Unveiling the Human Gastrointestinal Tract Microbiome: The Past, Present, and Future of Metagenomics
Abstract
1. Introduction
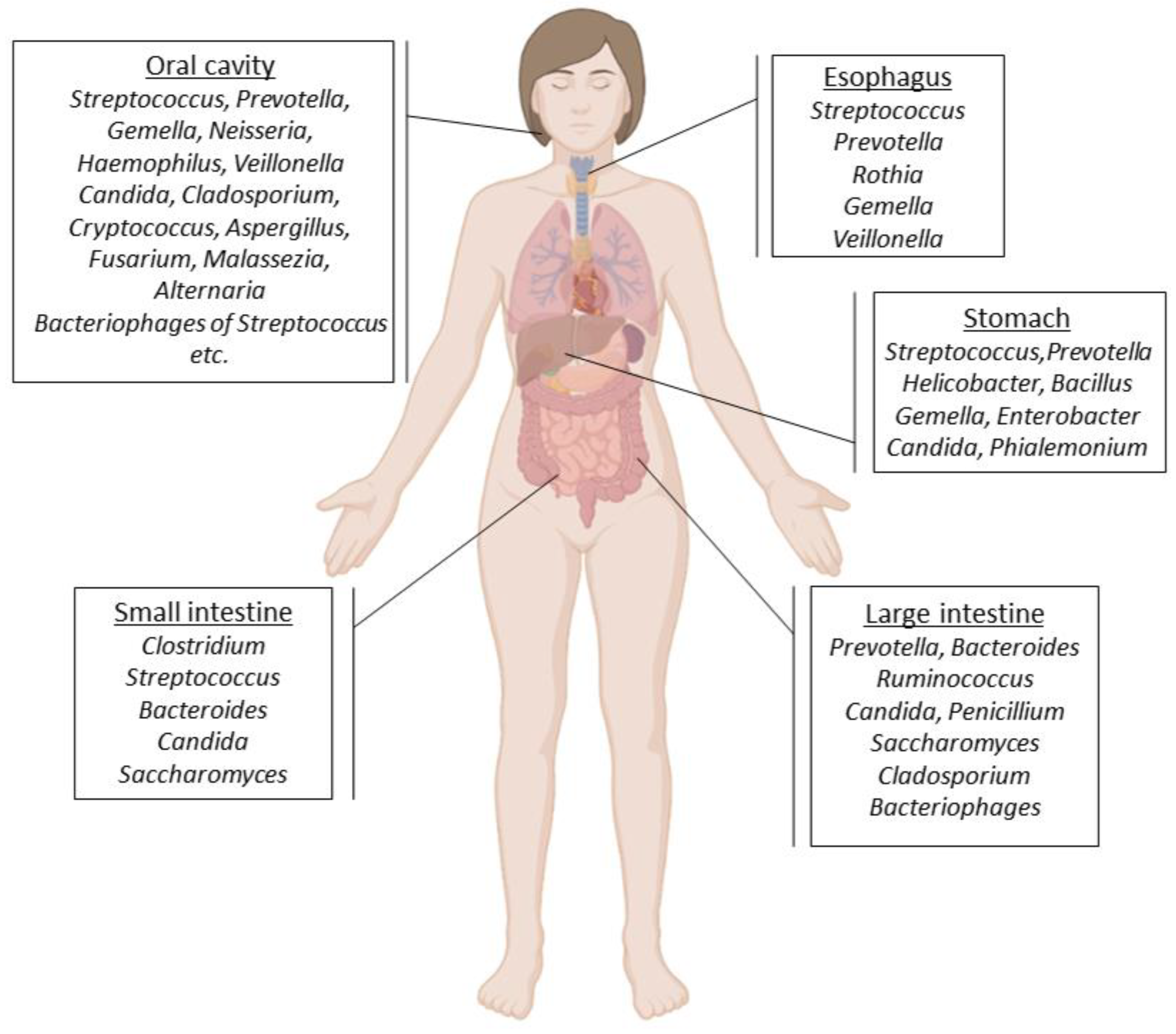
2. The Role of the Gastrointestinal Tract Microbiota in Human Health and Disease
| Human Disease | Related Microorganisms | Reference |
|---|---|---|
| Atopic dermatitis | Staphylococcucus aureus, Cutibacterium, Streptococcus, Acinetobacter, Gemella | [47,48] |
| Cystic fibrosis | Streptococcus species | [49] |
| Depression | Coprococcus, Sellimonas, Clostridium, Hungatella | [50,51] |
| Autism | Clostridium bolteae | [52] |
| Asthma | Clostridia, Proteobacteria | [53,54] |
| Obesity | Actinobacteria, Bacteroidetes | [55,56] |
| Tuberculosis | Mycobacterium tuberculosis, Bacteroides fragilis, Prevotella, Enterococcus | [57] |
| Periodontal diseases | Spirochaetes, Synergistetes, Bacteroidetes | [58] |
| Dental caries | Streptococcus mutans, Lactobacillus spp., Candida albicans | [35] |
| Oral cancer | Streptococcus species | [59] |
| Esophageal cancer | Tannerella forsythia, Porphyromonas gingivalis | [60] |
| Cardiovascular disease | Campylobacter rectus, Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia | [61] |
| Rheumatoid arthritis | Veillonella, Atopobium, Prevotella, Leptotrichia | [62,63,64] |
| Parkinson’s disease | Lachnospiraceae, Faecalibacterium, Lactobacillus, Akkermansia, Bifidobacterium | [65] |
| Alzheimer’s disease | Spirochaetes | [66] |
| Diabetes | Aggregatibacter, Neisseria, Gemella, Selenomonas, Actinomyces, Fusobacterium, Streptococcus | [67,68,69] |
3. Detecting Microbes
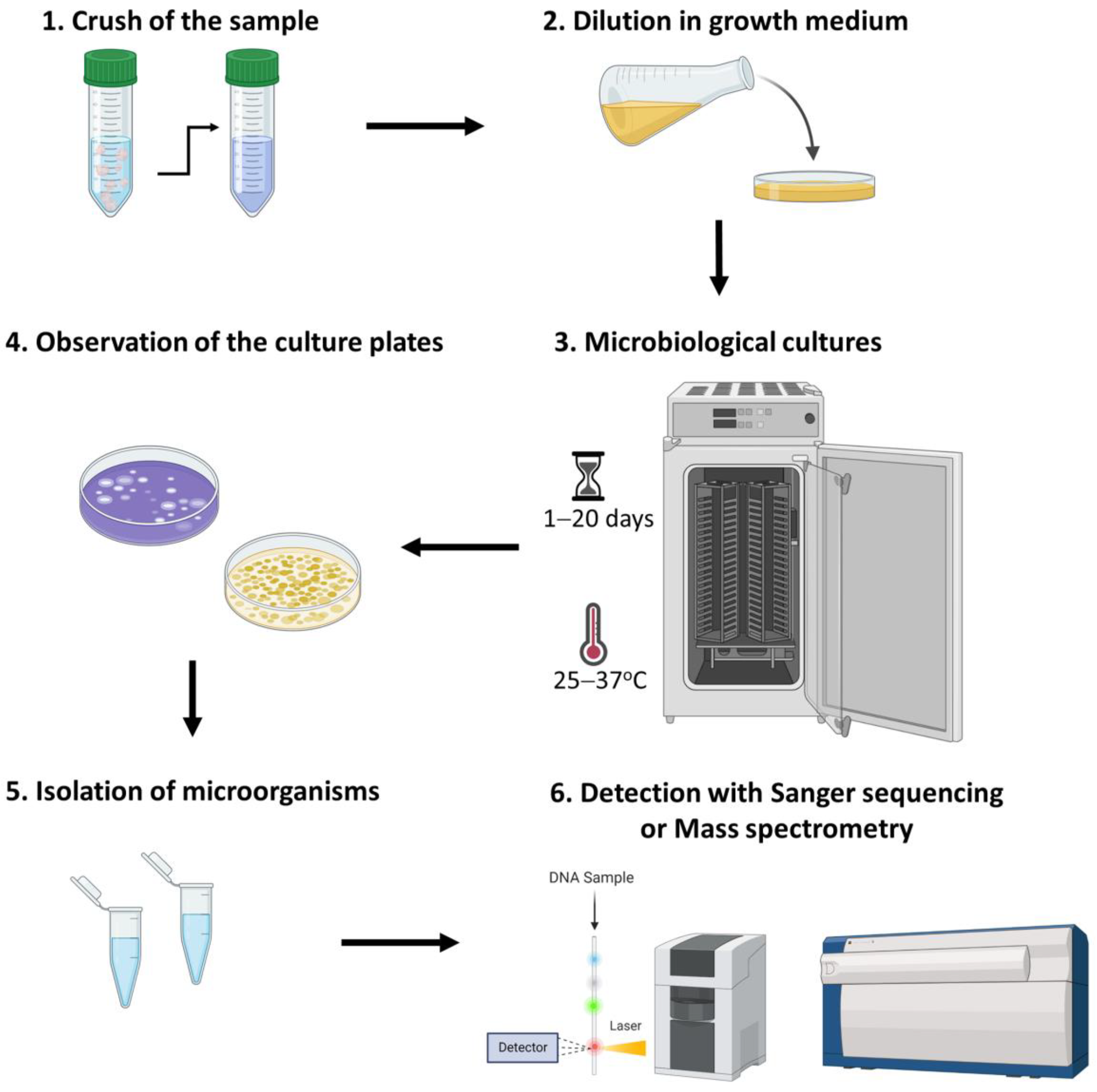
3.1. Culturomics
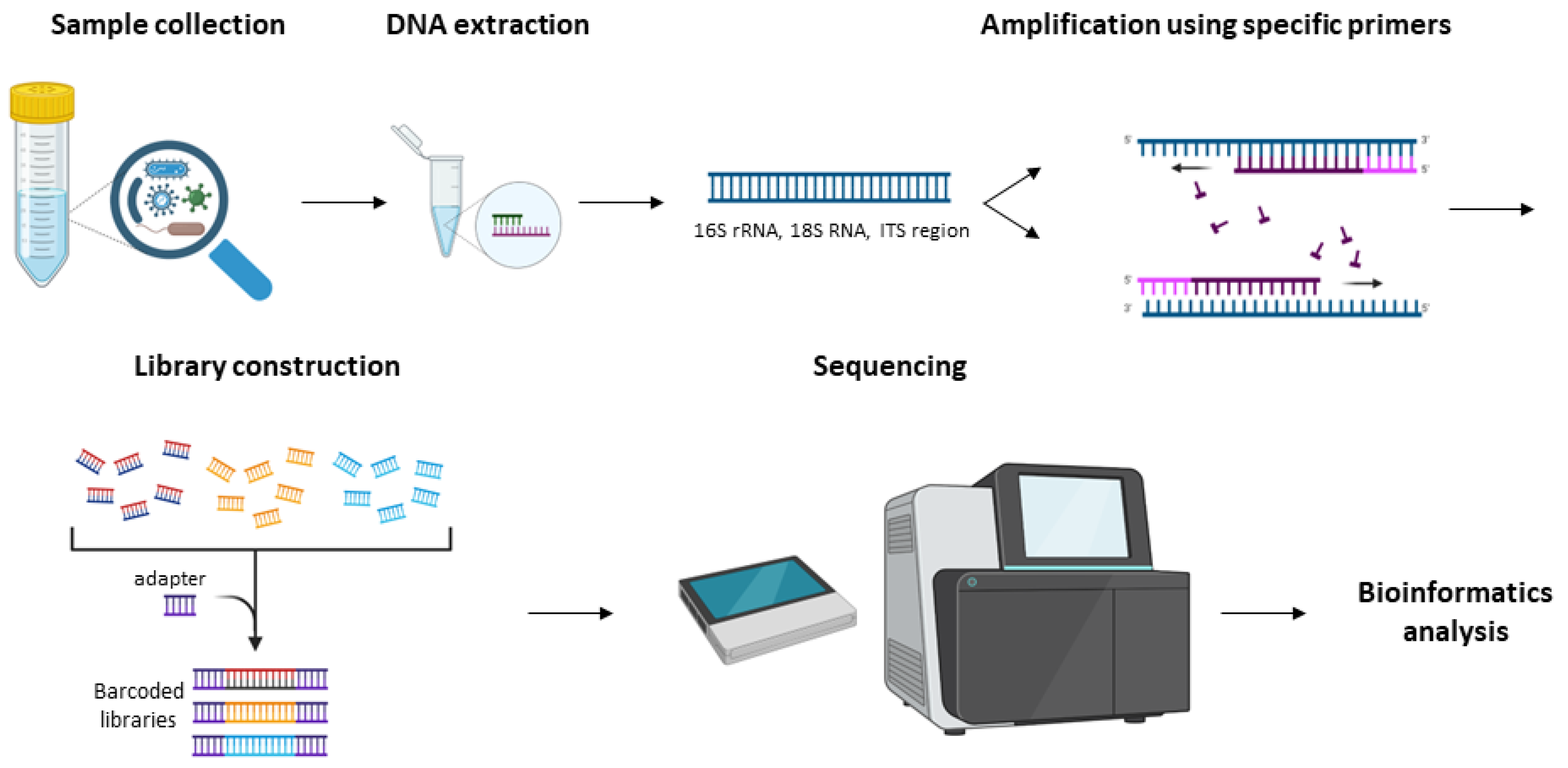
3.2. Amplicon Sequencing Analysis
3.3. Metagenomics
3.4. Metatranscriptomics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Verges, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Cardenas, J.A.; Manzano-Agugliaro, F. The metagenomics worldwide research. Curr. Genet. 2017, 63, 819–829. [Google Scholar] [CrossRef]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T. Colonic microflora: Nutrition and health. Nutrition 1997, 13, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 2021, 12, 360–373. [Google Scholar] [CrossRef]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700. [Google Scholar] [CrossRef]
- Boddy, S.L.; Giovannelli, I.; Sassani, M.; Cooper-Knock, J.; Snyder, M.P.; Segal, E.; Elinav, E.; Barker, L.A.; Shaw, P.J.; McDermott, C.J. The gut microbiome: A key player in the complexity of amyotrophic lateral sclerosis (ALS). BMC Med. 2021, 19, 13. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef] [PubMed]
- Ogobuiro, I.; Gonzales, J.; Tuma, F. Physiology, Gastrointestinal. In StatPearls; StatPerals Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral. Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018, 6, 5–6. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An Overview of the Carbonic Anhydrases from Two Pathogens of the Oral Cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr. Top Med. Chem. 2016, 16, 2359–2368. [Google Scholar] [CrossRef]
