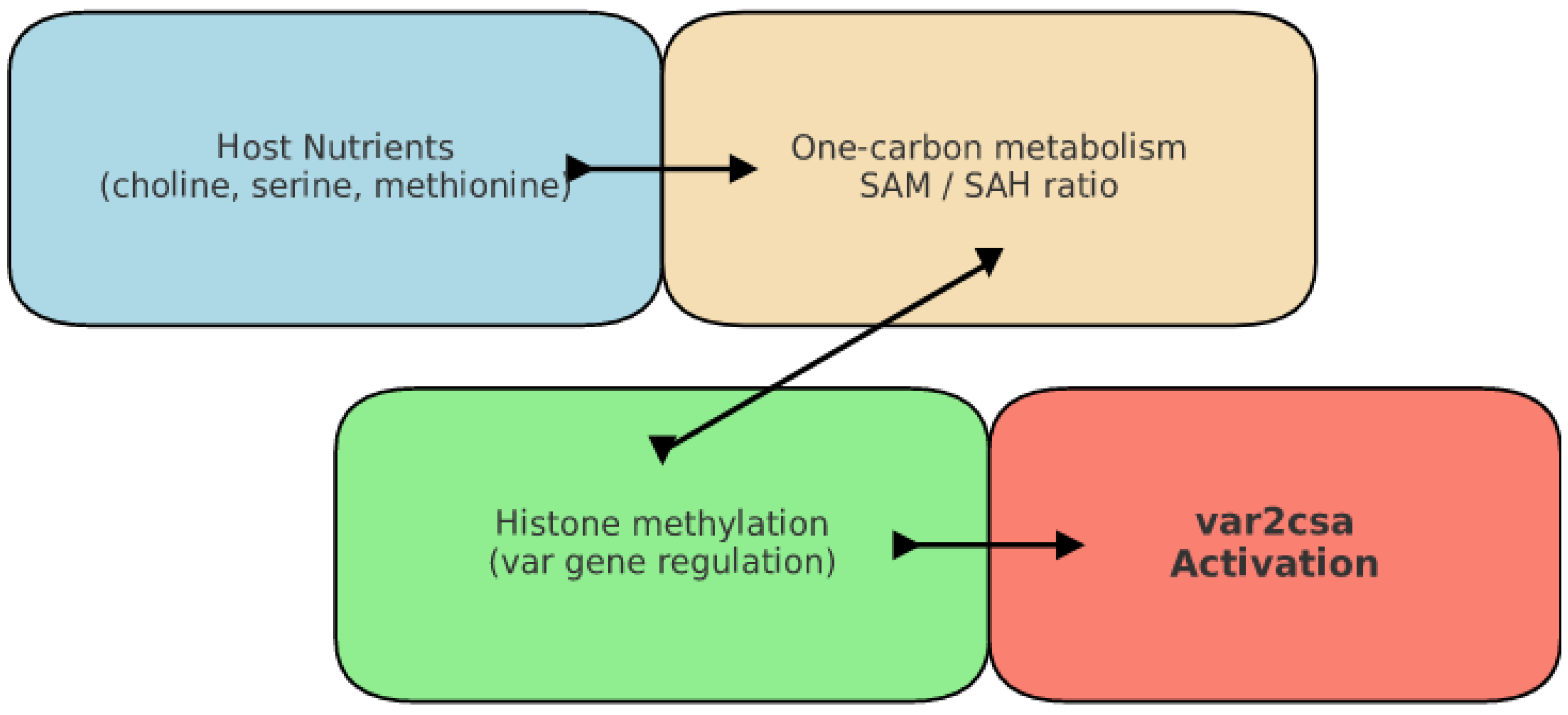
3.1. Epigenetic Regulation of Var Gene Expression
3.2. Metabolic Modulation and Experimental Evidence in Var Switching
3.3. Implications for Host–Parasite Interactions and Therapeutic Approaches
4. Co-Adaptive Dynamics with Human and Mosquito Hosts
4.1. Human Host Adaptations and Immune Dynamics
4.2. Mosquito Vector Adaptations and Insecticide Resistance
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). World Malaria Report 2022; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef]
- Reid, A.J.; Talman, A.M.; Bennett, H.M.; Gomes, A.R.; Sanders, M.J.; Illingworth, C.J.R.; Billker, O.; Berriman, M.; Lawniczak, M.K.N. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. eLife 2018, 7, e33105. [Google Scholar] [CrossRef]
- Waller, R.F.; McFadden, G.I. The apicoplast: A review of the derived plastid of apicomplexan parasites. Curr. Issues Mol. Biol. 2005, 7, 57–79. [Google Scholar] [CrossRef]
- Janouškovec, J.; Horák, A.; Oborník, M.; Lukeš, J.; Keeling, P.J. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 2010, 107, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.H.; Liao, P.C. Tracing evolutionary relicts of positive selection on eight malaria-related immune genes in mammals. Innate Immun. 2015, 21, 463–476. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Lahr, D.J.G.; Knoll, A.H.; Katz, L.A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. USA 2011, 108, 13624–13629. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.D.; Wilinski, D.; Assefa, S.; Keane, T.M.; Sarry, L.R.; Böhme, U.; Lemieux, J.; Barrell, B.; Pain, A.; Berriman, M.; et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010, 76, 12–24. [Google Scholar] [CrossRef]
- McFadden, G.I.; Waller, R.F. Plastids in parasites of humans. BioEssays 1997, 19, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.-C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. [Google Scholar] [CrossRef]
- Woo, Y.H.; Ansari, H.; Otto, T.D.; Klinger, C.M.; Kolisko, M.; Michálek, J.; Saxena, A.; Shanmugam, D.; Tayyrov, A.; Veluchamy, A.; et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Genome Biol. Evol. 2015, 7, 2449–2462. [Google Scholar]
- Manske, M.; Miotto, O.; Campino, S.; Auburn, S.; Almagro-Garcia, J.; Maslen, G.; O’bRien, J.; Djimde, A.; Doumbo, O.; Zongo, I.; et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 2012, 487, 375–379. [Google Scholar] [CrossRef]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2129. [Google Scholar] [CrossRef]
- Xie, S.C.; Dogovski, C.; Hanssen, E.; Chiu, F.; Yang, T.; Crespo, M.P.; Stafford, C.; Batinovic, S.; Teguh, S.; Charman, S.; et al. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J. Cell Sci. 2016, 129, 406–416. [Google Scholar] [CrossRef]
