How CD4+ T Cells Transcriptional Profile Is Affected by Culture Conditions: Towards the Design of Optimal In Vitro HIV Reactivation Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Participants
2.3. Sample Collection
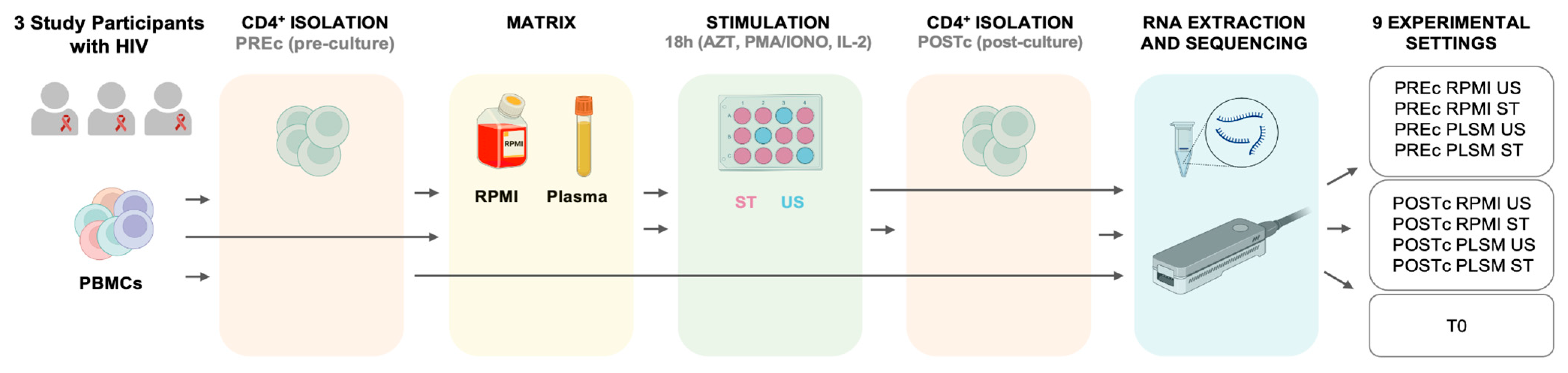
2.4. Study Design and In Vitro Stimulation
2.5. RNA Isolation
2.6. RNA Sequencing
2.7. HIV-1 DNA Quantification on CD4 T Cells by Droplet Digital PCR
2.8. Quantitative CA-HIV-1 RNA Assay (qRT-PCR)
2.9. Data Analysis
2.9.1. RNA-Seq Raw Data Processing
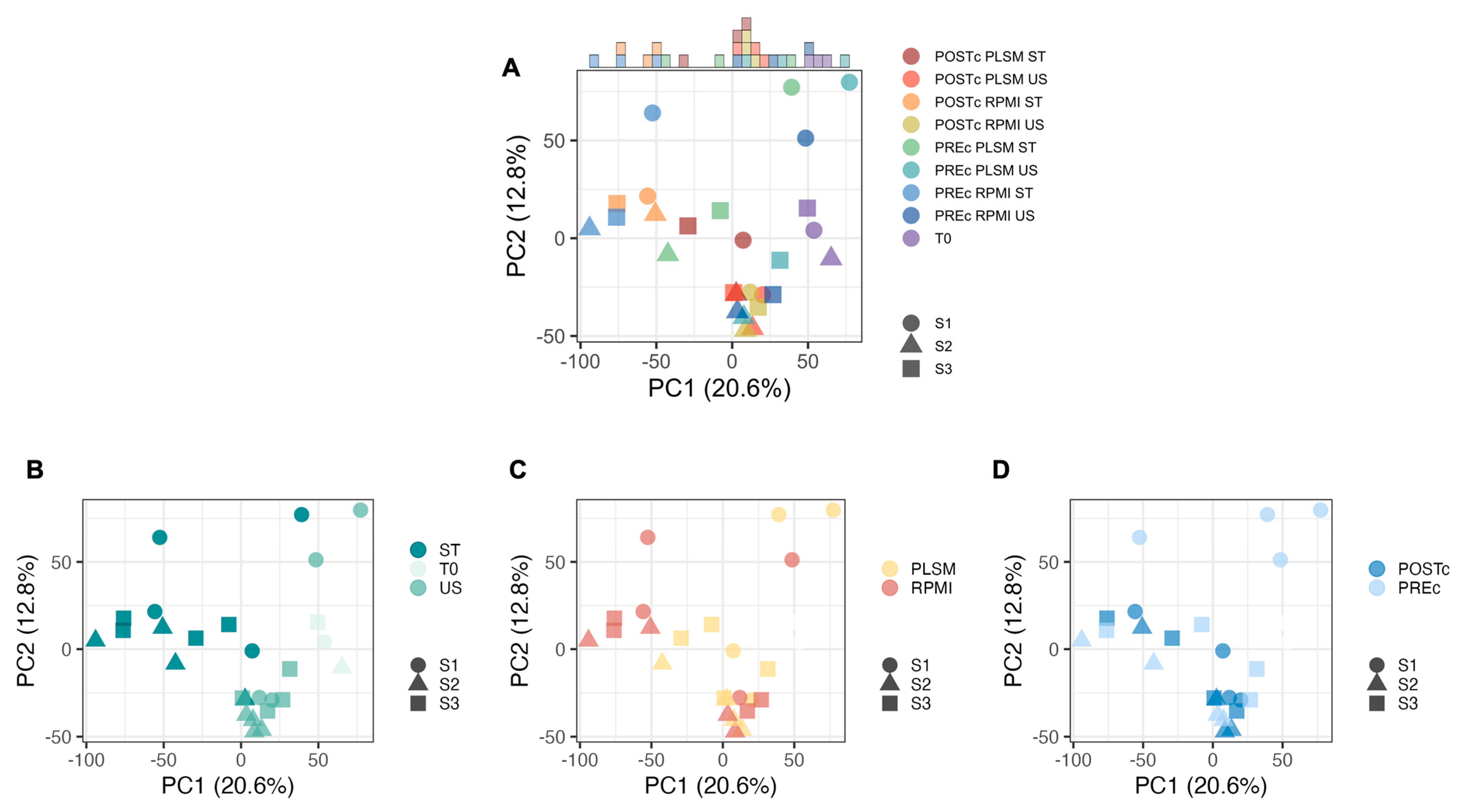
2.9.2. Principal Component Analysis
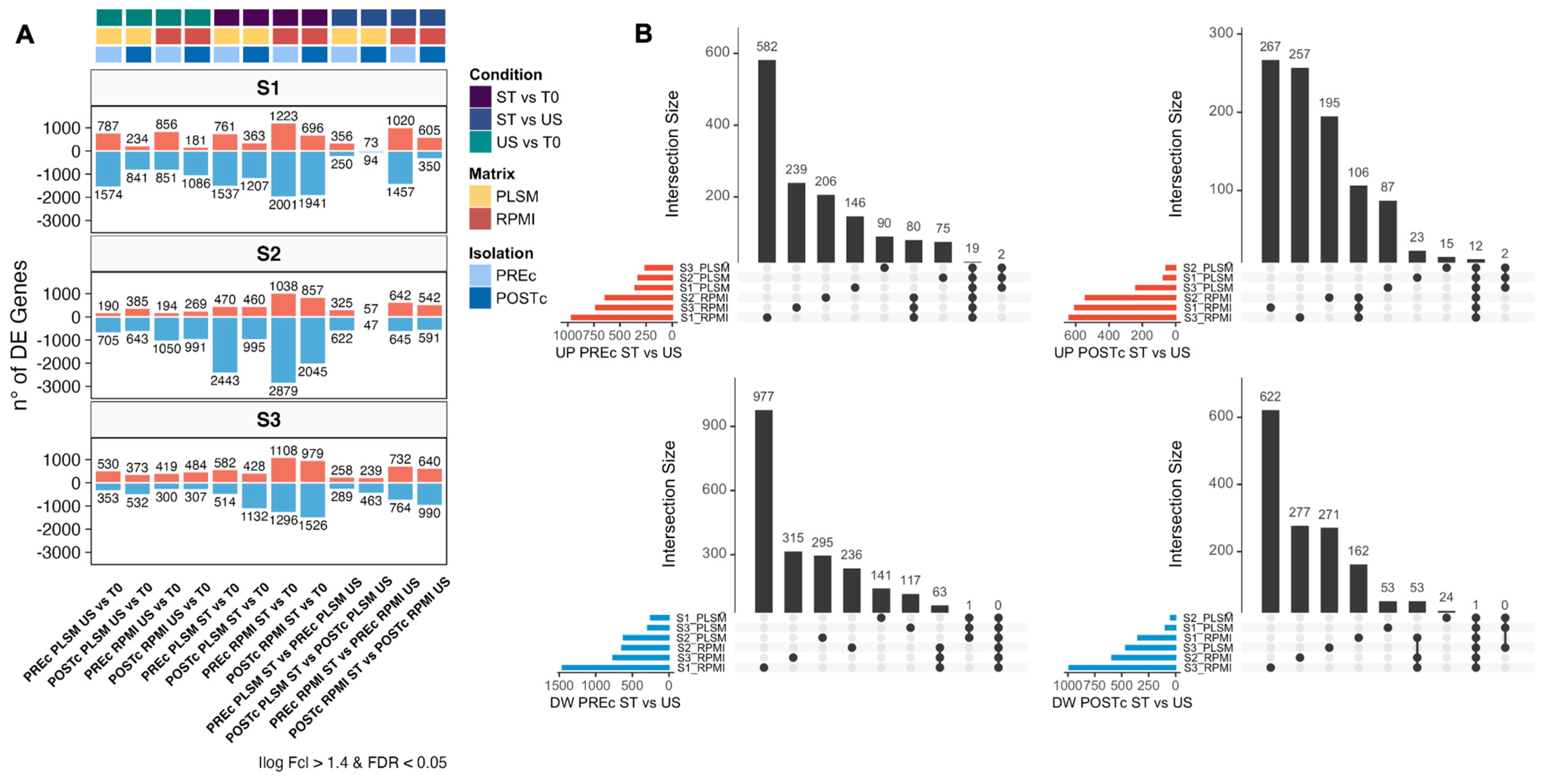
2.9.3. Differential Expression Analysis
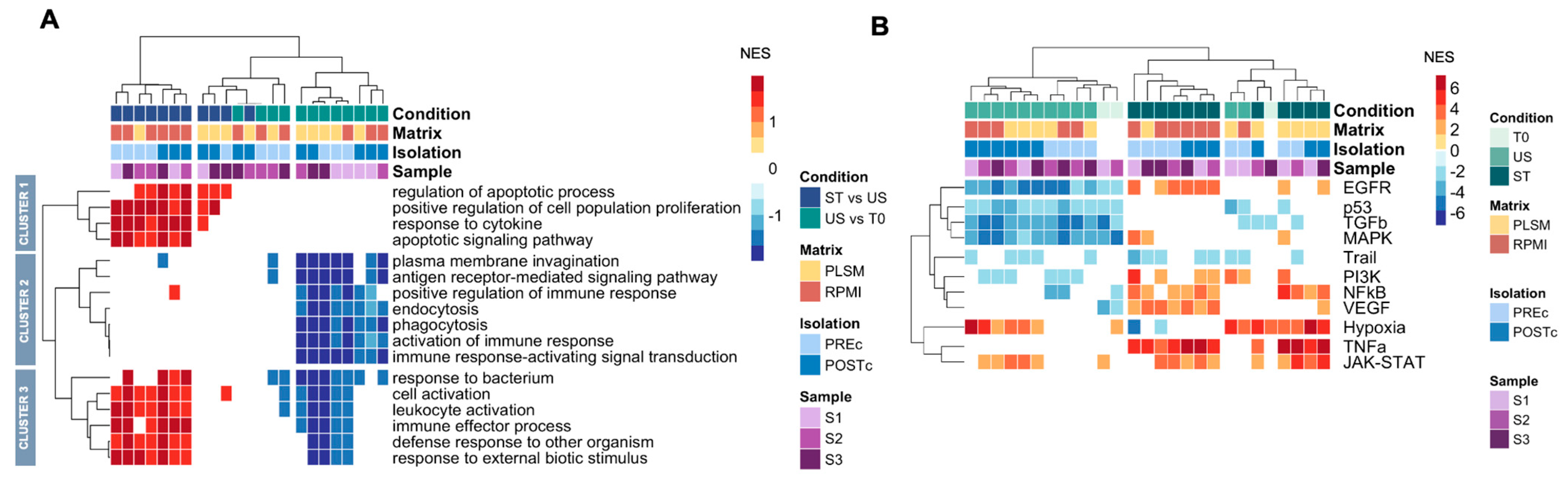
2.9.4. Gene Set Enrichment Analysis
2.9.5. Virtual Inference of Protein-Activity by Enriched Regulon Analysis (VIPER)
2.9.6. Differential Analysis of Genes Downstream the TCR Signaling Cascade
3. Results
3.1. Overview of Transcriptional Profiles in Different Conditions
3.2. Identification of Differentially Expressed Genes
3.3. Functional Phenotype in Different Conditions
3.4. TCR Signaling Cascade before and after Stimulation
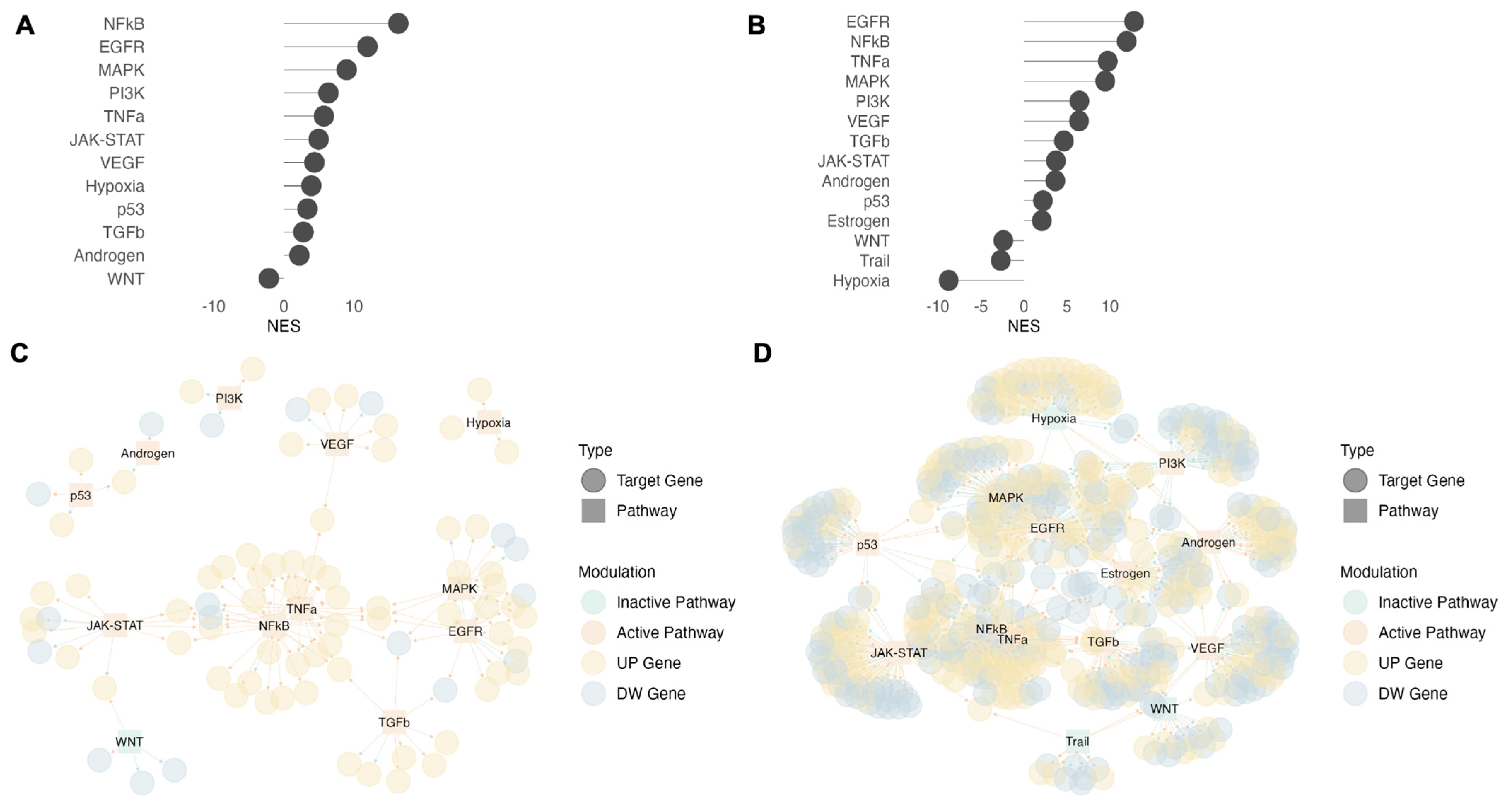
3.5. Hypoxia and JAK-STAT Pathways Are Activated in CD4+ T Cells Cultured in PBMCs
4. Discussion
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.K.; Hezareh, M.; Gunthard, H.F.; Havlir, D.V.; Ignacio, C.C.; Spina, C.A.; Richman, D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997, 278, 1291–1295. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, M.R.; Joseph, S.B.; Garrett, N.; Tyers, L.; Moeser, M.; Archin, N.; Council, O.D.; Matten, D.; Zhou, S.; Doolabh, D.; et al. The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci. Transl. Med. 2019, 11, eaaw5589. [Google Scholar] [CrossRef] [PubMed]
- Enick, P.N.; Brooker, J.P.; Tumiotto, C.M.; Staines, B.T.; Eron, J.J.; McMahon, D.K.; Gandhi, R.T.; Mellors, J.W.; Sobolewski, M.D. Comparison of methods to quantify inducible HIV-1 outgrowth. J. Virus Erad. 2021, 7, 100043. [Google Scholar] [CrossRef] [PubMed]
