MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization
Abstract
1. Introduction
2. MiRNAs and Cancer
3. Macrophages and Their Polarization
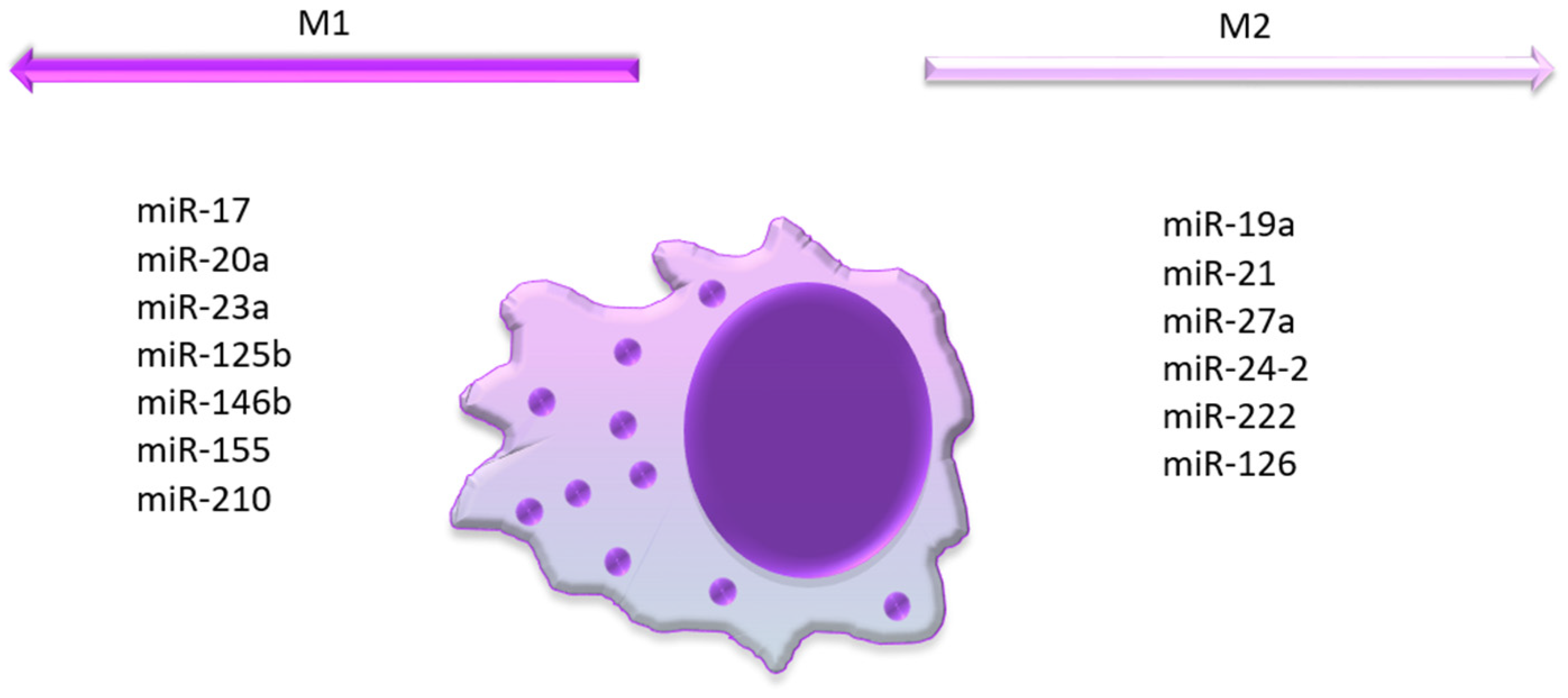
4. MicroRNAs Associated with a Bad Prognosis of AML and M1 Polarization
5. MicroRNAs Associated with a Bad Prognosis in AML and M2 Polarization
6. MicroRNAs Associated with a Bad Prognosis in AML and Unknown Macrophage Polarization
7. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Cobb, M. 60 years ago, Francis Crick changed the logic of biology. PLoS Biol. 2017, 15, e2003243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Jin, J.-P.; Wang, J.-Q.; Zhang, Z.-G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef]
- Tang, X.; Tang, G.; Ozcan, S. Role of microRNAs in diabetes. Biochim. Biophys. Acta 2008, 1779, 697–701. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef]
- Romano, N.; Macino, G. Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef]
- Neaga, A.; Bagacean, C.; Tempescul, A.; Jimbu, L.; Mesaros, O.; Blag, C.; Tomuleasa, C.; Bocsan, C.; Gaman, M.; Zdrenghea, M. MicroRNAs Associated with a Good Prognosis of Acute Myeloid Leukemia and Their Effect on Macrophage Polarization. Front. Immunol. 2020, 11, 582915. [Google Scholar] [CrossRef]
- Zojer, N.; Königsberg, R.; Ackermann, J.; Fritz, E.; Dallinger, S.; Krömer, E.; Kaufmann, H.; Riedl, L.; Gisslinger, H.; Schreiber, S.; et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood 2000, 95, 1925–1930. [Google Scholar] [CrossRef]
- Chung, C.-Y.; Kantarjian, H.; Haidar, M.; Starostik, P.; Manshouri, T.; Gidel, C.; Freireich, E.; Keating, M.; Albitar, M. Deletions in the 13q14 locus in adult lymphoblastic leukemia. Cancer 2000, 88, 1359–1364. [Google Scholar] [CrossRef]
- Rosenwald, A.; Ott, G.; Krumdiek, A.K.; Dreyling, M.H.; Katzenberger, T.; Kalla, J.; Roth, S.; Ott, M.M.; Müller-Hermelink, H.K. A biological role for deletions in chromosomal band 13q14 in mantle cell and peripheral t-cell lymphomas? Genes Chromosomes Cancer 1999, 26, 210–214. [Google Scholar] [CrossRef]
- Rouault, A.; Banneau, G.; MacGrogan, G.; Jones, N.; Elarouci, N.; Barouk-Simonet, E.; Venat, L.; Coupier, I.; Letouzé, E.; de Reyniès, A.; et al. Deletion of Chromosomes 13q and 14q Is a Common Feature of Tumors with BRCA2 Mutations. PLoS ONE 2012, 7, e52079. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.-S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Dixon-McIver, A.; East, P.; Mein, C.A.; Cazier, J.B.; Molloy, G.; Chaplin, T.; Andrew Lister, T.; Young, B.D.; Debernardi, S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS ONE 2008, 3, e2141. [Google Scholar] [CrossRef] [PubMed]
- Jongen-Lavrencic, M.; Sun, S.M.; Dijkstra, M.K.; Valk, P.J.M.; Löwenberg, B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008, 111, 5078–5085. [Google Scholar] [CrossRef]
- Liao, Q.; Wang, B.; Li, X.; Jiang, G. miRNAs in acute myeloid leukemia. Oncotarget 2017, 8, 3666–3682. [Google Scholar] [CrossRef]
- Hoang, D.H.; Zhao, D.; Branciamore, S.; Maestrini, D.; Rodriguez, I.R.; Kuo, Y.H.; Rockne, R.; Khaled, S.K.; Zhang, B.; Nguyen, L.X.T.; et al. MicroRNA networks in FLT3-ITD acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2112482119. [Google Scholar] [CrossRef]
- Gadewal, N.; Kumar, R.; Aher, S.; Gardane, A.; Gaur, T.; Varma, A.K.; Khattry, N.; Hasan, S.K. miRNA-mRNA Profiling Reveals Prognostic Impact of SMC1A Expression in Acute Myeloid Leukemia. Oncol. Res. 2020, 28, 321–330. [Google Scholar] [CrossRef]