- Thompson, J.; Janse, C.J.; Waters, A.P. Comparative genomics in Plasmodium: A tool for the identification of genes and functional analysis. Mol. Biochem. Parasitol. 2001, 118, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Hall, N.; Carlton, J. Comparative genomics of malaria parasites. Curr. Opin. Genet. Dev. 2005, 15, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Coppens, I. Contribution of host lipids to Toxoplasma pathogenesis. Cell. Microbiol. 2006, 8, 1–9. [Google Scholar] [CrossRef]
- van Dooren, G.G.; Stimmler, L.M.; McFadden, G.I. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 2006, 30, 596–630. [Google Scholar] [CrossRef] [PubMed]
- Counihan, N.A.; Modak, J.K.; de Koning-Ward, T.F. How Malaria Parasites Acquire Nutrients From Their Host. Front. Cell Dev. Biol. 2021, 9, 649184. [Google Scholar] [CrossRef]
- Ralph, S.A.; van Dooren, G.G.; Waller, R.F.; Crawford, M.J.; Fraunholz, M.J.; Foth, B.J.; Tonkin, C.J.; Roos, D.S.; McFadden, G.I. Tropical infectious diseases: Metabolic maps and management. Nat. Rev. Microbiol. 2007, 5, 837–840. [Google Scholar]
- Ay, F.; Bunnik, E.M.; Varoquaux, N.; Bol, S.M.; Prudhomme, J.; Vert, J.-P.; Noble, W.S.; Le Roch, K.G. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014, 24, 974–988. [Google Scholar] [CrossRef]
- Lopez-Rubio, J.J.; Mancio-Silva, L.; Scherf, A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 2009, 5, 179–190. [Google Scholar] [CrossRef]
- Duraisingh, M.T.; Skillman, K.M. Epigenetic Variation and Regulation in Malaria Parasites. Annu. Rev. Microbiol. 2018, 72, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Recker, M.; Buckee, C.O.; Serazin, A.; Kyes, S.; Pinches, R.; Christodoulou, Z.; Springer, A.L.; Gupta, S.; Newbold, C.I.; Kim, K. Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 2011, 7, e1001306. [Google Scholar] [CrossRef]
- Gómez-Díaz, E.; Yerbanga, R.S.; Lefèvre, T.; Cohuet, A.; Rowley, M.J.; Ouedraogo, J.B.; Corces, V.G. Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae. Sci. Rep. 2017, 7, 40655. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.; Deitsch, K.W. Malaria Epigenetics. Cold Spring Harb. Perspect. Med. 2017, 7, a025528. [Google Scholar] [CrossRef]
- Reyser, T.; Paloque, L.; Augereau, J.-M.; Di Stefano, L.; Benoit-Vical, F. Epigenetic regulation as a therapeutic target in the malaria parasite Plasmodium falciparum. Malar. J. 2024, 23, 44. [Google Scholar] [CrossRef]
- Boddey, J.A.; Cowman, A.F. Plasmodium nesting: Remaking the erythrocyte from the inside out. Annu. Rev. Microbiol. 2013, 67, 243–269. [Google Scholar] [CrossRef]
- Day, C.J.; Favuzza, P.; Bielfeld, S.; Haselhorst, T.; Seefeldt, L.; Hauser, J.; Shewell, L.K.; Flueck, C.; Poole, J.; Jen, F.E.-C.; et al. The essential malaria protein PfCyRPA targets glycans to invade erythrocytes. Cell Rep. 2024, 43, 114012. [Google Scholar] [CrossRef]