- Lepp, P.W.; Brinig, M.M.; Ouverney, C.C.; Palm, K.; Armitage, G.C.; Relman, D.A. Methanogenic Archaea and human periodontal disease. Proc. Natl. Acad. Sci. USA 2004, 101, 6176–6181. [Google Scholar] [CrossRef]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Patel, M. Oral Cavity and Candida albicans: Colonisation to the Development of Infection. Pathogens 2022, 11, 335. [Google Scholar] [CrossRef] [PubMed]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral. Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Heng, N.C.; Burton, J.P.; Chilcott, C.N.; Tagg, J.R. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. 2009, 4, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Bastos, J.A.; Diniz, C.G.; Bastos, M.G.; Vilela, E.M.; Silva, V.L.; Chaoubah, A.; Souza-Costa, D.C.; Andrade, L.C. Identification of periodontal pathogens and severity of periodontitis in patients with and without chronic kidney disease. Arch Oral Biol. 2011, 56, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Andrei, E.C.; Banita, I.M.; Munteanu, M.C.; Busuioc, C.J.; Mateescu, G.O.; Malin, R.D.; Pisoschi, C.G. Oral Papillomatosis: Its Relation with Human Papilloma Virus Infection and Local Immunity-An Update. Medicina 2022, 58, 1103. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef]
- Park, C.H.; Lee, S.K. Exploring Esophageal Microbiomes in Esophageal Diseases: A Systematic Review. J. Neurogastroenterol Motil. 2020, 26, 171–179. [Google Scholar] [CrossRef]
- Nardone, G.; Compare, D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 2015, 3, 255–260. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe. 2018, 23, 458–469.e455. [Google Scholar] [CrossRef] [PubMed]
- Kastl, A.J., Jr.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6, 116. [Google Scholar] [CrossRef] [PubMed]
- Struzycka, I. The oral microbiome in dental caries. Pol. J. Microbiol. 2014, 63, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Mombelli, A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol 2000 2018, 76, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.B.; Carroll, M.P.; Serisier, D.J.; Hockey, P.M.; Jones, G.; Kehagia, V.; Connett, G.J.; Bruce, K.D. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J. Clin. Microbiol. 2006, 44, 2601–2604. [Google Scholar] [CrossRef]
- Gao, Z.; Kang, Y.; Yu, J.; Ren, L. Human pharyngeal microbiome may play a protective role in respiratory tract infections. Genom. Proteom. Bioinform. 2014, 12, 144–150. [Google Scholar] [CrossRef]
- Kononen, E.; Gursoy, U.K. Oral Prevotella Species and Their Connection to Events of Clinical Relevance in Gastrointestinal and Respiratory Tracts. Front. Microbiol. 2021, 12, 798763. [Google Scholar] [CrossRef]
- Brooks, L.R.K.; Mias, G.I. Streptococcus pneumoniae’s Virulence and Host Immunity: Aging, Diagnostics, and Prevention. Front. Immunol. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Yolken, R.H.; Jones-Brando, L.; Dunigan, D.D.; Kannan, G.; Dickerson, F.; Severance, E.; Sabunciyan, S.; Talbot, C.C., Jr.; Prandovszky, E.; Gurnon, J.R.; et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc. Natl. Acad. Sci. USA 2014, 111, 16106–16111. [Google Scholar] [CrossRef]