- Ghazaryan, A.; Wallace, J.A.; Tang, W.W.; Barba, C.; Lee, S.-H.; Bauer, K.M.; Nelson, M.C.; Kim, C.N.; Stubben, C.; Voth, W.P.; et al. miRNA-1 promotes acute myeloid leukemia cell pathogenesis through metabolic regulation. Front. Genet. 2023, 14, 1192799. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, R.a.; Zhang, Y.; Fan, W.; Xiao, F.; Yan, X. Low expression of circulating microRNA-328 is associated with poor prognosis in patients with acute myeloid leukemia. Diagn. Pathol. 2015, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, Y.; Shang, L.; Chen, H.; Yue, Y.; Dong, W.; Guo, Y.; Yang, H.; Yang, X.; Liu, Y.; et al. Overexpression of miR-17 predicts adverse prognosis and disease recurrence for acute myeloid leukemia. Int. J. Clin. Oncol. 2022, 27, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Macha, M.A.; Seshacharyulu, P.; Krishn, S.R.; Pai, P.; Rachagani, S.; Jain, M.; Batra, S.K. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr. Pharm. Des. 2014, 20, 5287–5297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Xiao, D.; Wang, J. A 16-miRNA Prognostic Model to Predict Overall Survival in Neuroblastoma. Front. Genet. 2022, 13, 827842. [Google Scholar] [CrossRef]
- Gahlawat, A.W.; Fahed, L.; Witte, T.; Schott, S. Total circulating microRNA level as an independent prognostic marker for risk stratification in breast cancer. Br. J. Cancer 2022, 127, 156–162. [Google Scholar] [CrossRef]
- Sanchez-Espiridion, B.; Montalban, C.; Guisado, P.; Rodriguez, M.E.; Canales, M.; Alves, J.; Vega, F.; Morente, M.; Tomás, J.F.; Piris, M.A.; et al. Mirna EXPRESSION SIGNATURES as PROGNOSTIC Markers IN ADVANCED Classical HODGKIN LYMPHOMA. Blood 2010, 116, 4157. [Google Scholar] [CrossRef]
- Li, L.; Sun, P.; Zhang, C.; Li, Z.; Cui, K.; Zhou, W. MiR-98 modulates macrophage polarization and suppresses the effects of tumor-associated macrophages on promoting invasion and epithelial–mesenchymal transition of hepatocellular carcinoma. Cancer Cell Int. 2018, 18, 95. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, L.; Hu, Y.; Huang, Y.; Zhang, Y.; Zheng, X.; Wang, S.; Wang, Y.; Yu, Y.; Zhang, M.; et al. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci. Rep. 2016, 6, 25602. [Google Scholar] [CrossRef]
- Iurca, I.; Tirpe, A.; Zimta, A.-A.; Moldovan, C.; Gulei, D.; Slabý, O.; Condorelli, G.; Berindan-Neagoe, I. Macrophages Interaction and MicroRNA Interplay in the Modulation of Cancer Development and Metastasis. Front. Immunol. 2020, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.; Stölzel, F.; Wang, K.W.; Röllig, C.; Tursky, M.L.; Molloy, T.J.; Ma, D.D. miR-10a as a therapeutic target and predictive biomarker for MDM2 inhibition in acute myeloid leukemia. Leukemia 2021, 35, 1933–1948. [Google Scholar] [CrossRef] [PubMed]
- Issa, H.; Bhayadia, R.; Winkler, R.; Swart, L.E.; Heckl, D.; Klusmann, J.-H. Preclinical testing of miRNA-193b-3p mimic in acute myeloid leukemias. Leukemia 2023, 37, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Shahid, W.; Shaheen, J.; Akhtar, M.W.; Sadaf, S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 22783. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-L.; Wang, X.; Mann, M.; Adamus, T.P.; Wang, D.; Moreira, D.F.; Zhang, Z.; Ouyang, C.; He, X.; Zhang, B.; et al. Myeloid cell–targeted miR-146a mimic inhibits NF-κB–driven inflammation and leukemia progression in vivo. Blood 2020, 135, 167–180. [Google Scholar] [CrossRef]
- Chen, P.; Price, C.; Li, Z.; Li, Y.; Cao, D.; Wiley, A.; He, C.; Gurbuxani, S.; Kunjamma, R.B.; Huang, H.; et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 11511–11516. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Mesaros, O.; Jimbu, L.; Neaga, A.; Popescu, C.; Berceanu, I.; Tomuleasa, C.; Fetica, B.; Zdrenghea, M. Macrophage Polarization in Chronic Lymphocytic Leukemia: Nurse-Like Cells Are the Caretakers of Leukemic Cells. Biomedicines 2020, 8, 516. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.H.; Jin, L. Macrophage Polarization in Physiological and Pathological Pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Boutilier, A.J.; Elsawa, S.F. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guan, N.; Zhang, X.; Liu, Y.; Gao, X.; Wang, L. Study on the imbalance of M1/M2 macrophage polarization in severe chronic periodontitis. Technol. Health Care 2023, 31, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef] [PubMed]