- Siliciano, J.D.; Siliciano, R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005, 304, 3–15. [Google Scholar] [CrossRef]
- Pettengill, M.A.; van Haren, S.D.; Levy, O. Soluble mediators regulating immunity in early life. Front. Immunol. 2014, 5, 457. [Google Scholar] [CrossRef]
- Jones, G.M.; Busby, E.; Garson, J.A.; Grant, P.R.; Nastouli, E.; Devonshire, A.S.; Whale, A.S. Digital PCR dynamic range is approaching that of real-time quantitative PCR. Biomol. Detect. Quantif. 2016, 10, 31–33. [Google Scholar] [CrossRef]
- Sozzi, G.; Conte, D.; Leon, M.; Ciricione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2003, 21, 3902–3908. [Google Scholar] [CrossRef]
- Rinaldi, S.; Pallikkuth, S.; Cameron, M.; de Armas, L.R.; Cotugno, N.; Dinh, V.; Pahwa, R.; Richardson, B.; Saini, S.R.; Rocca, S.; et al. Impact of Early Antiretroviral Therapy Initiation on HIV-Specific CD4 and CD8 T Cell Function in Perinatally Infected Children. J. Immunol. 2020, 204, 540–549. [Google Scholar] [CrossRef]
- Busby, E.; Whale, A.S.; Ferns, R.B.; Grant, P.R.; Morley, G.; Campbell, J.; Foy, C.A.; Nastouli, E.; Huggett, J.F.; Garson, J.A. Instability of 8E5 calibration standard revealed by digital PCR risks inaccurate quantification of HIV DNA in clinical samples by qPCR. Sci. Rep. 2017, 7, 1209. [Google Scholar] [CrossRef]
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G.; Hall, E. edgeR: Differential analysis of sequence read count data User’s Guide. R Packag. 2020, 1–121. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Badia-i-Mompel, P.; Vélez Santiago, J.; Braunger, J.; Geiss, C.; Dimitrov, D.; Müller-Dott, S.; Taus, P.; Dugourd, A.; Holland, C.H.; Ramirez Flores, R.O.; et al. decoupleR: Ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2022, 2, vbac016. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Iorio, F.; Matchan, A.; Fonseca, N.; Jaaks, P.; Peat, G.; Pignatelli, M.; Falcone, F.; Benes, C.H.; Dunham, I.; et al. Transcription Factor Activities Enhance Markers of Drug Sensitivity in Cancer. Cancer Res. 2018, 78, 769–780. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef]