- May, M.; Abrams, J.A. Emerging Insights into the Esophageal Microbiome. Curr. Treat Options Gastroenterol. 2018, 16, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Ru, J.; Xue, J.; Schulz, S.; Mirzaei, M.K.; Janssen, K.P.; Quante, M.; Deng, L. Differences in Gut Virome Related to Barrett Esophagus and Esophageal Adenocarcinoma. Microorganisms 2021, 9, 1701. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, Z.; Lin, Y.; Chen, Y.; Peng, X.E.; He, F.; Liu, S.; Yan, S.; Huang, L.; Lu, W.; et al. Streptococcus and Prevotella are associated with the prognosis of oesophageal squamous cell carcinoma. J. Med. Microbiol. 2018, 67, 1058–1068. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.; Haboubi, H.N.; Attwood, S.E.; Auth, M.K.H.; Dunn, J.M.; Sweis, R.; Morris, D.; Epstein, J.; Novelli, M.R.; Hunter, H.; et al. British Society of Gastroenterology (BSG) and British Society of Paediatric Gastroenterology, Hepatology and Nutrition (BSPGHAN) joint consensus guidelines on the diagnosis and management of eosinophilic oesophagitis in children and adults. Gut 2022, 71, 1459–1487. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, X.; Liu, X.; Ling, Z.; Ji, F. Role of the Gastric Microbiome in Gastric Cancer: From Carcinogenesis to Treatment. Front. Microbiol. 2021, 12, 641322. [Google Scholar] [CrossRef]
- Paller, A.S.; Kong, H.H.; Seed, P.; Naik, S.; Scharschmidt, T.C.; Gallo, R.L.; Luger, T.; Irvine, A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 26–35. [Google Scholar] [CrossRef]
- Park, D.H.; Kim, J.W.; Park, H.J.; Hahm, D.H. Comparative Analysis of the Microbiome across the Gut-Skin Axis in Atopic Dermatitis. Int. J. Mol. Sci. 2021, 22, 4228. [Google Scholar] [CrossRef]
- Scott, J.E.; O’Toole, G.A. The Yin and Yang of Streptococcus Lung Infections in Cystic Fibrosis: A Model for Studying Polymicrobial Interactions. J. Bacteriol. 2019, 201, e00115-19. [Google Scholar] [CrossRef]
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef]
- Peirce, J.M.; Alvina, K. The role of inflammation and the gut microbiome in depression and anxiety. J. Neurosci. Res. 2019, 97, 1223–1241. [Google Scholar] [CrossRef]
- Pequegnat, B.; Sagermann, M.; Valliani, M.; Toh, M.; Chow, H.; Allen-Vercoe, E.; Monteiro, M.A. A vaccine and diagnostic target for Clostridium bolteae, an autism-associated bacterium. Vaccine 2013, 31, 2787–2790. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut(-)Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef]
- Liu, B.N.; Liu, X.T.; Liang, Z.H.; Wang, J.H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Aoun, A.; Darwish, F.; Hamod, N. The Influence of the Gut Microbiome on Obesity in Adults and the Role of Probiotics, Prebiotics, and Synbiotics for Weight Loss. Prev. Nutr. Food Sci. 2020, 25, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Wu, C. Microbiota and Tuberculosis: A Potential Role of Probiotics, and Postbiotics. Front. Nutr. 2021, 8, 626254. [Google Scholar] [CrossRef]
