Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification
Abstract
1. Introduction
2. DNA Methylation and Its Role in Melanoma
2.1. DNA Methylation, Tumor Suppressor Gene Silencing and miRNA Genes in Melanoma
2.2. Demethylating Agents: DNMT Inhibitors
2.3. DNMT Inhibitors: Potential Role in Melanoma Therapy
3. Histone Modifications in Melanoma
3.1. Histone Acetylation and HDAC Inhibitors
3.2. Histone Methylation and EZH2 Inhibitors
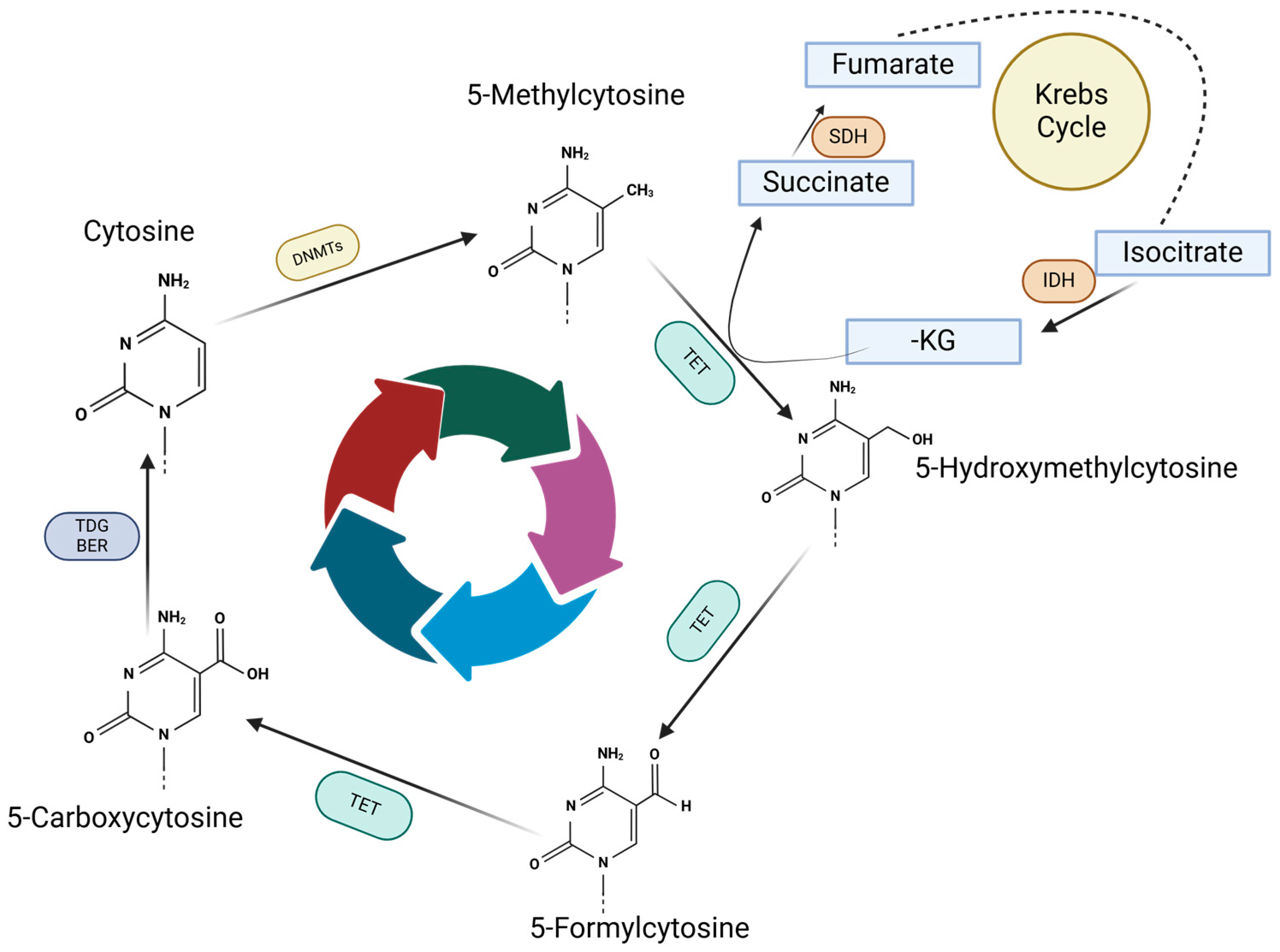
4. TET Enzymes and Their Role in Melanoma
4.1. TET Enzymes and DNA Demethylation
4.2. TET Modulators
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ingravallo, G.; Cazzato, G.; Fortarezza, F.; Federico, S. Epidemiology of Skin Cancer in 2024. In Skin Cancer–Past, Present and Future; Cazzato, G., Ingravallo, G., Eds.; IntechOpen: Rijeka, Croatia, 2025. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous melanoma. Lancet 2023, 402, 485–502. [Google Scholar] [CrossRef]
- Ferrara, G.; Argenziano, G. The WHO 2018 Classification of Cutaneous Melanocytic Neoplasms: Suggestions from Routine Practice. Front. Oncol. 2021, 11, 675296. [Google Scholar] [CrossRef]
- Valentini, R.; Quinn, J.; Murphy, M.J. Nevoid melanoma. Clin. Dermatol. 2025, 43, 29–35. [Google Scholar] [CrossRef]
- Kim, H.Y.; Yoon, J.H.; Cho, E.B.; Park, E.J.; Kim, K.H.; Kim, K.J. A case of spitzoid melanoma. Ann. Dermatol. 2015, 27, 206–209. [Google Scholar] [CrossRef]
- Sohail, A.; Kavaklieva, S. Identifying the clinical and histopathological characteristics of amelanotic melanoma: A case series. Oxf. Med. Case Rep. 2024, 2024, omae029. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Sondak, V.K.; Smalley, K.S. A brief history of melanoma: From mummies to mutations. Melanoma Res. 2012, 22, 114–122. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Breslow, A. Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann. Surg. 1970, 172, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Gerami, P.; Busam, K.; Cochran, A.; Cook, M.G.; Duncan, L.M.; Elder, D.E.; Fullen, D.R.; Guitart, J.; LeBoit, P.E.; Mihm, M.C., Jr.; et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am. J. Surg. Pathol. 2014, 38, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, E.K.; Karakousis, G.C. Current staging and prognostic factors in melanoma. Surg. Oncol. Clin. N. Am. 2015, 24, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Timar, J.; Ladanyi, A. Molecular Pathology of Skin Melanoma: Epidemiology, Differential Diagnostics, Prognosis and Therapy Prediction. Int. J. Mol. Sci. 2022, 23, 5384. [Google Scholar] [CrossRef]
- Teixido, C.; Castillo, P.; Martinez-Vila, C.; Arance, A.; Alos, L. Molecular Markers and Targets in Melanoma. Cells 2021, 10, 2320. [Google Scholar] [CrossRef]
- Timis, T.; Buruiana, S.; Dima, D.; Nistor, M.; Muresan, X.M.; Cenariu, D.; Tigu, A.B.; Tomuleasa, C. Advances in Cell and Immune Therapies for Melanoma. Biomedicines 2025, 13, 98. [Google Scholar] [CrossRef]
- Klempner, S.J.; Fabrizio, D.; Bane, S.; Reinhart, M.; Peoples, T.; Ali, S.M.; Sokol, E.S.; Frampton, G.; Schrock, A.B.; Anhorn, R.; et al. Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist 2020, 25, e147–e159. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Butter, R.; Spijker, R.; Medema, J.P.; van Laarhoven, H.W.M. Clinical and biological effects of demethylating agents on solid tumours—A systematic review. Cancer Treat. Rev. 2017, 54, 10–23. [Google Scholar] [CrossRef]
- Munteanu, R.A.; Moldovan, C.S.; Tigu, A.B.; Drula, R.; Feder, R.; Magdo, L.; Jurj, A.; Raduly, L.; Budisan, L.; Pirlog, R.; et al. 5-Azacytidine treatment inhibits the development of lung cancer models via epigenetic reprogramming and activation of cellular pathways with anti-tumor activity. Med. Pharm. Rep. 2024, 97, 488–506. [Google Scholar] [CrossRef]