- Otto, T.D.; Assefa, S.A.; Böhme, U.; Sanders, M.J.; Kwiatkowski, D.P.; Pf3k consortium; Berriman, M.; Newbold, C. Evolutionary analysis of the most polymorphic gene family in falciparum malaria. Wellcome Open Res. 2019, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Deitsch, K.W.; Dzikowski, R. Variant gene expression and antigenic variation by malaria parasites. Annu. Rev. Microbiol. 2017, 71, 625–641. [Google Scholar] [CrossRef]
- Andradi-Brown, C.; Wichers-Misterek, J.S.; von Thien, H.; Höppner, Y.D.; Scholz, J.A.M.; Hansson, H.S.; Hocke, E.F.; Gilberger, T.W.; Duffy, M.; Lavstsen, T.; et al. A novel computational pipeline for var gene expression augments the discovery of changes in the Plasmodium falciparum transcriptome during transition from in vivo to short-term in vitro culture. eLife 2024, 12, RP87726. [Google Scholar] [CrossRef]
- Jiang, L.; Mu, J.; Zhang, Q.; Ni, T.; Srinivasan, P.; Rayavara, K.; Yang, W.; Turner, L.; Lavstsen, T.; Theander, T.G.; et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 2013, 499, 223–227. [Google Scholar] [CrossRef]
- Ukaegbu, U.E.; Kishore, S.P.; Kwiatkowski, D.L.; Pandarinath, C.; Dahan-Pasternak, N.; Dzikowski, R.; Deitsch, K.W.; Sullivan, W.J. Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog. 2014, 10, e1003854. [Google Scholar] [CrossRef] [PubMed]
- Volz, J.C.; Bártfai, R.; Petter, M.; Langer, C.; Josling, G.A.; Tsuboi, T.; Schwach, F.; Baum, J.; Rayner, J.C.; Stunnenberg, H.G.; et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 2012, 11, 7–18. [Google Scholar] [CrossRef]
- Bancells, C.; Deitsch, K.W. A molecular switch in the efficiency of translation reinitiation controls expression of var2csa, a gene implicated in pregnancy-associated malaria. Mol. Microbiol. 2013, 90, 472–488. [Google Scholar] [CrossRef]
- Musabyimana, J.-P.; Musa, S.; Manti, J.; Distler, U.; Tenzer, S.; Ngwa, C.J.; Pradel, G. The Plasmodium falciparum histone methyltransferase PfSET10 participates in a chromatin modulation network crucial for intraerythrocytic development. mSphere 2024, 9, e00495-24. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, A.; Bruske, E.; Krumkamp, R.; Turner, L.; Wichers, J.S.; Petter, M.; Held, J.; Duffy, M.F.; Sim, B.K.L.; Hoffman, S.L.; et al. Controlled human malaria infection with Plasmodium falciparum demonstrates impact of naturally acquired immunity on virulence gene expression. PLoS Pathog. 2019, 15, e1007906. [Google Scholar] [CrossRef]
- Ukaegbu, U.E.; Zhang, X.; Heinberg, A.R.; Wele, M.; Chen, Q.; Deitsch, K.W.; Wahlgren, M. A unique virulence gene occupies a principal position in immune evasion by the malaria parasite Plasmodium falciparum. PLoS Genet. 2015, 11, e1005234. [Google Scholar] [CrossRef]
- Zhang, X.; Florini, F.; Visone, J.E.; Lionardi, I.; Gross, M.R.; Patel, V.; Deitsch, K.W. A coordinated transcriptional switching network mediates antigenic variation of human malaria parasites. Elife 2022, 11, e83840. [Google Scholar] [CrossRef]