- Lourenvarsigmao, T.G.B.; Spencer, S.J.; Alm, E.J.; Colombo, A.P.V. Defining the gut microbiota in individuals with periodontal diseases: An exploratory study. J. Oral Microbiol. 2018, 10, 1487741. [Google Scholar] [CrossRef]
- Chocolatewala, N.; Chaturvedi, P.; Desale, R. The role of bacteria in oral cancer. Indian J. Med. Paediatr. Oncol. 2010, 31, 126–131. [Google Scholar] [CrossRef]
- Baba, Y.; Iwatsuki, M.; Yoshida, N.; Watanabe, M.; Baba, H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann. Gastroenterol. Surg. 2017, 1, 99–104. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Zhao, T.; Wei, Y.; Zhu, Y.; Xie, Z.; Hai, Q.; Li, Z.; Qin, D. Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front. Immunol. 2022, 13, 1007165. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Role of Gut Microbiota in Rheumatoid Arthritis. J. Clin. Med. 2017, 6, 60. [Google Scholar] [CrossRef]
- Chen, B.; Zhao, Y.; Li, S.; Yang, L.; Wang, H.; Wang, T.; Bin, S.; Gai, Z.; Heng, X.; Zhang, C.; et al. Variations in oral microbiome profiles in rheumatoid arthritis and osteoarthritis with potential biomarkers for arthritis screening. Sci. Rep. 2018, 8, 17126. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Varesi, A.; Pierella, E.; Romeo, M.; Piccini, G.B.; Alfano, C.; Bjorklund, G.; Oppong, A.; Ricevuti, G.; Esposito, C.; Chirumbolo, S.; et al. The Potential Role of Gut Microbiota in Alzheimer’s Disease: From Diagnosis to Treatment. Nutrients 2022, 14, 668. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Yu, D.; Zhu, C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front. Cell Infect. Microbiol. 2022, 12, 834485. [Google Scholar] [CrossRef] [PubMed]
- Saad, E.; Tummala, A.; Agab, M.; Rodriguez-Nava, G. Gemella morbillorum as the Culprit Organism of Post-Colonoscopy Necrotizing Perineal Soft Tissue Infection in a Diabetic Patient With Crohn’s Disease. J. Med. Cases 2022, 13, 99–103. [Google Scholar] [CrossRef]
- Keogh, R.A.; Doran, K.S. Group B Streptococcus and diabetes: Finding the sweet spot. PLoS Pathog. 2023, 19, e1011133. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S.S. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef]
- Tiamani, K.; Luo, S.; Schulz, S.; Xue, J.; Costa, R.; Khan Mirzaei, M.; Deng, L. The role of virome in the gastrointestinal tract and beyond. FEMS Microbiol. Rev. 2022, 46, fuac027. [Google Scholar] [CrossRef]
- Chibani, C.M.; Mahnert, A.; Borrel, G.; Almeida, A.; Werner, A.; Brugere, J.F.; Gribaldo, S.; Finn, R.D.; Schmitz, R.A.; Moissl-Eichinger, C. A catalogue of 1,167 genomes from the human gut archaeome. Nat. Microbiol. 2022, 7, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.D.; Chang, H.W.; Kim, K.H.; Roh, S.W.; Kim, M.S.; Jung, M.J.; Lee, S.W.; Kim, J.Y.; Yoon, J.H.; Bae, J.W. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J. Microbiol. 2008, 46, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Seck, E.H.; Senghor, B.; Merhej, V.; Bachar, D.; Cadoret, F.; Robert, C.; Azhar, E.I.; Yasir, M.; Bibi, F.; Jiman-Fatani, A.A.; et al. Salt in stools is associated with obesity, gut halophilic microbiota and Akkermansia muciniphila depletion in humans. Int. J. Obes. 2019, 43, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Bernstein, C.N.; Tremlett, H.; Van Domselaar, G.; Knox, N.C. A Fungal World: Could the Gut Mycobiome Be Involved in Neurological Disease? Front. Microbiol. 2018, 9, 3249. [Google Scholar] [CrossRef]