- Takeshima, H.; Yoda, Y.; Wakabayashi, M.; Hattori, N.; Yamashita, S.; Ushijima, T. Low-dose DNA demethylating therapy induces reprogramming of diverse cancer-related pathways at the single-cell level. Clin. Epigenet. 2020, 12, 142. [Google Scholar] [CrossRef]
- Zhong, F.; Lin, Y.; Zhao, L.; Yang, C.; Ye, Y.; Shen, Z. Reshaping the tumour immune microenvironment in solid tumours via tumour cell and immune cell DNA methylation: From mechanisms to therapeutics. Br. J. Cancer 2023, 129, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.; Edmiston, S.N.; Vondras, A.; Reiner, A.; Corcoran, D.L.; Shen, R.; Parrish, E.A.; Hao, H.; Lin, L.; Kenney, J.M.; et al. DNA Methylation Classes of Stage II and III Primary Melanomas and Their Clinical and Prognostic Significance. JCO Precis. Oncol. 2024, 8, e2400375. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.B.; Houy, A.; Ganier, O.; Ozemek, B.; Vanhuele, S.; Vincent-Salomon, A.; Cassoux, N.; Mariani, P.; Pierron, G.; Leyvraz, S.; et al. Base-excision repair pathway shapes 5-methylcytosine deamination signatures in pan-cancer genomes. Nat. Commun. 2024, 15, 9864. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Lienert, F.; Wirbelauer, C.; Som, I.; Dean, A.; Mohn, F.; Schubeler, D. Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 2011, 43, 1091–1097. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef]
- Sonar, S.; Nyahatkar, S.; Kalele, K.; Adhikari, M.D. Role of DNA methylation in cancer development and its clinical applications. Clin. Transl. Discov. 2024, 4, e279. [Google Scholar] [CrossRef]
- Stefancu, A.; Moisoiu, V.; Desmirean, M.; Iancu, S.D.; Tigu, A.B.; Petrushev, B.; Jurj, A.; Cozan, R.G.; Budisan, L.; Fetica, B.; et al. SERS-based DNA methylation profiling allows the differential diagnosis of malignant lymphadenopathy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 264, 120216. [Google Scholar] [CrossRef]
- Moisoiu, V.; Sas, V.; Stefancu, A.; Iancu, S.D.; Jurj, A.; Pasca, S.; Iluta, S.; Zimta, A.A.; Tigu, A.B.; Teodorescu, P.; et al. SERS-Based Evaluation of the DNA Methylation Pattern Associated with Progression in Clonal Leukemogenesis of Down Syndrome. Front. Bioeng. Biotechnol. 2021, 9, 703268. [Google Scholar] [CrossRef]
- Moisoiu, V.; Stefancu, A.; Iancu, S.D.; Moisoiu, T.; Loga, L.; Dican, L.; Alecsa, C.D.; Boros, I.; Jurj, A.; Dima, D.; et al. SERS assessment of the cancer-specific methylation pattern of genomic DNA: Towards the detection of acute myeloid leukemia in patients undergoing hematopoietic stem cell transplantation. Anal. Bioanal. Chem. 2019, 411, 7907–7913. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wu, H.; Zhang, H.; Lian, C.G.; Lu, Q. DNA methylation/hydroxymethylation in melanoma. Oncotarget 2017, 8, 78163–78173. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Murphy, G.F.; Lian, C.G. Melanoma epigenetics: Novel mechanisms, markers, and medicines. Lab. Investig. 2014, 94, 822–838. [Google Scholar] [CrossRef] [PubMed]
- Karpf, A.R.; Matsui, S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005, 65, 8635–8639. [Google Scholar] [CrossRef]
- Schinke, C.; Mo, Y.; Yu, Y.; Amiri, K.; Sosman, J.; Greally, J.; Verma, A. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010, 20, 253–265. [Google Scholar] [CrossRef]
- Mezzanotte, J.J.; Hill, V.; Schmidt, M.L.; Shinawi, T.; Tommasi, S.; Krex, D.; Schackert, G.; Pfeifer, G.P.; Latif, F.; Clark, G.J. RASSF6 exhibits promoter hypermethylation in metastatic melanoma and inhibits invasion in melanoma cells. Epigenetics 2014, 9, 1496–1503. [Google Scholar] [CrossRef]