- Dahlbäck, M.; Nielsen, M.A.; Salanti, A. Can any lessons be learned from the ambiguous glycan binding of PfEMP1 domains? Trends Parasitol. 2010, 26, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Mentch, S.J.; Mehrmohamadi, M.; Huang, L.; Liu, X.; Gupta, D.; Mattocks, D.; Padilla, P.G.; Ables, G.; Bamman, M.M.; Thalacker-Mercer, A.E.; et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015, 22, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Mentch, S.J.; Locasale, J.W. One-carbon metabolism and epigenetics: Understanding the specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef]
- Gao, X.; Reid, M.A.; Kong, M.; Locasale, J.W. Metabolic interactions with cancer epigenetics. Mol. Asp. Med. 2017, 54, 50–57. [Google Scholar]
- Locasale, J.W.; Cantley, L.C. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011, 14, 443–451. [Google Scholar] [CrossRef]
- Sadhu, M.J.; Guan, Q.; Li, F.; Sales-Lee, J.; Iavarone, A.T.; Hammond, M.C.; Cande, W.Z.; Rine, J. Nutritional control of epigenetic processes in yeast and human cells. Genetics 2013, 195, 831–844. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013, 339, 222–226. [Google Scholar] [CrossRef]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodelling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef]
- Ye, C.; Sutter, B.M.; Wang, Y.; Kuang, Z.; Tu, B.P. A metabolic function for phospholipid and histone methylation. Mol. Cell 2017, 66, 180–193.e8. [Google Scholar] [CrossRef] [PubMed]
- Haws, S.A.; Yu, D.; Ye, C.; Wille, C.K.; Nguyen, L.C.; Krautkramer, K.A.; Tomasiewicz, J.L.; Yang, S.E.; Miller, B.R.; Liu, W.H.; et al. Methyl-metabolite depletion elicits adaptive responses to support heterochromatin stability and epigenetic persistence. Mol. Cell 2020, 78, 210–223.e218. [Google Scholar] [CrossRef] [PubMed]
- Beri, D.; Balan, B.; Chaubey, S.; Subramaniam, S.; Surendra, B.; Tatu, U. A disrupted transsulphuration pathway results in accumulation of redox metabolites and induction of gametocytogenesis in malaria. Sci. Rep. 2017, 7, 40213. [Google Scholar] [CrossRef]
- Brancucci, N.M.; Gerdt, J.P.; Wang, C.; De Niz, M.; Philip, N.; Adapa, S.R.; Zhang, M.; Hitz, E.; Niederwieser, I.; Boltryk, S.D.; et al. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite Plasmodium falciparum. Cell 2017, 171, 1532–1544.e15. [Google Scholar] [CrossRef]
- Mancio-Silva, L.; Slavic, K.; Ruivo, M.T.G.; Grosso, A.R.; Modrzynska, K.K.; Vera, I.M.; Sales-Dias, J.; MacPherson, C.R.; Crozet, P.; Adamo, M.; et al. Nutrient sensing modulates malaria parasite virulence. Nature 2017, 547, 213–216. [Google Scholar] [CrossRef]