- Moreau, J.M.; Velegraki, M.; Bolyard, C.; Rosenblum, M.D.; Li, Z. Transforming growth factor-beta1 in regulatory T cell biology. Sci. Immunol. 2022, 7, eabi4613. [Google Scholar] [CrossRef]
- Travis, M.A.; Sheppard, D. TGF-beta activation and function in immunity. Annu. Rev. Immunol. 2014, 32, 51–82. [Google Scholar] [CrossRef]
- Zaiss, D.M.; van Loosdregt, J.; Gorlani, A.; Bekker, C.P.; Grone, A.; Sibilia, M.; van Bergen en Henegouwen, P.M.; Roovers, R.C.; Coffer, P.J.; Sijts, A.J. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 2013, 38, 275–284. [Google Scholar] [CrossRef]
- MacDonald, F.; Zaiss, D.M.W. The Immune System’s Contribution to the Clinical Efficacy of EGFR Antagonist Treatment. Front. Pharmacol. 2017, 8, 575. [Google Scholar] [CrossRef]
- Huang, W.; August, A. The signaling symphony: T cell receptor tunes cytokine-mediated T cell differentiation. J. Leukoc. Biol. 2015, 97, 477–485. [Google Scholar] [CrossRef]
- Takatori, H.; Kawashima, H.; Suzuki, K.; Nakajima, H. Role of p53 in systemic autoimmune diseases. Crit. Rev. Immunol. 2014, 34, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.R.; Byeon, Y.; Kim, D.; Park, S.G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020, 52, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Brignall, R.; Cauchy, P.; Bevington, S.L.; Gorman, B.; Pisco, A.O.; Bagnall, J.; Boddington, C.; Rowe, W.; England, H.; Rich, K.; et al. Integration of Kinase and Calcium Signaling at the Level of Chromatin Underlies Inducible Gene Activation in T Cells. J. Immunol. 2017, 199, 2652–2667. [Google Scholar] [CrossRef]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef]
- Bettelli, E.; Dastrange, M.; Oukka, M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5138–5143. [Google Scholar] [CrossRef] [PubMed]
