Difference in Methylation and Expression of Brain-Derived Neurotrophic Factor in Alzheimer’s Disease and Mild Cognitive Impairment
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Gene Expression
2.2.1. Blood Sample Collection
2.2.2. RNA Isolation and Reverse Transcription
2.2.3. Real-Time PCR Analysis
2.2.4. Statistical Analysis
2.3. DNA Methylation
2.3.1. DNA Isolation and Bisulfite Conversion
2.3.2. Primer Design
2.3.3. Amplicon Generation and Sequencing
2.3.4. Library Preparation and Sequencing
2.3.5. Bioinformatic and Statistical Analysis
3. Results
3.1. Participants
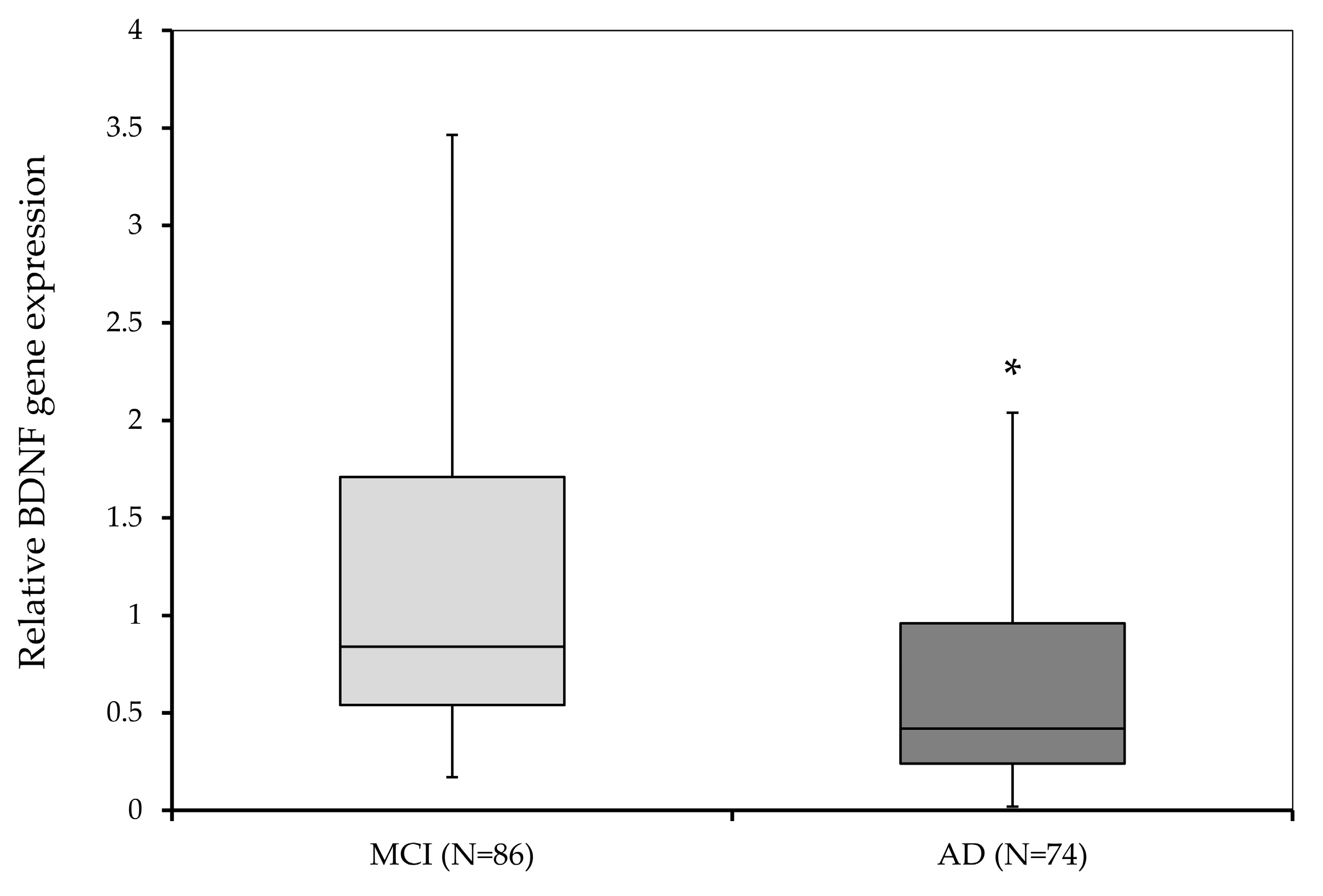
3.2. BDNF Gene Expression
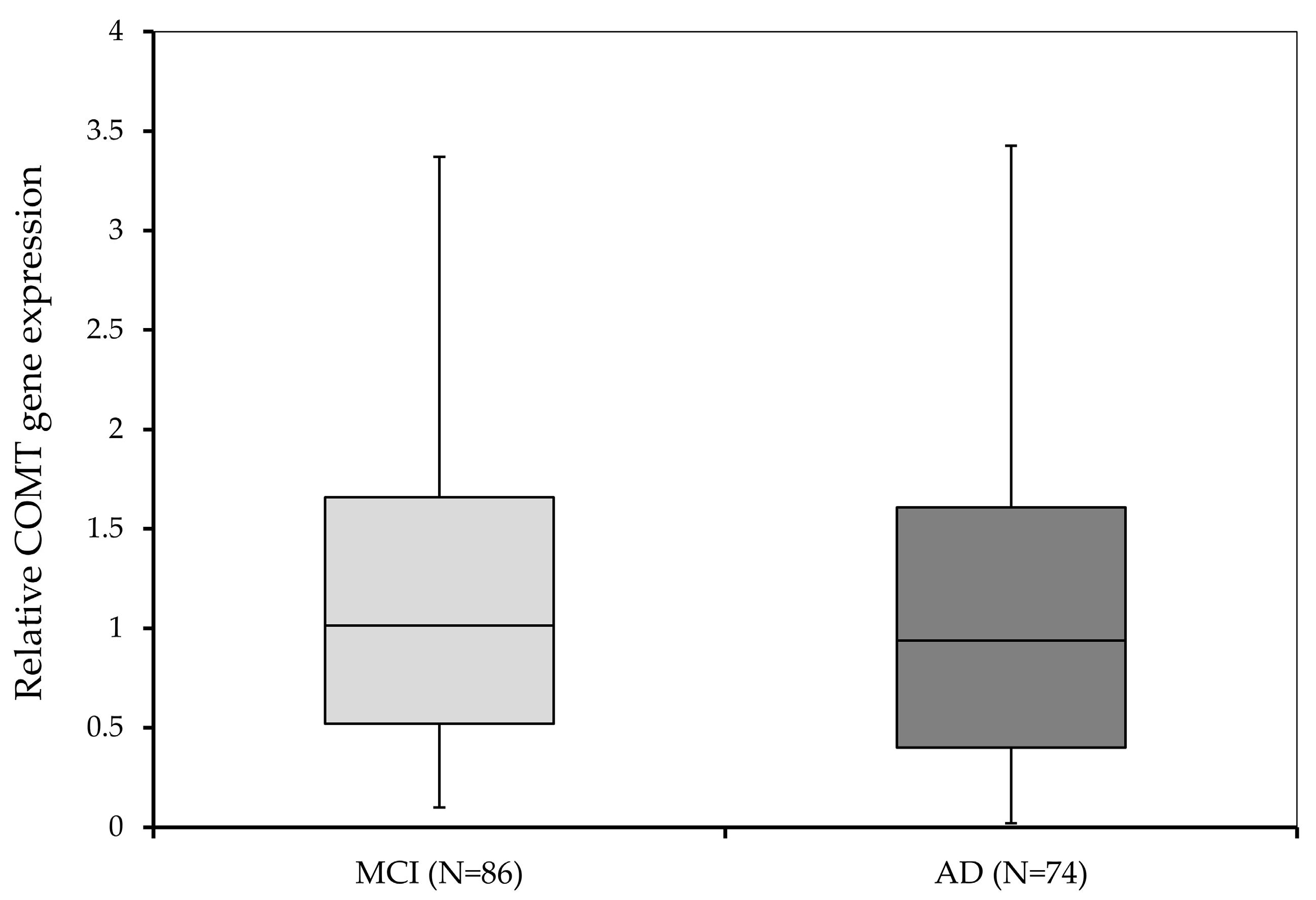
3.3. COMT Gene Expression
3.4. DNA Methylation
4. Discussion
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2022 Alzheimer’s Disease Facts and Figures; Alzheimer’s Association: Chicago, IL, USA, 2022. [Google Scholar]
- Qiu, C.; Kivipelto, M.; Von Strauss, E. Epidemiology of alzheimer’s disease: Occurrence, determinants, and strategies toward intervention. Dialogues Clin. Neurosci. 2009, 11, 111–128. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Publishing, Inc.: Arlington, TX, USA, 1994; p. 886. [Google Scholar]