- intó-Font, E.; Michel-Todó, L.; Russell, T.J.; Casas-Vila, N.; Conway, D.J.; Bozdech, Z.; Llinás, M.; Cortés, A. A heat-shock response regulated by the PfAP2-HS transcription factor protects human malaria parasites from febrile temperatures. Nat. Microbiol. 2021, 6, 1163–1174. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, C.; Oberstaller, J.; Thomas, P.; Otto, T.D.; Casandra, D.; Boyapalle, S.; Adapa, S.R.; Xu, S.; Button-Simons, K.; et al. The apicoplast link to fever-survival and artemisinin-resistance in the malaria parasite. Nat. Commun. 2021, 12, 4563. [Google Scholar] [CrossRef] [PubMed]
- Mathews, E.S.; Jezewski, A.J.; John, A.R.O.; Sibley, L.D. Protein prenylation and Hsp40 in thermotolerance of Plasmodium falciparum malaria parasites. mBio 2021, 12, e0076021. [Google Scholar] [CrossRef] [PubMed]
- Kilian, N.; Choi, J.-Y.; Voelker, D.R.; Ben Mamoun, C. Role of phospholipid synthesis in the development and differentiation of malaria parasites in the blood. J. Biol. Chem. 2018, 293, 17308–17316. [Google Scholar] [CrossRef]
- Bobenchik, A.M.; Witola, W.H.; Augagneur, Y.; Lochlainn, L.N.; Garg, A.; Pachikara, N.; Choi, J.-Y.; Zhao, Y.O.; Usmani-Brown, S.; Lee, A.; et al. Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 18262–18267. [Google Scholar] [CrossRef]
- Ben Mamoun, C.; Prigge, S.T.; Vial, H. Targeting the lipid metabolic pathways for the treatment of Malaria. Drug Dev. Res. 2010, 71, 44–55. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.; Dziedziech, A.; Kirkman, L.A.; Deitsch, K.W.; Ankarklev, J. A histone methyltransferase inhibitor can reverse epigenetically acquired drug resistance in the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 2020, 64, e02021-19. [Google Scholar] [CrossRef]
- Chan, S.; Frasch, A.; Mandava, C.S.; Ch’NG, J.-H.; Quintana, M.d.P.; Vesterlund, M.; Ghorbal, M.; Joannin, N.; Franzén, O.; Lopez-Rubio, J.-J.; et al. Regulation of PfEMP1-VAR2CSA translation by a Plasmodium translation-enhancing factor. Nat. Microbiol. 2017, 2, 17068. [Google Scholar] [CrossRef]
- Kwiatkowski, D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005, 77, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Riley, E.M. T-cell unresponsiveness in human malaria. Immunol. Lett. 1998, 62, 163–170. [Google Scholar]
- Doolan, D.L.; Dobaño, C.; Baird, J.K. Acquired immunity to malaria. Clin. Microbiol. Rev. 2009, 22, 13–36. [Google Scholar] [CrossRef] [PubMed]
- Bull, P.C.; Marsh, K. The role of var genes in malaria pathogenesis. Trends Microbiol. 2002, 10, 31–36. [Google Scholar]
- Scherf, A.; Lopez-Rubio, J.J.; Riviere, L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 2008, 62, 445–470. [Google Scholar] [CrossRef]
- Pikor, D.; Hurła, M.; Banaszek-Hurła, N.; Drelichowska, A.; Paul, M. Neurovascular Pathophysiology and Emerging Biomarkers in Cerebral Malaria: An Integrative Perspective. Neurol. Int. 2025, 17, 149. [Google Scholar] [CrossRef]