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Mat Zin, A.A.; Ang, K.C.; Ch’ng, E.S. Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front. Oncol. 2019, 9, 1512. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Lu, L.; McCurdy, S.; Huang, S.; Zhu, X.; Peplowska, K.; Tiirikainen, M.; Boisvert, W.A.; Garmire, L.X. Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci. Rep. 2016, 6, 37446. [Google Scholar] [CrossRef]
- Feketea, G.; Bocsan, C.I.; Popescu, C.; Gaman, M.; Stanciu, L.A.; Zdrenghea, M.T. A Review of Macrophage MicroRNAs’ Role in Human Asthma. Cells 2019, 8, 420. [Google Scholar] [CrossRef]
- Mian, Y.A.; Zeleznik-Le, N.J. The miR-17∼92 cluster contributes to MLL leukemia through the repression of MEIS1 competitor PKNOX1. Leuk. Res. 2016, 46, 51–60. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Y.; Yang, X.; Chen, H.; Yang, H.; Liu, Y.; Gu, W. High expression of miR-107 and miR-17 predicts poor prognosis and guides treatment selection in acute myeloid leukemia. Transl. Cancer Res. 2023, 12, 913–927. [Google Scholar] [CrossRef]
- Kuo, G.; Wu, C.-Y.; Yang, H.-Y. MiR-17-92 cluster and immunity. J. Formos. Med. Assoc. 2019, 118, 2–6. [Google Scholar] [CrossRef]
- Huang, D.; Peng, Y.; Ma, K.; Deng, X.; Tang, L.; Jing, D.; Shao, Z. MiR-20a, a novel promising biomarker to predict prognosis in human cancer: A meta-analysis. BMC Cancer 2018, 18, 1189. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gu, H.; Wang, S.; Qian, H.; Zhu, W.; Zhang, L.; Zhao, C.; Tao, Y.; Xu, W. Circulating miR-17-5p and miR-20a: Molecular markers for gastric cancer. Mol. Med. Rep. 2012, 5, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Zhao, S.; Tang, H.; Zhang, D.; Sun, H.; Yu, F.; Jiang, W.; Yue, B.; Wang, J.; Zhang, M.; et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget 2016, 7, 45199–45213. [Google Scholar] [CrossRef] [PubMed]
- Selven, H.; Andersen, S.; Pedersen, M.I.; Lombardi, A.P.G.; Busund, L.-T.R.; Kilvær, T.K. High expression of miR-17-5p and miR-20a-5p predicts favorable disease-specific survival in stage I-III colon cancer. Sci. Rep. 2022, 12, 7080. [Google Scholar] [CrossRef]
- Fan, M.-Q.; Huang, C.-B.; Gu, Y.; Xiao, Y.; Sheng, J.-X.; Zhong, L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2013, 32, 21. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Zhang, L.; Pei, X.; Lian, C.; Liu, Y.; Tan, H.; Lei, P. MiR-20a-5p functions as a potent tumor suppressor by targeting PPP6C in acute myeloid leukemia. PLoS ONE 2021, 16, e0256995. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Pan, C.; Li, L.; Bian, Z.; Lv, Z.; Shi, L.; Zhang, J.; Li, D.; Gu, H.; Zhang, C.-Y.; et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α. J. Allergy Clin. Immunol. 2013, 132, 426–436. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef]
- Shaham, L.; Binder, V.; Gefen, N.; Borkhardt, A.; Izraeli, S. MiR-125 in normal and malignant hematopoiesis. Leukemia 2012, 26, 2011–2018. [Google Scholar] [CrossRef]
- Klusmann, J.H.; Li, Z.; Böhmer, K.; Maroz, A.; Koch, M.L.; Emmrich, S.; Godinho, F.J.; Orkin, S.H.; Reinhardt, D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010, 24, 478–490. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; So, A.Y.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Harris, M.H.; Zhou, B.; Lodish, H.F. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 21558–21563. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Gibson, W.S.; Balazs, A.B.; Baltimore, D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA 2010, 107, 14235–14240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Bai, H.; Wang, C.; Wei, D.; Qin, Y.; Xu, X. microRNA-125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis. Mol. Med. Rep. 2014, 9, 1909–1916. [Google Scholar] [CrossRef] [PubMed]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Su, Y.; Zheng, Z.; Qi, J.; Wang, W.; Wang, C. miR-146b-5p promotes colorectal cancer progression by targeting TRAF6. Exp. Ther. Med. 2022, 23, 231. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhou, Z.; Li, J.; Xiang, R. miR-146b Functions as an Oncogene in Oral Squamous Cell Carcinoma by Targeting HBP1. Technol. Cancer Res. Treat. 2020, 19, 1533033820959404. [Google Scholar] [CrossRef]