- Lewin, S.R.; Vesanen, M.; Kostrikis, L.; Hurley, A.; Duran, M.; Zhang, L.; Ho, D.D.; Markowitz, M. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 1999, 73, 6099–6103. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruel, T.; Fujimoto, K.; Hatano, H.; Yukl, S.; Eller, L.A.; Liegler, T.; Kamya, M.; Gassasira, A.; Dorsey, G.; et al. Novel application of Locked Nucleic Acid chemistry for a Taqman assay for measuring diverse human immunodeficiency virus type 1 subtypes. J. Virol. Methods 2010, 170, 115–120. [Google Scholar] [CrossRef]
- Palmer, S.; Wiegand, A.P.; Maldarelli, F.; Bazmi, H.; Mican, J.M.; Polis, M.; Dewar, R.L.; Planta, A.; Liu, S.; Metcalf, J.A.; et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 2003, 41, 4531–4536. [Google Scholar] [CrossRef] [PubMed]
- Vandergeeten, C.; Fromentin, R.; Merlini, E.; Lawani, M.B.; DaFonseca, S.; Bakeman, W.; McNulty, A.; Ramgopal, M.; Michael, N.; Kim, J.H.; et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 2014, 88, 12385–12396. [Google Scholar] [CrossRef]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef]
- Procopio, F.A.; Fromentin, R.; Kulpa, D.A.; Brehm, J.H.; Bebin, A.G.; Strain, M.C.; Richman, D.D.; O’Doherty, U.; Palmer, S.; Hecht, F.M.; et al. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine 2015, 2, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Pinzone, M.R.; O’Doherty, U. Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained. Retrovirology 2018, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Gohil, Y.; Mehta, K.; D’Silva, A.; Amanullah, A.; Selvam, D.; Pargain, N.; Nala, N.; Sanjeeva, G.N.; Ranga, U. An Optimized Tat/Rev Induced Limiting Dilution Assay for the Characterization of HIV-1 Latent Reservoirs. Bio-Protocol 2022, 12, e4391. [Google Scholar] [CrossRef] [PubMed]
- Avettand-Fenoel, V.; Hocqueloux, L.; Ghosn, J.; Cheret, A.; Frange, P.; Melard, A.; Viard, J.P.; Rouzioux, C. Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clin. Microbiol. Rev. 2016, 29, 859–880. [Google Scholar] [CrossRef]