- Helmbold, P.; Richter, A.M.; Walesch, S.; Skorokhod, A.; Marsch, W.; Enk, A.; Dammann, R.H. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J. Investig. Dermatol. 2012, 132, 687–694. [Google Scholar] [CrossRef]
- De Araujo, E.S.; Kashiwabara, A.Y.; Achatz, M.I.; Moredo, L.F.; De Sa, B.C.; Duprat, J.P.; Rosenberg, C.; Carraro, D.M.; Krepischi, A.C. LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res. 2015, 25, 173–177. [Google Scholar] [CrossRef]
- Muthusamy, V.; Duraisamy, S.; Bradbury, C.M.; Hobbs, C.; Curley, D.P.; Nelson, B.; Bosenberg, M. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006, 66, 11187–11193. [Google Scholar] [CrossRef]
- Curry, J.L.; Richards, H.W.; Huttenbach, Y.T.; Medrano, E.E.; Reed, J.A. Different expression patterns of p27 and p57 proteins in benign and malignant melanocytic neoplasms and in cultured human melanocytes. J. Cutan. Pathol. 2009, 36, 197–205. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Biondo, C.; Oteri, R.; Agliano, F.; Morabito, S.; Teti, D.; Venza, I. Epigenetic marks responsible for cadmium-induced melanoma cell overgrowth. Toxicol. In Vitro 2015, 29, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ren, S.; Howell, P.; Fodstad, O.; Riker, A.I. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment. Cell Melanoma Res. 2008, 21, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.G.; Xu, Y.; Ceol, C.; Wu, F.; Larson, A.; Dresser, K.; Xu, W.; Tan, L.; Hu, Y.; Zhan, Q.; et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012, 150, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Trerotola, M.; Relli, V.; Simeone, P.; Alberti, S. Epigenetic inheritance and the missing heritability. Hum. Genom. 2015, 9, 17. [Google Scholar] [CrossRef]
- Geissler, F.; Nesic, K.; Kondrashova, O.; Dobrovic, A.; Swisher, E.M.; Scott, C.L.; Wakefield, M.J. The role of aberrant DNA methylation in cancer initiation and clinical impacts. Ther. Adv. Med. Oncol. 2024, 16, 17588359231220511. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Yuan, Y.; Dong, C.; Yang, M. APC Promoter Methylation in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 653222. [Google Scholar] [CrossRef]
- Barrow, T.M.; Klett, H.; Toth, R.; Bohm, J.; Gigic, B.; Habermann, N.; Scherer, D.; Schrotz-King, P.; Skender, S.; Abbenhardt-Martin, C.; et al. Smoking is associated with hypermethylation of the APC 1A promoter in colorectal cancer: The ColoCare Study. J. Pathol. 2017, 243, 366–375. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Cunningham, J.M.; Cicek, M.S.; Larson, N.B.; Davila, J.; Wang, C.; Larson, M.C.; Song, H.; Dicks, E.M.; Harrington, P.; Wick, M.; et al. Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Sci. Rep. 2014, 4, 4026. [Google Scholar] [CrossRef]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 3970. [Google Scholar] [CrossRef]
- Stefansson, O.A.; Hilmarsdottir, H.; Olafsdottir, K.; Tryggvadottir, L.; Sverrisdottir, A.; Johannsson, O.T.; Jonasson, J.G.; Eyfjord, J.E.; Sigurdsson, S. BRCA1 Promoter Methylation Status in 1031 Primary Breast Cancers Predicts Favorable Outcomes Following Chemotherapy. JNCI Cancer Spectr. 2020, 4, pkz100. [Google Scholar] [CrossRef] [PubMed]
- Nesic, K.; Kondrashova, O.; Hurley, R.M.; McGehee, C.D.; Vandenberg, C.J.; Ho, G.Y.; Lieschke, E.; Dall, G.; Bound, N.; Shield-Artin, K.; et al. Acquired RAD51C Promoter Methylation Loss Causes PARP Inhibitor Resistance in High-Grade Serous Ovarian Carcinoma. Cancer Res. 2021, 81, 4709–4722. [Google Scholar] [CrossRef] [PubMed]