- Mitsumura, T.; Ito, Y.; Chiba, T.; Matsushima, T.; Kurimoto, R.; Tanaka, Y.; Kato, T.; Uchida, K.; Ito, T.; Yamamoto, K.; et al. Ablation of miR-146b in mice causes hematopoietic malignancy. Blood Adv. 2018, 2, 3483–3491. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; He, C.; Wang, D.; Yuan, X.; Chen, J.; Jin, J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol. Dis. 2010, 44, 191–197. [Google Scholar] [CrossRef]
- Zhu, R.; Zhao, W.; Fan, F.; Tang, L.; Liu, J.; Luo, T.; Deng, J.; Hu, Y. A 3-miRNA signature predicts prognosis of pediatric and adolescent cytogenetically normal acute myeloid leukemia. Oncotarget 2017, 8, 38902–38913. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Hur, E.H.; Moon, J.H.; Goo, B.K.; Choi, D.R.; Lee, J.H. Expression and prognostic significance of microRNAs in Korean patients with myelodysplastic syndrome. Korean J. Intern. Med. 2019, 34, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Zhang, H.; Hao, Y.; Xu, F.; Yang, J.; Zhang, R.; Lu, G.; Zheng, Z.; Cui, M.; Qi, C.F.; et al. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine 2016, 14, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Kalkusova, K.; Taborska, P.; Stakheev, D.; Smrz, D. The Role of miR-155 in Antitumor Immunity. Cancers 2022, 14, 5414. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Hua, L.-M.; Guo, M.; Luo, J.-M. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol. Rep. 2014, 32, 2253–2259. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Yang, F.; Qin, Z.; Jing, X.; Shu, Y.; Shen, H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Guan, Y.; Sun, Y.; Wang, X.; Lively, K.; Wang, Y.; Luo, M.; Kim, J.A.; Murphy, E.A.; et al. Breast cancer cell–derived microRNA-155 suppresses tumor progression via enhancing immune cell recruitment and antitumor function. J. Clin. Investig. 2022, 132, e157248. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Nguyen, M.-H.T.; Luo, Y.-H.; Li, A.-L.; Tsai, J.-C.; Wu, K.-L.; Chung, P.-J.; Ma, N. miRNA as a Modulator of Immunotherapy and Immune Response in Melanoma. Biomolecules 2021, 11, 1648. [Google Scholar] [CrossRef]
- Hu, X.L.; Tang, A.P. Expression of miR-155 in Acute Myeloid Leukemia and Its Clinical Significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2016, 24, 980–984. [Google Scholar] [CrossRef]
- Tang, L.; Peng, Y.Z.; Li, C.G.; Jiang, H.W.; Mei, H.; Hu, Y. Prognostic and Clinicopathological Significance of MiR-155 in Hematologic Malignancies: A Systematic Review and Meta-analysis. J. Cancer 2019, 10, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.S.; Metzeler, K.H.; Volinia, S.; Wu, Y.Z.; Mrózek, K.; Nicolet, D.; Kohlschmidt, J.; Whitman, S.P.; Mendler, J.H.; et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J. Clin. Oncol. 2013, 31, 2086–2093. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, R.; Hughes, M.; Morris, V.; Bolouri, H.; Gerbing, R.B.; Wang, Y.-C.; Loken, M.R.; Raimondi, S.C.; Hirsch, B.A.; Gamis, A.S.; et al. miR-155 expression and correlation with clinical outcome in pediatric AML: A report from Children’s Oncology Group. Pediatr. Blood Cancer 2016, 63, 2096–2103. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.A.; Kagele, D.A.; Eiring, A.M.; Kim, C.N.; Hu, R.; Runtsch, M.C.; Alexander, M.; Huffaker, T.B.; Lee, S.H.; Patel, A.B.; et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood 2017, 129, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.; Staffas, A.; Röhner, L.; Krowiorz, K.; Heuser, M.; Döhner, K.; Bullinger, L.; Döhner, H.; Fogelstrand, L.; Rouhi, A.; et al. MicroRNA-155 is upregulated in MLL-rearranged AML but its absence does not affect leukemia development. Exp. Hematol. 2016, 44, 1166–1171. [Google Scholar] [CrossRef]