- Graf, E.H.; O’Doherty, U. Quantitation of integrated proviral DNA in viral reservoirs. Curr. Opin. HIV AIDS 2013, 8, 100–105. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10, eaap9927. [Google Scholar] [CrossRef]
- Moron-Lopez, S.; Kim, P.; Sogaard, O.S.; Tolstrup, M.; Wong, J.K.; Yukl, S.A. Characterization of the HIV-1 transcription profile after romidepsin administration in ART-suppressed individuals. AIDS 2019, 33, 425–431. [Google Scholar] [CrossRef]
- Einkauf, K.B.; Osborn, M.R.; Gao, C.; Sun, W.; Sun, X.; Lian, X.; Parsons, E.M.; Gladkov, G.T.; Seiger, K.W.; Blackmer, J.E.; et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 2022, 185, 266–282.e15. [Google Scholar] [CrossRef]
- Laird, G.M.; Eisele, E.E.; Rabi, S.A.; Lai, J.; Chioma, S.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013, 9, e1003398. [Google Scholar] [CrossRef]
- Bertoldi, A.; D’Urbano, V.; Bon, I.; Verbon, A.; Rokx, C.; Boucher, C.; van Kampen, J.J.A.; Gruters, R.A.; Gallinella, G.; Calza, L.; et al. Development of C-TILDA: A modified TILDA method for reservoir quantification in long term treated patients infected with subtype C HIV-1. J. Virol. Methods 2020, 276, 113778. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Cao, W.; Huang, L.; Zhou, J.; Sheng, L. Influence of various medium environment to in vitro human T cell culture. In Vitro Cell. Dev. Biol. Anim. 2018, 54, 559–566. [Google Scholar] [CrossRef]
- Leney-Greene, M.A.; Boddapati, A.K.; Su, H.C.; Cantor, J.R.; Lenardo, M.J. Human Plasma-like Medium Improves T Lymphocyte Activation. iScience 2020, 23, 100759. [Google Scholar] [CrossRef] [PubMed]
- van Haren, S.D.; Ganapathi, L.; Bergelson, I.; Dowling, D.J.; Banks, M.; Samuels, R.C.; Reed, S.G.; Marshall, J.D.; Levy, O. In vitro cytokine induction by TLR-activating vaccine adjuvants in human blood varies by age and adjuvant. Cytokine 2016, 83, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Taneja, V. Sex Hormones Determine Immune Response. Front. Immunol. 2018, 9, 1931. [Google Scholar] [CrossRef]