- Helland, A.; Borresen-Dale, A.L.; Peltomaki, P.; Hektoen, M.; Kristensen, G.B.; Nesland, J.M.; de la Chapelle, A.; Lothe, R.A. Microsatellite instability in cervical and endometrial carcinomas. Int. J. Cancer 1997, 70, 499–501. [Google Scholar] [CrossRef]
- Li, K.; Luo, H.; Huang, L.; Luo, H.; Zhu, X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020, 20, 16. [Google Scholar] [CrossRef]
- Castillejo, A.; Hernandez-Illan, E.; Rodriguez-Soler, M.; Perez-Carbonell, L.; Egoavil, C.; Barbera, V.M.; Castillejo, M.I.; Guarinos, C.; Martinez-de-Duenas, E.; Juan, M.J.; et al. Prevalence of MLH1 constitutional epimutations as a cause of Lynch syndrome in unselected versus selected consecutive series of patients with colorectal cancer. J. Med. Genet. 2015, 52, 498–502. [Google Scholar] [CrossRef]
- Ligtenberg, M.J.; Kuiper, R.P.; Chan, T.L.; Goossens, M.; Hebeda, K.M.; Voorendt, M.; Lee, T.Y.; Bodmer, D.; Hoenselaar, E.; Hendriks-Cornelissen, S.J.; et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009, 41, 112–117. [Google Scholar] [CrossRef]
- Imai, K.; Yamamoto, H. Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis 2008, 29, 673–680. [Google Scholar] [CrossRef]
- Micevic, G.; Theodosakis, N.; Bosenberg, M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin. Epigenet. 2017, 9, 34. [Google Scholar] [CrossRef]
- Koga, Y.; Pelizzola, M.; Cheng, E.; Krauthammer, M.; Sznol, M.; Ariyan, S.; Narayan, D.; Molinaro, A.M.; Halaban, R.; Weissman, S.M. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009, 19, 1462–1470. [Google Scholar] [CrossRef]
- Conway, K.; Edmiston, S.N.; Khondker, Z.S.; Groben, P.A.; Zhou, X.; Chu, H.; Kuan, P.F.; Hao, H.; Carson, C.; Berwick, M.; et al. DNA-methylation profiling distinguishes malignant melanomas from benign nevi. Pigment. Cell Melanoma Res. 2011, 24, 352–360. [Google Scholar] [CrossRef]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Korner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef]
- Dong, F.; Lou, D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol. Vis. 2012, 18, 537–546. [Google Scholar]
- Asangani, I.A.; Harms, P.W.; Dodson, L.; Pandhi, M.; Kunju, L.P.; Maher, C.A.; Fullen, D.R.; Johnson, T.M.; Giordano, T.J.; Palanisamy, N.; et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget 2012, 3, 1011–1025. [Google Scholar] [CrossRef]
- Micevic, G.; Muthusamy, V.; Damsky, W.; Theodosakis, N.; Liu, X.; Meeth, K.; Wingrove, E.; Santhanakrishnan, M.; Bosenberg, M. DNMT3b Modulates Melanoma Growth by Controlling Levels of mTORC2 Component RICTOR. Cell Rep. 2016, 14, 2180–2192. [Google Scholar] [CrossRef]
- Lopez-Serra, P.; Esteller, M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012, 31, 1609–1622. [Google Scholar] [CrossRef]
- Cyrta, J.; Masliah-Planchon, J.; Hoare, O.; Brillet, R.; Andrianteranagna, M.; Sohier, P.; Cardoen, L.; Bouchoucha, Y.; Filser, M.; Goncalves, A.; et al. SMARCB1-deficient malignant melanocytic uveal tumours: A new neural crest-derived tumour entity with SMARCB1-related germline predisposition. J. Pathol. 2025, 265, 357–371. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Shinojima, T.; Yu, Q.; Huang, S.K.; Li, M.; Mizuno, R.; Liu, E.T.; Hoon, D.S.; Lessard, L. Heterogeneous epigenetic regulation of TIMP3 in prostate cancer. Epigenetics 2012, 7, 1279–1289. [Google Scholar] [CrossRef]
- Tigu, A.B.; Bancos, A. The Role of Epigenetic Modifier Mutations in Peripheral T-Cell Lymphomas. Curr. Issues Mol. Biol. 2023, 45, 8974–8988. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: Mechanism and clinical application. Clin. Epigenet. 2021, 13, 166. [Google Scholar] [CrossRef]
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.G.; Fandy, T.E. DNA methylation inhibitors: Retrospective and perspective view. Adv. Cancer Res. 2021, 152, 205–223. [Google Scholar] [CrossRef]