- Rauh, M.J.; Sly, L.M.; Kalesnikoff, J.; Hughes, M.R.; Cao, L.P.; Lam, V.; Krystal, G. The role of SHIP1 in macrophage programming and activation. Biochem. Soc. Trans. 2004, 32, 785–788. [Google Scholar] [CrossRef]
- Li, G.; Hao, W.; Hu, W. Transcription factor PU.1 and immune cell differentiation (Review). Int. J. Mol. Med. 2020, 46, 1943–1950. [Google Scholar] [CrossRef]
- Alachkar, H.; Santhanam, R.; Harb, J.G.; Lucas, D.M.; Oaks, J.J.; Hickey, C.J.; Pan, L.; Kinghorn, A.D.; Caligiuri, M.A.; Perrotti, D.; et al. Silvestrol exhibits significant in vivo and in vitro antileukemic activities and inhibits FLT3 and miR-155 expressions in acute myeloid leukemia. J. Hematol. Oncol. 2013, 6, 21. [Google Scholar] [CrossRef]
- Cencic, R.; Carrier, M.; Trnkus, A.; Porco, J.A., Jr.; Minden, M.; Pelletier, J. Synergistic effect of inhibiting translation initiation in combination with cytotoxic agents in acute myelogenous leukemia cells. Leuk. Res. 2010, 34, 535–541. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, P.; Li, J.; Huang, Y.; Wen, J.; Wu, Z.; Chen, Y.; Hu, J. Loss of MiR-155 Sensitizes FLT3-ITD(+)AML to Chemotherapy and FLT3 Inhibitors via Glycolysis Blocking by Targeting PIK3R1. J. Cancer 2023, 14, 99–113. [Google Scholar] [CrossRef]
- Moussa Agha, D.; Rouas, R.; Najar, M.; Bouhtit, F.; Naamane, N.; Fayyad-Kazan, H.; Bron, D.; Meuleman, N.; Lewalle, P.; Merimi, M. Identification of Acute Myeloid Leukemia Bone Marrow Circulating MicroRNAs. Int. J. Mol. Sci. 2020, 21, 7065. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, L.; Yan, X.; Li, Y.; Xiong, Y.; Zhou, X. Overexpression of miR-210 is Associated with Poor Prognosis of Acute Myeloid Leukemia. Med. Sci. Monit. 2015, 21, 3427–3433. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Futami, M.; Carroll, M.; Feng, Y.; Wang, Z.; Fernandez, M.; Whichard, Z.; Chen, Y.; Kornblau, S.; Shpall, E.J.; et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene 2012, 31, 4085–4094. [Google Scholar] [CrossRef] [PubMed]
- Virga, F.; Cappellesso, F.; Stijlemans, B.; Henze, A.-T.; Trotta, R.; Van Audenaerde, J.; Mirchandani, A.S.; Sanchez-Garcia, M.A.; Vandewalle, J.; Orso, F.; et al. Macrophage miR-210 induction and metabolic reprogramming in response to pathogen interaction boost life-threatening inflammation. Sci. Adv. 2021, 7, eabf0466. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, A.; Calabrese, G.; Campolo, M.; Filippone, A.; Giuffrida, D.; Esposito, F.; Colarossi, C.; Cuzzocrea, S.; Esposito, E.; Paterniti, I. Role of miRNA-19a in Cancer Diagnosis and Poor Prognosis. Int. J. Mol. Sci. 2021, 22, 4697. [Google Scholar] [CrossRef]
- Gantier, M.P.; Stunden, H.J.; McCoy, C.E.; Behlke, M.A.; Wang, D.; Kaparakis-Liaskos, M.; Sarvestani, S.T.; Yang, Y.H.; Xu, D.; Corr, S.C.; et al. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 2012, 40, 8048–8058. [Google Scholar] [CrossRef]
- Zhang, T.-j.; Lin, J.; Zhou, J.-d.; Li, X.-x.; Zhang, W.; Guo, H.; Xu, Z.-j.; Yan, Y.; Ma, J.-c.; Qian, J. High bone marrow miR-19b level predicts poor prognosis and disease recurrence in de novo acute myeloid leukemia. Gene 2018, 640, 79–85. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, Q.; Zou, J.; Wang, B.; Zhang, Z.; Wei, R.; Zhao, L.; Zhang, Y.; Chu, C.; Fu, X.; et al. MiR-19a-3p Suppresses M1 Macrophage Polarization by Inhibiting STAT1/IRF1 Pathway. Front. Pharmacol. 2021, 12, 614044. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Zhang, D.; Sun, X.; Wu, Y.; Wang, J.; Li, Q.; Jiang, G. The macrophage polarization by miRNAs and its potential role in the treatment of tumor and inflammation (Review). Oncol. Rep. 2023, 50, 190. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wan, X.; Ruan, Q. The MicroRNA-21 in Autoimmune Diseases. Int. J. Mol. Sci. 2016, 17, 864. [Google Scholar] [CrossRef] [PubMed]