- Morrocchi, E.; Pighi, C.; Pascucci, G.R.; Cotugno, N.; Medri, C.; Amodio, D.; Colagrossi, L.; Ruggiero, A.; Manno, E.C.; Casamento Tumeo, C.; et al. Perinatally Human Immunodeficiency Virus-Infected Adolescents and Young Adults Demonstrate Distinct BNT162b2 Messenger RNA Coronavirus Disease 2019 Vaccine Immunogenicity. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2022, 75, S51–S60. [Google Scholar] [CrossRef] [PubMed]
- Vitali, F.; Li, Q.; Schissler, A.G.; Berghout, J.; Kenost, C.; Lussier, Y.A. Developing a ‘personalome’ for precision medicine: Emerging methods that compute interpretable effect sizes from single-subject transcriptomes. Brief. Bioinform. 2019, 20, 789–805. [Google Scholar] [CrossRef]
- Rachid Zaim, S.; Kenost, C.; Zhang, H.H.; Lussier, Y.A. Personalized beyond Precision: Designing Unbiased Gold Standards to Improve Single-Subject Studies of Personal Genome Dynamics from Gene Products. J. Pers. Med. 2020, 11, 24. [Google Scholar] [CrossRef]
- Dhummakupt, A.; Rubens, J.H.; Anderson, T.; Powell, L.; Nonyane, B.A.; Siems, L.V.; Collinson-Streng, A.; Nilles, T.; Jones, R.B.; Tepper, V.; et al. Differences in inducibility of the latent HIV reservoir in perinatal and adult infection. JCI Insight 2020, 5, e134105. [Google Scholar] [CrossRef]
- Majowicz, A.; van der Marel, S.; te Velde, A.A.; Meijer, S.L.; Petry, H.; van Deventer, S.J.; Ferreira, V. Murine CD4(+)CD25(−) cells activated in vitro with PMA/ionomycin and anti-CD3 acquire regulatory function and ameliorate experimental colitis in vivo. BMC Gastroenterol. 2012, 12, 172. [Google Scholar] [CrossRef]
- Agosto, L.M.; Henderson, A.J. CD4(+) T Cell Subsets and Pathways to HIV Latency. AIDS Res. Hum. Retrovir. 2018, 34, 780–789. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 2017, 4, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Ruelas, D.S.; Greene, W.C. An integrated overview of HIV-1 latency. Cell 2013, 155, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Liu, Z.; Zandi, E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997, 9, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Radbruch, A.; Chang, H.D. A Ca(2+) concentration of 1.5 mM, as present in IMDM but not in RPMI, is critical for maximal response of Th cells to PMA/ionomycin. Eur. J. Immunol. 2015, 45, 1270–1273. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Montazeri Aliabadi, H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Dela Pena-Ponce, M.G.; Rodriguez-Nieves, J.; Bernhardt, J.; Tuck, R.; Choudhary, N.; Mengual, M.; Mollan, K.R.; Hudgens, M.G.; Peter-Wohl, S.; De Paris, K. Increasing JAK/STAT Signaling Function of Infant CD4(+) T Cells during the First Year of Life. Front. Pediatr. 2017, 5, 15. [Google Scholar] [CrossRef]
- Zhang, H.; Watanabe, R.; Berry, G.J.; Tian, L.; Goronzy, J.J.; Weyand, C.M. Inhibition of JAK-STAT Signaling Suppresses Pathogenic Immune Responses in Medium and Large Vessel Vasculitis. Circulation 2018, 137, 1934–1948. [Google Scholar] [CrossRef]
- Dudley, A.C.; Thomas, D.; Best, J.; Jenkins, A. The STATs in cell stress-type responses. Cell Commun. Signal. CCS 2004, 2, 8. [Google Scholar] [CrossRef]
- Roger, I.; Milara, J.; Montero, P.; Cortijo, J. The Role of JAK/STAT Molecular Pathway in Vascular Remodeling Associated with Pulmonary Hypertension. Int. J. Mol. Sci. 2021, 22, 4980. [Google Scholar] [CrossRef]
- Malekan, M.; Ebrahimzadeh, M.A.; Sheida, F. The role of Hypoxia-Inducible Factor-1alpha and its signaling in melanoma. Biomed. Pharmacother. 2021, 141, 111873. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Dudley, A.C.; Thomas, D.; Best, J.; Jenkins, A. A VEGF/JAK2/STAT5 axis may partially mediate endothelial cell tolerance to hypoxia. Biochem. J. 2005, 390, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Takata, H.; Kessing, C.; Sy, A.; Lima, N.; Sciumbata, J.; Mori, L.; Jones, R.B.; Chomont, N.; Michael, N.L.; Valente, S. Modeling HIV-1 Latency Using Primary CD4+ T Cells from Virally Suppressed HIV-1-Infected Individuals on Antiretroviral Therapy. J Virol. 2019, 93, 11. [Google Scholar] [CrossRef]