- Belvoncikova, P.; Splichalova, P.; Videnska, P.; Gardlik, R. The Human Mycobiome: Colonization, Composition and the Role in Health and Disease. J. Fungi 2022, 8, 1046. [Google Scholar] [CrossRef]
- Haque, R. Human intestinal parasites. J. Health Popul. Nutr. 2007, 25, 387–391. [Google Scholar]
- Petri, W.A., Jr.; Haque, R.; Lyerly, D.; Vines, R.R. Estimating the impact of amebiasis on health. Parasitol Today 2000, 16, 320–321. [Google Scholar] [CrossRef]
- Davis, A.N.; Haque, R.; Petri, W.A., Jr. Update on protozoan parasites of the intestine. Curr. Opin. Gastroenterol. 2002, 18, 10–14. [Google Scholar] [CrossRef]
- Wright, J.E.; Werkman, M.; Dunn, J.C.; Anderson, R.M. Current epidemiological evidence for predisposition to high or low intensity human helminth infection: A systematic review. Parasit Vectors 2018, 11, 65. [Google Scholar] [CrossRef]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. EBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef]
- Hoyles, L.; McCartney, A.L.; Neve, H.; Gibson, G.R.; Sanderson, J.D.; Heller, K.J.; van Sinderen, D. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 2014, 165, 803–812. [Google Scholar] [CrossRef]
- Spencer, L.; Olawuni, B.; Singh, P. Gut Virome: Role and Distribution in Health and Gastrointestinal Diseases. Front. Cell Infect. Microbiol. 2022, 12, 836706. [Google Scholar] [CrossRef]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [PubMed]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung-gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal. Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Sorboni, S.G.; Moghaddam, H.S.; Jafarzadeh-Esfehani, R.; Soleimanpour, S. A Comprehensive Review on the Role of the Gut Microbiome in Human Neurological Disorders. Clin. Microbiol. Rev. 2022, 35, e0033820. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.I.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Tripathi, P. Gut microbiome and type 2 diabetes: Where we are and where to go? J. Nutr. Biochem. 2019, 63, 101–108. [Google Scholar] [CrossRef]
- Schneeberger, M.; Everard, A.; Gomez-Valades, A.G.; Matamoros, S.; Ramirez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Hu, Y.; Wang, X.; Shen, L.; Liao, X.; Zhu, Y.; Yu, J.; Zhao, F.; Zhou, Y.; Shen, H.; et al. Microbiome in cancer: An exploration of carcinogenesis, immune responses and immunotherapy. Front. Immunol. 2022, 13, 877939. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Gotschlich, E.C.; Colbert, R.A.; Gill, T. Methods in microbiome research: Past, present, and future. Best Pract. Res. Clin. Rheumatol. 2019, 33, 101498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.X.; Guo, X.; Qin, Y.; Garrido-Oter, R.; Schulze-Lefert, P.; Bai, Y. High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 2021, 16, 988–1012. [Google Scholar] [CrossRef]