- Nie, J.; Liu, L.; Li, X.; Han, W. Decitabine, a new star in epigenetic therapy: The clinical application and biological mechanism in solid tumors. Cancer Lett. 2014, 354, 12–20. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, G.; Li, Y.; Lei, D.; Xiang, J.; Ouyang, L.; Wang, Y.; Yang, J. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 2022, 13, 1072651. [Google Scholar] [CrossRef]
- Hollenbach, P.W.; Nguyen, A.N.; Brady, H.; Williams, M.; Ning, Y.; Richard, N.; Krushel, L.; Aukerman, S.L.; Heise, C.; MacBeth, K.J. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS ONE 2010, 5, e9001. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Matei, D.; Fang, F.; Shen, C.; Schilder, J.; Arnold, A.; Zeng, Y.; Berry, W.A.; Huang, T.; Nephew, K.P. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012, 72, 2197–2205. [Google Scholar] [CrossRef]
- Hagemann, S.; Heil, O.; Lyko, F.; Brueckner, B. Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS ONE 2011, 6, e17388. [Google Scholar] [CrossRef]
- Kagan, A.B.; Garrison, D.A.; Anders, N.M.; Webster, J.A.; Baker, S.D.; Yegnasubramanian, S.; Rudek, M.A. DNA methyltransferase inhibitor exposure-response: Challenges and opportunities. Clin. Transl. Sci. 2023, 16, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Giunta, E.F.; Arrichiello, G.; Curvietto, M.; Pappalardo, A.; Bosso, D.; Rosanova, M.; Diana, A.; Giordano, P.; Petrillo, A.; Federico, P.; et al. Epigenetic Regulation in Melanoma: Facts and Hopes. Cells 2021, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Aleotti, V.; Catoni, C.; Poggiana, C.; Rosato, A.; Facchinetti, A.; Scaini, M.C. Methylation Markers in Cutaneous Melanoma: Unravelling the Potential Utility of Their Tracking by Liquid Biopsy. Cancers 2021, 13, 6217. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.; Chhabra, G.; Singh, C.K.; Guzman-Perez, G.; Shirley, C.A.; Ahmad, N. Mechanisms of Immunotherapy Resistance in Cutaneous Melanoma: Recognizing a Shapeshifter. Front. Oncol. 2022, 12, 880876. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Iles, M.M.; Law, M.H.; Stacey, S.N.; Han, J.; Fang, S.; Pfeiffer, R.; Harland, M.; Macgregor, S.; Taylor, J.C.; Aben, K.K.; et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat. Genet. 2013, 45, 428–432. [Google Scholar] [CrossRef]
- Lim, S.Y.; Shklovskaya, E.; Lee, J.H.; Pedersen, B.; Stewart, A.; Ming, Z.; Irvine, M.; Shivalingam, B.; Saw, R.P.M.; Menzies, A.M.; et al. The molecular and functional landscape of resistance to immune checkpoint blockade in melanoma. Nat. Commun. 2023, 14, 1516. [Google Scholar] [CrossRef]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef]
- Hanly, A.; Gibson, F.; Nocco, S.; Rogers, S.; Wu, M.; Alani, R.M. Drugging the Epigenome: Overcoming Resistance to Targeted and Immunotherapies in Melanoma. JID Innov. 2022, 2, 100090. [Google Scholar] [CrossRef]
- Zakharia, Y.; Monga, V.; Swami, U.; Bossler, A.D.; Freesmeier, M.; Frees, M.; Khan, M.; Frydenlund, N.; Srikantha, R.; Vanneste, M.; et al. Targeting epigenetics for treatment of BRAF mutated metastatic melanoma with decitabine in combination with vemurafenib: A phase lb study. Oncotarget 2017, 8, 89182–89193. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Beumer, J.H.; Tarhini, A.A.; Moschos, S.; Buch, S.C.; Egorin, M.J.; Lin, Y.; Christner, S.; Kirkwood, J.M. Safety and efficacy of decitabine in combination with temozolomide in metastatic melanoma: A phase I/II study and pharmacokinetic analysis. Ann. Oncol. 2013, 24, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Ahn, A.; Stockwell, P.A.; Parry, M.; Motwani, J.; Gallagher, S.J.; Shklovskaya, E.; Tiffen, J.; Eccles, M.R.; et al. Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma. iScience 2018, 4, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Xu, T.; Lee, J.J.; Murphy, G.F.; Lian, C.G. Epigenetic markers in melanoma. Melanoma Manag. 2015, 2, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Tiffen, J.C.; Hersey, P. Histone Modifications, Modifiers and Readers in Melanoma Resistance to Targeted and Immune Therapy. Cancers 2015, 7, 1959–1982. [Google Scholar] [CrossRef]