- Barletta, A.B.F.; Barillas-Mury, C.; Molina-Cruz, A. Mosquito immune responses to Plasmodium parasites that limit malaria transmission. Cell. Mol. Life Sci. 2025, 82, 143. [Google Scholar] [CrossRef] [PubMed]
- Riehle, M.M.; Srinivasan, P.; Jacobs-Lorena, M. Deconstructing mosquito immunity to malaria parasites. Insect Biochem. Mol. Biol. 2006, 36, 728–737. [Google Scholar]
- Lambrechts, L.; Morlais, I.; Cohuet, A. Genetic architecture of vector competence in mosquitoes. Evol. Biol. 2005, 35, 1–18. [Google Scholar]
- Hemingway, J.; Ranson, H.; Magill, A.; Kolaczinski, J.; Fornadel, C.; Gimnig, J.; Coetzee, M.; Simard, F.; Dabiré, R.K.; Hinzoumbe, C.K.; et al. Averting a malaria disaster: Will insecticide resistance derail malaria control? Lancet. 2016, 387, 1785–1788. [Google Scholar] [CrossRef]
- Feyereisen, R. Insecticide resistance by metabolism: Detoxication enzymes and insecticide resistance. Pestic. Biochem. Physiol. 2011, 100, 186–198. [Google Scholar]
- Martinez-Torres, D.; Chandre, F.; Williamson, M.S.; Darriet, F.; Bergé, J.B.; Devonshire, A.L.; Guillet, P.; Pasteur, N.; Pauron, D. Molecular characterization of pyrethroid knockdown resistance (kdr) in Anopheles gambiae s.s. Insect Mol. Biol. 1998, 7, 179–189. [Google Scholar] [CrossRef]
- Trape, J.F.; Tall, A.; Diagne, N.; Ndiath, O.; Ly, A.B.; Faye, J.; Dieye-Ba, F.; Roucher, C.; Bouganali, C.; Badiane, A. Malaria morbidity and pyrethroid resistance after the introduction of insecticide-treated bed nets and artemisinin-based combination therapies: A longitudinal study. Lancet Infect. Dis. 2011, 11, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Russell, T.L.; Govella, N.J.; Azizi, S.; Drakeley, C.J.; Kachur, S.P.; Killeen, G.F. Increased proportions of outdoor feeding among residual malaria vector populations following increased use of insecticide-treated nets in rural Tanzania. Malar. J. 2009, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, H.M.; Dornhaus, A. Ecology of mosquito larval dispersal and migration in relation to vector-borne disease. Trends Parasitol. 2011, 27, 531–537. [Google Scholar]
- Koppula, D.T.; Shriram, A.N.; Ramasamy, A.; Kumar, A.; Rahi, M. Evaluation of long lasting insecticidal nets in experimental huts and WHO PQTVCP compliance: A systematic review. PLoS ONE. 2025, 20, e0318673. [Google Scholar] [CrossRef]
- White, N.J. Antimalarial drug resistance. J. Antimicrob. Chemother. 2004, 54, i143–i148. [Google Scholar] [CrossRef] [PubMed]
- Hastings, I.M.; Donnelly, M.J. The impact of antimalarial drug resistance on the control of Plasmodium falciparum. Drug Resist. Updates 2005, 8, 43–50. [Google Scholar] [CrossRef]