- Liu, Y.X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Schwerdtfeger, L.A.; Nealon, N.J.; Ryan, E.P.; Tobet, S.A. Human colon function ex vivo: Dependence on oxygen and sensitivity to antibiotic. PLoS ONE 2019, 14, e0217170. [Google Scholar] [CrossRef]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.L.; Lagier, J.C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 9674. [Google Scholar] [CrossRef]
- Kim, R.; Attayek, P.J.; Wang, Y.; Furtado, K.L.; Tamayo, R.; Sims, C.E.; Allbritton, N.L. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 2019, 12, 015006. [Google Scholar] [CrossRef]
- Kaspar, U.; Kriegeskorte, A.; Schubert, T.; Peters, G.; Rudack, C.; Pieper, D.H.; Wos-Oxley, M.; Becker, K. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 2016, 18, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
- Matar, G.; Bilen, M. Culturomics, A potential approach paving the way toward bacteriotherapy. Curr. Opin. Microbiol. 2022, 69, 102194. [Google Scholar] [CrossRef]
- Greub, G. Culturomics: A new approach to study the human microbiome. Clin. Microbiol. Infect 2012, 18, 1157–1159. [Google Scholar] [CrossRef] [PubMed]
- Naud, S.; Khelaifia, S.; Mbogning Fonkou, M.D.; Dione, N.; Lagier, J.C.; Raoult, D. Proof of Concept of Culturomics Use of Time of Care. Front. Cell Infect Microbiol. 2020, 10, 524769. [Google Scholar] [CrossRef]
- Kellenberger, E. Exploring the unknown. The silent revolution of microbiology. EMBO Rep. 2001, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Rezasoltani, S.; Ahmadi Bashirzadeh, D.; Nazemalhosseini Mojarad, E.; Asadzadeh Aghdaei, H.; Norouzinia, M.; Shahrokh, S. Signature of Gut Microbiome by Conventional and Advanced Analysis Techniques: Advantages and Disadvantages. Middle East J. Dig. Dis. 2020, 12, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H.; et al. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding, C.; Fungal Barcoding Consortium Author, L. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Gupta, S.; Mortensen, M.S.; Schjorring, S.; Trivedi, U.; Vestergaard, G.; Stokholm, J.; Bisgaard, H.; Krogfelt, K.A.; Sorensen, S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019, 2, 291. [Google Scholar] [CrossRef]
- Rozas, M.; Brillet, F.; Callewaert, C.; Paetzold, B. MinION Nanopore Sequencing of Skin Microbiome 16S and 16S-23S rRNA Gene Amplicons. Front. Cell Infect Microbiol. 2021, 11, 806476. [Google Scholar] [CrossRef]
- Callahan, B.J.; Grinevich, D.; Thakur, S.; Balamotis, M.A.; Yehezkel, T.B. Ultra-accurate microbial amplicon sequencing with synthetic long reads. Microbiome 2021, 9, 130. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Niwa, H.; Uchida-Fujii, E.; Nukada, T. Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome. Sci. Rep. 2021, 11, 11884. [Google Scholar] [CrossRef]