- Martinez, D.R.; Richards, H.W.; Lin, Q.; Torres-Cabala, C.A.; Prieto, V.G.; Curry, J.L.; Medrano, E.E. H3K79me3T80ph is a Novel Histone Dual Modification and a Mitotic Indicator in Melanoma. J. Ski. Cancer 2012, 2012, 823534. [Google Scholar] [CrossRef]
- Yu, J.; Chai, P.; Xie, M.; Ge, S.; Ruan, J.; Fan, X.; Jia, R. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021, 22, 85. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Utility of histone H3K27me3 and H4K20me as diagnostic indicators of melanoma. Melanoma Res. 2020, 30, 159–165. [Google Scholar] [CrossRef]
- Fiziev, P.; Akdemir, K.C.; Miller, J.P.; Keung, E.Z.; Samant, N.S.; Sharma, S.; Natale, C.A.; Terranova, C.J.; Maitituoheti, M.; Amin, S.B.; et al. Systematic Epigenomic Analysis Reveals Chromatin States Associated with Melanoma Progression. Cell Rep. 2017, 19, 875–889. [Google Scholar] [CrossRef]
- Segura, M.F.; Fontanals-Cirera, B.; Gaziel-Sovran, A.; Guijarro, M.V.; Hanniford, D.; Zhang, G.; Gonzalez-Gomez, P.; Morante, M.; Jubierre, L.; Zhang, W.; et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013, 73, 6264–6276. [Google Scholar] [CrossRef]
- Daud, A.I.; Dawson, J.; DeConti, R.C.; Bicaku, E.; Marchion, D.; Bastien, S.; Hausheer, F.A., III; Lush, R.; Neuger, A.; Sullivan, D.M.; et al. Potentiation of a topoisomerase I inhibitor, karenitecin, by the histone deacetylase inhibitor valproic acid in melanoma: Translational and phase I/II clinical trial. Clin. Cancer Res. 2009, 15, 2479–2487. [Google Scholar] [CrossRef] [PubMed]
- Gore, L.; Rothenberg, M.L.; O’Bryant, C.L.; Schultz, M.K.; Sandler, A.B.; Coffin, D.; McCoy, C.; Schott, A.; Scholz, C.; Eckhardt, S.G. A phase I and pharmacokinetic study of the oral histone deacetylase inhibitor, MS-275, in patients with refractory solid tumors and lymphomas. Clin. Cancer Res. 2008, 14, 4517–4525. [Google Scholar] [CrossRef] [PubMed]
- Murray, K. The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry 1964, 3, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008, 647, 21–29. [Google Scholar] [CrossRef]
- Chien, Y.C.; Wu, J.Y.; Liu, L.C.; Yu, Y.L. Capsanthin inhibits migration and reduces N-linked glycosylation of PD-L1 via the EZH2-PD-L1 axis in triple-negative breast cancer brain metastasis. Cell Death Discov. 2025, 11, 85. [Google Scholar] [CrossRef]
- Li, C.; Wang, Z.; Yao, L.; Lin, X.; Jian, Y.; Li, Y.; Zhang, J.; Shao, J.; Tran, P.D.; Hagman, J.R.; et al. Mi-2β promotes immune evasion in melanoma by activating EZH2 methylation. Nat. Commun. 2024, 15, 2163. [Google Scholar] [CrossRef]
- Hou, Y.; Zak, J.; Shi, Y.; Pratumchai, I.; Dinner, B.; Wang, W.; Qin, K.; Weber, E.W.; Teijaro, J.R.; Wu, P. Transient EZH2 Suppression by Tazemetostat during In Vitro Expansion Maintains T-Cell Stemness and Improves Adoptive T-Cell Therapy. Cancer Immunol. Res. 2025, 13, 47–65. [Google Scholar] [CrossRef]