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.; Jenike, K.; et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019, 566, 7742. [Google Scholar] [CrossRef]
- Massanella, M.; Yek, C.; Lada, S.M.; Nakazawa, M.; Shefa, N.; Huang, K.; Richman, D.D. Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine 2018, 36, 113–121. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascucci, G.R.; Morrocchi, E.; Pighi, C.; Rotili, A.; Neri, A.; Medri, C.; Olivieri, G.; Sanna, M.; Rasi, G.; Persaud, D.; et al. How CD4+ T Cells Transcriptional Profile Is Affected by Culture Conditions: Towards the Design of Optimal In Vitro HIV Reactivation Assays. Biomedicines 2023, 11, 888. https://doi.org/10.3390/biomedicines11030888
Pascucci GR, Morrocchi E, Pighi C, Rotili A, Neri A, Medri C, Olivieri G, Sanna M, Rasi G, Persaud D, et al. How CD4+ T Cells Transcriptional Profile Is Affected by Culture Conditions: Towards the Design of Optimal In Vitro HIV Reactivation Assays. Biomedicines. 2023; 11(3):888. https://doi.org/10.3390/biomedicines11030888
Chicago/Turabian StylePascucci, Giuseppe Rubens, Elena Morrocchi, Chiara Pighi, Arianna Rotili, Alessia Neri, Chiara Medri, Giulio Olivieri, Marco Sanna, Gianmarco Rasi, Deborah Persaud, and et al. 2023. "How CD4+ T Cells Transcriptional Profile Is Affected by Culture Conditions: Towards the Design of Optimal In Vitro HIV Reactivation Assays" Biomedicines 11, no. 3: 888. https://doi.org/10.3390/biomedicines11030888
APA StylePascucci, G. R., Morrocchi, E., Pighi, C., Rotili, A., Neri, A., Medri, C., Olivieri, G., Sanna, M., Rasi, G., Persaud, D., Chahroudi, A., Lichterfeld, M., Nastouli, E., Cancrini, C., Amodio, D., Rossi, P., Cotugno, N., & Palma, P. (2023). How CD4+ T Cells Transcriptional Profile Is Affected by Culture Conditions: Towards the Design of Optimal In Vitro HIV Reactivation Assays. Biomedicines, 11(3), 888. https://doi.org/10.3390/biomedicines11030888