- Kameoka, S.; Motooka, D.; Watanabe, S.; Kubo, R.; Jung, N.; Midorikawa, Y.; Shinozaki, N.O.; Sawai, Y.; Takeda, A.K.; Nakamura, S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1-V2 and V3-V4 primer sets. BMC Genom. 2021, 22, 527. [Google Scholar] [CrossRef] [PubMed]
- Alcon-Giner, C.; Dalby, M.J.; Caim, S.; Ketskemety, J.; Shaw, A.; Sim, K.; Lawson, M.A.E.; Kiu, R.; Leclaire, C.; Chalklen, L.; et al. Microbiota Supplementation with Bifidobacterium and Lactobacillus Modifies the Preterm Infant Gut Microbiota and Metabolome: An Observational Study. Cell Rep. Med. 2020, 1, 100077. [Google Scholar] [CrossRef] [PubMed]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella copri Isolates. Cell Host Microbe. 2019, 26, 680–690.e685. [Google Scholar] [CrossRef] [PubMed]
- Allali, I.; Arnold, J.W.; Roach, J.; Cadenas, M.B.; Butz, N.; Hassan, H.M.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R.; et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017, 17, 194. [Google Scholar] [CrossRef]
- Mortensen, M.S.; Brejnrod, A.D.; Roggenbuck, M.; Abu Al-Soud, W.; Balle, C.; Krogfelt, K.A.; Stokholm, J.; Thorsen, J.; Waage, J.; Rasmussen, M.A.; et al. The developing hypopharyngeal microbiota in early life. Microbiome 2016, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Sanschagrin, S.; Yergeau, E. Next-generation sequencing of 16S ribosomal RNA gene amplicons. J. Vis. Exp. 2014, 90, 51709. [Google Scholar] [CrossRef]
- Nygaard, A.B.; Tunsjo, H.S.; Meisal, R.; Charnock, C. A preliminary study on the potential of Nanopore MinION and Illumina MiSeq 16S rRNA gene sequencing to characterize building-dust microbiomes. Sci. Rep. 2020, 10, 3209. [Google Scholar] [CrossRef]
- Kumar Awasthi, M.; Ravindran, B.; Sarsaiya, S.; Chen, H.; Wainaina, S.; Singh, E.; Liu, T.; Kumar, S.; Pandey, A.; Singh, L.; et al. Metagenomics for taxonomy profiling: Tools and approaches. Bioengineered 2020, 11, 356–374. [Google Scholar] [CrossRef]
- Lam, K.N.; Cheng, J.; Engel, K.; Neufeld, J.D.; Charles, T.C. Current and future resources for functional metagenomics. Front. Microbiol. 2015, 6, 1196. [Google Scholar] [CrossRef] [PubMed]
- Leigh, D.M.; Schefer, C.; Cornejo, C. Determining the Suitability of MinION’s Direct RNA and DNA Amplicon Sequencing for Viral Subtype Identification. Viruses 2020, 12, 801. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef]
- Terron-Camero, L.C.; Gordillo-Gonzalez, F.; Salas-Espejo, E.; Andres-Leon, E. Comparison of Metagenomics and Metatranscriptomics Tools: A Guide to Making the Right Choice. Genes 2022, 13, 2280. [Google Scholar] [CrossRef]
- Brown, B.L.; Watson, M.; Minot, S.S.; Rivera, M.C.; Franklin, R.B. MinION nanopore sequencing of environmental metagenomes: A synthetic approach. Gigascience 2017, 6, 1–10. [Google Scholar] [CrossRef]

| Bacteria | Oxygen Requirement |
|---|---|
| Mycobacterium tuberculosis | Obligate aerobe |
| Micrococcus luteus | |
| Neisseria meningitidis | |
| Neisseria gonorrhoeae | |
| Bacteroidetes | Obligate anaerobe |
| Porphyromonas sp. | |
| Prevotella sp. | |
| Clostridium spp. | |
| Staphylococci | Facultative anaerobes |
| Gemella sp. | |
| Enterobacteriaceae | |