- Jin, N.; George, T.L.; Otterson, G.A.; Verschraegen, C.; Wen, H.; Carbone, D.; Herman, J.; Bertino, E.M.; He, K. Advances in epigenetic therapeutics with focus on solid tumors. Clin. Epigenet. 2021, 13, 83. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Smitheman, K.N.; Kamat, C.D.; Soong, D.; Federowicz, K.E.; Van Aller, G.S.; Schneck, J.L.; Carson, J.D.; Liu, Y.; Butticello, M.; et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015, 28, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, C.; Wang, X. TET (Ten-eleven translocation) family proteins: Structure, biological functions and applications. Signal Transduct. Target. Ther. 2023, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Epigenetic regulation in human melanoma: Past and future. Epigenetics 2015, 10, 103–121. [Google Scholar] [CrossRef] [PubMed]
- Bonvin, E.; Radaelli, E.; Bizet, M.; Luciani, F.; Calonne, E.; Putmans, P.; Nittner, D.; Singh, N.K.; Santagostino, S.F.; Petit, V.; et al. TET2-Dependent Hydroxymethylome Plasticity Reduces Melanoma Initiation and Progression. Cancer Res. 2019, 79, 482–494. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Gong, F.; Guo, Y.; Niu, Y.; Jin, J.; Zhang, X.; Shi, X.; Zhang, L.; Li, R.; Chen, L.; Ma, R.Z. Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma. Oncotarget 2017, 8, 315–328. [Google Scholar] [CrossRef]
- Xu, Y.P.; Lv, L.; Liu, Y.; Smith, M.D.; Li, W.C.; Tan, X.M.; Cheng, M.; Li, Z.; Bovino, M.; Aube, J.; et al. Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J. Clin. Investig. 2019, 129, 4316–4331. [Google Scholar] [CrossRef]
- Cheng, M.; Chu, A.K.Y.; Li, Z.; Yang, S.; Smith, M.D.; Zhang, Q.; Brown, N.G.; Marzluff, W.F.; Bardeesy, N.; Milner, J.J.; et al. TET2 promotes tumor antigen presentation and T cell IFN-γ, which is enhanced by vitamin C. JCI Insight 2024, 9, e175098. [Google Scholar] [CrossRef]
- Mills, C.N.; Joshi, S.S.; Niles, R.M. Expression and function of hypoxia inducible factor-1 alpha in human melanoma under non-hypoxic conditions. Mol. Cancer 2009, 8, 104. [Google Scholar] [CrossRef]
- Fischer, A.P.; Miles, S.L. Silencing HIF-1α induces TET2 expression and augments ascorbic acid induced 5-hydroxymethylation of DNA in human metastatic melanoma cells. Biochem. Biophys. Res. Commun. 2017, 490, 176–181. [Google Scholar] [CrossRef]
- Bray, J.K.; Dawlaty, M.M.; Verma, A.; Maitra, A. Roles and Regulations of TET Enzymes in Solid Tumors. Trends Cancer 2021, 7, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, J.M.; Ghosh, S.; Lackford, B.L.; Yellaboina, S.; Zheng, X.; Li, R.; Cuddapah, S.; Wade, P.A.; Hu, G.; Jothi, R. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012, 40, 3364–3377. [Google Scholar] [CrossRef] [PubMed]
- Moyon, S.; Frawley, R.; Marechal, D.; Huang, D.; Marshall-Phelps, K.L.H.; Kegel, L.; Bostrand, S.M.K.; Sadowski, B.; Jiang, Y.H.; Lyons, D.A.; et al. TET1-mediated DNA hydroxymethylation regulates adult remyelination in mice. Nat. Commun. 2021, 12, 3359. [Google Scholar] [CrossRef]
- Mimouni, N.E.H.; Paiva, I.; Barbotin, A.L.; Timzoura, F.E.; Plassard, D.; Le Gras, S.; Ternier, G.; Pigny, P.; Catteau-Jonard, S.; Simon, V.; et al. Polycystic ovary syndrome is transmitted via a transgenerational epigenetic process. Cell Metab. 2021, 33, 513–530.e8. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, O.; Mullally, A.; Hedvat, C.; Garcia-Manero, G.; Patel, J.; Wadleigh, M.; Malinge, S.; Yao, J.; Kilpivaara, O.; Bhat, R.; et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 2009, 114, 144–147. [Google Scholar] [CrossRef]
- Kosmider, O.; Delabesse, E.; de Mas, V.M.; Cornillet-Lefebvre, P.; Blanchet, O.; Delmer, A.; Recher, C.; Raynaud, S.; Bouscary, D.; Viguie, F.; et al. TET2 mutations in secondary acute myeloid leukemias: A French retrospective study. Haematologica 2011, 96, 1059–1063. [Google Scholar] [CrossRef]