- Zahm, A.M.; Thayu, M.; Hand, N.J.; Horner, A.; Leonard, M.B.; Friedman, J.R. Circulating microRNA is a biomarker of pediatric Crohn disease. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Riccioni, R.; Lulli, V.; Castelli, G.; Biffoni, M.; Tiberio, R.; Pelosi, E.; Lo-Coco, F.; Testa, U. miR-21 is overexpressed in NPM1-mutant acute myeloid leukemias. Leuk. Res. 2015, 39, 221–228. [Google Scholar] [CrossRef]
- Vandewalle, V.; Essaghir, A.; Bollaert, E.; Lenglez, S.; Graux, C.; Schoemans, H.; Saussoy, P.; Michaux, L.; Valk, P.J.M.; Demoulin, J.B.; et al. miR-15a-5p and miR-21-5p contribute to chemoresistance in cytogenetically normal acute myeloid leukaemia by targeting PDCD4, ARL2 and BTG2. J. Cell Mol. Med. 2021, 25, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, X.; Ma, H.; Liu, Y.; Cheng, S.; Wang, H.; Sun, J. Upregulation of serum exosomal miR-21 was associated with poor prognosis of acute myeloid leukemia patients. Food Sci. Technol. 2022, 42, e51621. [Google Scholar] [CrossRef]
- Velu, C.S.; Chaubey, A.; Phelan, J.D.; Horman, S.R.; Wunderlich, M.; Guzman, M.L.; Jegga, A.G.; Zeleznik-Le, N.J.; Chen, J.; Mulloy, J.C.; et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Investig. 2014, 124, 222–236. [Google Scholar] [CrossRef]
- Ren, W.; Hou, J.; Yang, C.; Wang, H.; Wu, S.; Wu, Y.; Zhao, X.; Lu, C. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J. Exp. Clin. Cancer Res. 2019, 38, 62. [Google Scholar] [CrossRef]
- Yao, M.; Cui, B.; Zhang, W.; Ma, W.; Zhao, G.; Xing, L. Exosomal miR-21 secreted by IL-1β-primed-mesenchymal stem cells induces macrophage M2 polarization and ameliorates sepsis. Life Sci. 2021, 264, 118658. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.Y.; Feng, Y.G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef]
- Guo, Y.X.; Wang, N.; Wu, W.C.; Li, C.Q.; Chen, R.H.; Zhang, Y.; Li, X. The Role of miR-23b in Cancer and Autoimmune Disease. J. Oncol. 2021, 2021, 6473038. [Google Scholar] [CrossRef] [PubMed]
- Boucher, A.; Klopfenstein, N.; Hallas, W.M.; Skibbe, J.; Appert, A.; Jang, S.H.; Pulakanti, K.; Rao, S.; Cowden Dahl, K.D.; Dahl, R. The miR-23a~27a~24-2 microRNA Cluster Promotes Inflammatory Polarization of Macrophages. J. Immunol. 2021, 206, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Sun, L.; Zhu, J.; Li, N.; Gu, H.; Zhang, Y.; Li, M.; Xu, J. MicroRNA-23a-3p promotes macrophage M1 polarization and aggravates lipopolysaccharide-induced acute lung injury by regulating PLK1/STAT1/STAT3 signalling. Int. J. Exp. Pathol. 2022, 103, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, M.; Ding, L.; Tang, J. MiR-27a: A Novel Biomarker and Potential Therapeutic Target in Tumors. J. Cancer 2019, 10, 2836–2848. [Google Scholar] [CrossRef] [PubMed]
- Hatzl, S.; Perfler, B.; Wurm, S.; Uhl, B.; Quehenberger, F.; Ebner, S.; Troppmair, J.; Reinisch, A.; Wölfler, A.; Sill, H.; et al. Increased Expression of Micro-RNA-23a Mediates Chemoresistance to Cytarabine in Acute Myeloid Leukemia. Cancers 2020, 12, 496. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.Y.; Tang, Q.; Qian, W.; Qian, J.; Lin, J.; Wen, X.M.; Zhou, J.D.; Zhang, Y.Y.; Zhu, X.W.; Deng, Z.Q. Increased expression of miR-24 is associated with acute myeloid leukemia with t(8;21). Int. J. Clin. Exp. Pathol. 2014, 7, 8032–8038. [Google Scholar] [PubMed]
- Organista-Nava, J.; Gómez-Gómez, Y.; Illades-Aguiar, B.; Del Carmen Alarcón-Romero, L.; Saavedra-Herrera, M.V.; Rivera-Ramírez, A.B.; Garzón-Barrientos, V.H.; Leyva-Vázquez, M.A. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol. Rep. 2015, 33, 1639–1649. [Google Scholar] [CrossRef]
- Mi, S.; Lu, J.; Sun, M.; Li, Z.; Zhang, H.; Neilly, M.B.; Wang, Y.; Qian, Z.; Jin, J.; Zhang, Y.; et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar] [CrossRef]