- Flegg, J.A.; Metcalf, C.J.E.; Gharbi, M.; Venkatesan, M.; Shewchuk, T.; Sibley, C.H.; Guerin, P.J. Trends in antimalarial drug use in Africa. Am. J. Trop. Med. Hyg. 2013, 89, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Menard, D.; Dondorp, A. Antimalarial drug resistance: A threat to malaria elimination. Cold Spring Harb. Perspect. Med. 2017, 7, a025619. [Google Scholar] [CrossRef] [PubMed]
- Mackinnon, M.J.; Read, A.F. Virulence in malaria parasites: Integrating genetic, ecological and evolutionary approaches. Philosophical Transactions of the Royal Society of London. Ser. B Biol. Sci. 2004, 359, 965–986. [Google Scholar] [CrossRef] [PubMed]
- Healy, S.A.; Sagara, I.; Assadou, M.H.; Katile, A.; Kone, M.; Imeru, A.; Kwan, J.L.; Swihart, B.J.; Fintzi, J.; Potter, G.E.; et al. A vaccine to block Plasmodium falciparum transmission. NEJM Evid. 2025, 4, EVIDoa2400188. [Google Scholar] [CrossRef]

| Metabolic Pathway | Products/Functions | Therapeutic Relevance |
|---|---|---|
| Fatty acid biosynthesis | Precursors for membrane lipids | Inhibitors block parasite growth in liver stage |
| Isoprenoid biosynthesis | Essential for protein prenylation | Fosmidomycin targets this pathway |
| Haem biosynthesis | Cofactor for electron transport | Loss impairs survival under stress conditions |
| tRNA and protein synthesis | Supports apicoplast-encoded functions | Disruption → delayed death phenotype |
| Epigenetic Mark/Regulator | Role in var Gene Regulation | Connection to Metabolism |
|---|---|---|
| H3K9me3 (trimethylation) | Maintains silent heterochromatin at subtelomeric var loci | Requires SAM as methyl donor |
| H3K9ac (acetylation) | Marks active var promoter regions | Balance shifts with nutrient availability |
| HP1 (Heterochromatin Protein 1) | Mediates clustering of silent var genes at nuclear periphery | Sensitive to chromatin methylation state |
| PfSET10 (histone methyltransferase) | Maintains active var gene in poised state | Activity depends on SAM pool |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pikor, D.; Hurla, M.; Drelichowska, A.; Paul, M. Evolutionary Trajectory of Plasmodium falciparum: From Autonomous Phototroph to Dedicated Parasite. Biomedicines 2025, 13, 2287. https://doi.org/10.3390/biomedicines13092287
Pikor D, Hurla M, Drelichowska A, Paul M. Evolutionary Trajectory of Plasmodium falciparum: From Autonomous Phototroph to Dedicated Parasite. Biomedicines. 2025; 13(9):2287. https://doi.org/10.3390/biomedicines13092287
Chicago/Turabian StylePikor, Damian, Mikołaj Hurla, Alicja Drelichowska, and Małgorzata Paul. 2025. "Evolutionary Trajectory of Plasmodium falciparum: From Autonomous Phototroph to Dedicated Parasite" Biomedicines 13, no. 9: 2287. https://doi.org/10.3390/biomedicines13092287
APA StylePikor, D., Hurla, M., Drelichowska, A., & Paul, M. (2025). Evolutionary Trajectory of Plasmodium falciparum: From Autonomous Phototroph to Dedicated Parasite. Biomedicines, 13(9), 2287. https://doi.org/10.3390/biomedicines13092287