- Antunes, C.; Da Silva, J.D.; Guerra-Gomes, S.; Alves, N.D.; Ferreira, F.; Loureiro-Campos, E.; Branco, M.R.; Sousa, N.; Reik, W.; Pinto, L.; et al. Tet3 ablation in adult brain neurons increases anxiety-like behavior and regulates cognitive function in mice. Mol. Psychiatry 2021, 26, 1445–1457. [Google Scholar] [CrossRef]
- Chen, B.; Du, Y.R.; Zhu, H.; Sun, M.L.; Wang, C.; Cheng, Y.; Pang, H.; Ding, G.; Gao, J.; Tan, Y.; et al. Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature 2022, 605, 761–766. [Google Scholar] [CrossRef]
- Gu, T.P.; Guo, F.; Yang, H.; Wu, H.P.; Xu, G.F.; Liu, W.; Xie, Z.G.; Shi, L.; He, X.; Jin, S.G.; et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011, 477, 606–610. [Google Scholar] [CrossRef]
- Jung, I.; An, J.; Ko, M. Epigenetic Regulators of DNA Cytosine Modification: Promising Targets for Cancer Therapy. Biomedicines 2023, 11, 654. [Google Scholar] [CrossRef]
- Agathocleous, M.; Meacham, C.E.; Burgess, R.J.; Piskounova, E.; Zhao, Z.; Crane, G.M.; Cowin, B.L.; Bruner, E.; Murphy, M.M.; Chen, W.; et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 2017, 549, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; He, X.; Zhu, Y.; Ding, Z.; Dong, H.; Feng, Y.; Du, J.; Wang, H.; Wu, X.; Zhang, L.; et al. SIRT1 Activation Disrupts Maintenance of Myelodysplastic Syndrome Stem and Progenitor Cells by Restoring TET2 Function. Cell Stem Cell 2018, 23, 355–369.e9. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.S.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018, 559, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Yen, K.; Travins, J.; Wang, F.; David, M.D.; Artin, E.; Straley, K.; Padyana, A.; Gross, S.; DeLaBarre, B.; Tobin, E.; et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov. 2017, 7, 478–493. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef]
- Chen, L.L.; Morcelle, C.; Cheng, Z.L.; Chen, X.; Xu, Y.; Gao, Y.; Song, J.; Li, Z.; Smith, M.D.; Shi, M.; et al. Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses. Nat. Cell Biol. 2022, 24, 353–363. [Google Scholar] [CrossRef]
- Zhang, D.; An, X.; Li, Z.; Zhang, S. Role of gene promoter methylation regulated by TETs and DNMTs in the overexpression of HLA-G in MCF-7 cells. Exp. Ther. Med. 2019, 17, 4709–4714. [Google Scholar] [CrossRef]
- Kaplanek, R.; Kejik, Z.; Hajduch, J.; Vesela, K.; Kucnirova, K.; Skalickova, M.; Venhauerova, A.; Hosnedlova, B.; Hromadka, R.; Dytrych, P.; et al. TET protein inhibitors: Potential and limitations. Biomed. Pharmacother. 2023, 166, 115324. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tigu, A.B.; Ivancuta, A.; Uhl, A.; Sabo, A.C.; Nistor, M.; Mureșan, X.-M.; Cenariu, D.; Timis, T.; Diculescu, D.; Gulei, D. Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification. Biomedicines 2025, 13, 1188. https://doi.org/10.3390/biomedicines13051188
Tigu AB, Ivancuta A, Uhl A, Sabo AC, Nistor M, Mureșan X-M, Cenariu D, Timis T, Diculescu D, Gulei D. Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification. Biomedicines. 2025; 13(5):1188. https://doi.org/10.3390/biomedicines13051188
Chicago/Turabian StyleTigu, Adrian Bogdan, Andrei Ivancuta, Andrada Uhl, Alexandru Cristian Sabo, Madalina Nistor, Ximena-Maria Mureșan, Diana Cenariu, Tanase Timis, Doru Diculescu, and Diana Gulei. 2025. "Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification" Biomedicines 13, no. 5: 1188. https://doi.org/10.3390/biomedicines13051188
APA StyleTigu, A. B., Ivancuta, A., Uhl, A., Sabo, A. C., Nistor, M., Mureșan, X.-M., Cenariu, D., Timis, T., Diculescu, D., & Gulei, D. (2025). Epigenetic Therapies in Melanoma—Targeting DNA Methylation and Histone Modification. Biomedicines, 13(5), 1188. https://doi.org/10.3390/biomedicines13051188