- Abak, A.; Amini, S.; Sakhinia, E.; Abhari, A. MicroRNA-221: Biogenesis, function and signatures in human cancers. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3094–3117. [Google Scholar] [CrossRef]
- Song, J.; Ouyang, Y.; Che, J.; Li, X.; Zhao, Y.; Yang, K.; Zhao, X.; Chen, Y.; Fan, C.; Yuan, W. Potential Value of miR-221/222 as Diagnostic, Prognostic, and Therapeutic Biomarkers for Diseases. Front. Immunol. 2017, 8, 56. [Google Scholar] [CrossRef]
- Pei, H.Z.; Peng, Z.; Zhuang, X.; Wang, X.; Lu, B.; Guo, Y.; Zhao, Y.; Zhang, D.; Xiao, Y.; Gao, T.; et al. miR-221/222 induce instability of p53 By downregulating deubiquitinase YOD1 in acute myeloid leukemia. Cell Death Discov. 2023, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Shen, N.; Yang, Y.; Yu, H.; Xu, S.; Yang, Y.-W.; Liu, S.; Meguellati, K.; Yan, F. Targeting epigenetic pathway with gold nanoparticles for acute myeloid leukemia therapy. Biomaterials 2018, 167, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Iravani Saadi, M.; Arandi, N.; Yaghobi, R.; Azarpira, N.; Geramizadeh, B.; Ramzi, M. Aberrant Expression of the miR-181b/miR-222 after Hematopoietic Stem Cell Transplantation in Patients with Acute Myeloid Leukemia. Indian J. Hematol. Blood Transfus. 2019, 35, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, L.; Dan, W.; Chu, X.; Liu, C.; Luo, X.; Zhang, Z.; Lu, Y.; Wan, P.; Wang, X.; et al. miRNA-222-3p enhances the proliferation and suppresses the apoptosis of acute myeloid leukemia cells by targeting Axin2 and modulating the Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2022, 620, 83–91. [Google Scholar] [CrossRef]
- Pons, A.; Nomdedeu, B.; Navarro, A.; Gaya, A.; Gel, B.; Diaz, T.; Valera, S.; Rozman, M.; Belkaid, M.; Montserrat, E.; et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk. Lymphoma 2009, 50, 1854–1859. [Google Scholar] [CrossRef]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef]
- Ito, I.; Finnerty, C.C.; Herndon, D.N.; Kobayashi, M.; Suzuki, F. miR-222 stimulates M2b macrophage polarization in severely burned mice through the degradation of long noncoding RNA GAS5. J. Immunol. 2019, 202, 187.25. [Google Scholar] [CrossRef]
- Lechman, E.R.; Gentner, B.; Ng, S.W.; Schoof, E.M.; van Galen, P.; Kennedy, J.A.; Nucera, S.; Ciceri, F.; Kaufmann, K.B.; Takayama, N.; et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 2016, 29, 214–228. [Google Scholar] [CrossRef]
- de Leeuw, D.C.; Denkers, F.; Olthof, M.C.; Rutten, A.P.; Pouwels, W.; Jan Schuurhuis, G.; Ossenkoppele, G.J.; Smit, L. Attenuation of microRNA-126 Expression That Drives CD34+38− Stem/Progenitor Cells in Acute Myeloid Leukemia Leads to Tumor Eradication. Cancer Res. 2014, 74, 2094–2105. [Google Scholar] [CrossRef]
- Dorrance, A.M.; Neviani, P.; Ferenchak, G.J.; Huang, X.; Nicolet, D.; Maharry, K.S.; Ozer, H.G.; Hoellarbauer, P.; Khalife, J.; Hill, E.B.; et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 2015, 29, 2143–2153. [Google Scholar] [CrossRef]
- Zhang, L.; Nguyen, L.X.T.; Chen, Y.-C.; Wu, D.; Cook, G.J.; Hoang, D.H.; Brewer, C.J.; He, X.; Dong, H.; Li, S.; et al. Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance. Nat. Commun. 2021, 12, 6154. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, P.; Su, R.; Li, Y.; Hu, C.; Wang, Y.; Arnovitz, S.; He, M.; Gurbuxani, S.; Zuo, Z.; et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood 2015, 126, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Shou, X.; Wang, Y.; Jiang, Q.; Chen, J.; Liu, Q. miR-126 promotes M1 to M2 macrophage phenotype switching via VEGFA and KLF4. PeerJ 2023, 11, e15180. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Lai, W.; Song, L.; Deng, J.; Li, C.; Jiang, S. MicroRNA-126 regulates macrophage polarization to prevent the resorption of alveolar bone in diabetic periodontitis. Arch. Oral Biol. 2023, 150, 105686. [Google Scholar] [CrossRef]