| Lactobacilli | Aerotolerant anaerobes |
| Campylobacter jejuni | Microaerophile |
| Related Microorganisms | Culture Media |
|---|---|
| Gram-positive Staphylococci | Baird-Parker agar |
| Streptococcus pyogenes | Crystal Violet Blood Agar |
| Gram-negative bacterial species | Hektoen Enteric Agar |
| Mycobacterium species | Lowenstein Jensen Medium |
| Gram-negative bacterial species | MacConkey’s Agar |
| Gram-positive bacterial species | Mannitol Salt Agar |
| Enterococcus species | Potassium Tellurite Medium |
| Pseudomonas aeruginosa | Pseudosel Agar |
| Neisseria gonorrhoeae | Thayer Martin Agar |
| Vibrio species | Thiosulfate Citrate Bile Salts Sucrose Agar |
| Salmonella & Shigella species | Salmonella-Shigella Agar |
| Salmonella species | Wilson and Blair’s Agar |
| Ectomycorrhizal fungi | BAF Medium |
| Ectomycorrhizal fungi | Modified Melin-Norkrans Medium |
| Certain fungi | Sabouraud Agar |
| Detection Method | Advantages | Drawbacks | Reference |
|---|---|---|---|
| Culturome | Visible colonies | Time-consuming and expensive | [106] |
| Microbial isolates | Sterile environmental conditions | ||
| Selection of the target | Laborious | ||
| Amplicon sequencing | Cost-effective | PCR biases | [107,108] |
| Easy and quick analysis | False-positive and false-negative samples | ||
| Selection of the target | No discrimination between dead and live microbes | ||
| Low-biomass requirement | Limited genus-level taxonomic resolution | ||
| Metagenomics | Culture-independent | Expensive | [109] |
| Species or strain level taxonomic resolution | Complex and time-consuming analysis | ||
| Captures all genomes present in a sample | No discrimination between dead and live microbes | ||
| Culture-independent | Host-derived contamination | ||
| Metatranscriptomics | Transcript level resolution | Expensive | [110] |
| Assessment of gene expression | Complex and time-consuming analysis | ||
| Discrimination between dead and live microbes | Snapshot of protein expression levels Host RNA contamination |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Athanasopoulou, K.; Adamopoulos, P.G.; Scorilas, A. Unveiling the Human Gastrointestinal Tract Microbiome: The Past, Present, and Future of Metagenomics. Biomedicines 2023, 11, 827. https://doi.org/10.3390/biomedicines11030827
Athanasopoulou K, Adamopoulos PG, Scorilas A. Unveiling the Human Gastrointestinal Tract Microbiome: The Past, Present, and Future of Metagenomics. Biomedicines. 2023; 11(3):827. https://doi.org/10.3390/biomedicines11030827
Chicago/Turabian StyleAthanasopoulou, Konstantina, Panagiotis G. Adamopoulos, and Andreas Scorilas. 2023. "Unveiling the Human Gastrointestinal Tract Microbiome: The Past, Present, and Future of Metagenomics" Biomedicines 11, no. 3: 827. https://doi.org/10.3390/biomedicines11030827
APA StyleAthanasopoulou, K., Adamopoulos, P. G., & Scorilas, A. (2023). Unveiling the Human Gastrointestinal Tract Microbiome: The Past, Present, and Future of Metagenomics. Biomedicines, 11(3), 827. https://doi.org/10.3390/biomedicines11030827