- Wei, Y.; Zhu, M.; Schober, A. Macrophage MicroRNAs as Therapeutic Targets for Atherosclerosis, Metabolic Syndrome, and Cancer. Int. J. Mol. Sci. 2018, 19, 1756. [Google Scholar] [CrossRef]
- Eisfeld, A.-K.; Marcucci, G.; Maharry, K.; Schwind, S.; Radmacher, M.D.; Nicolet, D.; Becker, H.; Mrózek, K.; Whitman, S.P.; Metzeler, K.H.; et al. miR-3151 interplays with its host gene BAALC and independently affects outcome of patients with cytogenetically normal acute myeloid leukemia. Blood 2012, 120, 249–258. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; Brunet, S.; Nomdedéu, J.; Cordeiro, A.; Tormo, M.; Escoda, L.; Ribera, J.M.; Arnan, M.; Heras, I.; Gallardo, D.; et al. The expression level of BAALC-associated microRNA miR-3151 is an independent prognostic factor in younger patients with cytogenetic intermediate-risk acute myeloid leukemia. Blood Cancer J. 2015, 5, e352. [Google Scholar] [CrossRef]
- Shakirova, A.I.; Barkhatov, I.M.; Churkina, A.I.; Mamaev, N.N.; Zubarovskaya, L.S.; Afanas’ev, B.V. Clinical Value of miR-3151 Overexpression in Synergistic Interaction with BAALC Host Gene in Patients with Acute Myeloid Leukemia after Allogeneic Hematopoietic Stem Cell Transplantation. Clin. Oncohematol. 2019, 12, 303–308. [Google Scholar] [CrossRef]
- Han, G.T.; Sun, Z.L. Up-regulation of serum miR-4262 predicts clinical outcome of patients with acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2172–2176. [Google Scholar]
- Qu, H.; Chen, Y.; Zeng, W.; Huang, X.; Cheng, S. Boosting effects of MiR-4262 on acute myeloid leukemia advancement via governing KLF6. Res. Sq. 2020, in press. [Google Scholar] [CrossRef]
| Name | Abbreviation | |
|---|---|---|
| housekeeping ncRNAs | ribosomal RNA | rRNA |
| transfer RNA | tRNA | |
| small nuclear RNA | snRNA | |
| small nucleolar RNA | snoRNA | |
| telomerase RNA | TERC | |
| tRNA halves | tiRNA | |
| tRNA-derived fragments | t-RF | |
| regulatory ncRNAs | microRNA | miRNA |
| small interfering RNA | siRNA | |
| piwi-interfering RNA | piRNA | |
| enhancer RNA | eRNA | |
| long non-coding RNAs | lncRNA | |
| circular RNA | circRNA | |
| YRNA | YRNA |
| miRNA | Pathway/Transcription Factor Targeted |
|---|---|
| miR-17-92 cluster | TP53, c-MYC, n-MYC, STAT3, MXI1, E2F1, E2F2, E2F3, NKX3.1, TGF-β receptor II, Smad2, Smad4, BCL2L11, TSP-1, CTGF, Isl-1 and Tbx1 |
| miR-20a | ENH1/Id1, MAPK/c-Myc, PTEN/PI3K/AKT, FBXL5/BTG3, the Sonic hedgehog pathways, and SIRPa |
| miR-125b | NF-κB, p53, PI3K/Akt/mTOR, ErbB2, Wnt, DICER1, IRF4, PUMA and GRK2 |
| miR-146 | NF-kB pathway, TRAF6, IRAK1, IRF5 |
| miR-155 | PI3K/Akt, PU.1, SHIP1 |
| miR-210 | SHIP1 |
| miR-19a | NF-κB, STAT1, IRF1, STAT3 |
| miR-21 | PDCD4, BTG2, KLF5 |
| miR-23a | TOP2B |
| miR-221 | TP53 |
| miR-222 | Wnt/β-catenin pathway, Axin2, |
| miR-126 | PI3K/AKT/MTOR |
| miR-4262 | KLF6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimbu, L.; Mesaros, O.; Joldes, C.; Neaga, A.; Zaharie, L.; Zdrenghea, M. MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization. Biomedicines 2024, 12, 121. https://doi.org/10.3390/biomedicines12010121
Jimbu L, Mesaros O, Joldes C, Neaga A, Zaharie L, Zdrenghea M. MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization. Biomedicines. 2024; 12(1):121. https://doi.org/10.3390/biomedicines12010121
Chicago/Turabian StyleJimbu, Laura, Oana Mesaros, Corina Joldes, Alexandra Neaga, Laura Zaharie, and Mihnea Zdrenghea. 2024. "MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization" Biomedicines 12, no. 1: 121. https://doi.org/10.3390/biomedicines12010121
APA StyleJimbu, L., Mesaros, O., Joldes, C., Neaga, A., Zaharie, L., & Zdrenghea, M. (2024). MicroRNAs Associated with a Bad Prognosis in Acute Myeloid Leukemia and Their Impact on Macrophage Polarization. Biomedicines, 12(1), 121. https://doi.org/10.3390/biomedicines12010121