- WHO. International Statistical Classification of Diseases and Related Health Problems; 10th Revision icd-10: Tabular List; World Health Organization: Geneva, Switzerland, 2016; Volume 1, pp. 332–345. [Google Scholar]
- Tanaka, M.; Vécsei, L. Editorial of special issue ‘dissecting neurological and neuropsychiatric diseases: Neurodegeneration and neuroprotection’. Int. J. Mol. Sci. 2022, 23, 6991. [Google Scholar] [CrossRef] [PubMed]
- Carrera-González, M.D.P.; Cantón-Habas, V.; Rich-Ruiz, M. Aging, depression and dementia: The inflammatory process. Adv. Clin. Exp. Med. 2022, 31, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S. Neurobiological advances of learned fear in humans. Adv. Clin. Exp. Med. 2022, 31, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Fox, N.C. The search for early markers of ad: Hippocampal atrophy and memory deficits. Int. Psychogeriatr. 2014, 26, 1065–1066. [Google Scholar] [CrossRef]
- Battaglia, S.; Harrison, B.J.; Fullana, M.A. Does the human ventromedial prefrontal cortex support fear learning, fear extinction or both? A commentary on subregional contributions. Mol. Psychiatry 2022, 27, 784–786. [Google Scholar] [CrossRef]
- Wenk, G.L. Neuropathologic changes in alzheimer’s disease. J. Clin. Psychiatry 2003, 64 (Suppl. S9), 7–10. [Google Scholar] [PubMed]
- Findeis, M.A. The role of amyloid beta peptide 42 in alzheimer’s disease. Pharmacol. Ther. 2007, 116, 266–286. [Google Scholar] [CrossRef]
- Strac, D.S.; Muck-Seler, D.; Pivac, N. Neurotransmitter measures in the cerebrospinal fluid of patients with alzheimer’s disease: A review. Psychiatr. Danub. 2015, 27, 14–24. [Google Scholar]
- Tanaka, M.; Szabó, Á.; Spekker, E.; Polyák, H.; Tóth, F.; Vécsei, L. Mitochondrial impairment: A common motif in neuropsychiatric presentation? The link to the tryptophan–kynurenine metabolic system. Cells 2022, 11, 2607. [Google Scholar]
- Kowalczyk, P.; Sulejczak, D.; Kleczkowska, P.; Bukowska-Ośko, I.; Kucia, M.; Popiel, M.; Wietrak, E.; Kramkowski, K.; Wrzosek, K.; Kaczyńska, K. Mitochondrial oxidative stress—a causative factor and therapeutic target in many diseases. Int. J. Mol. Sci. 2021, 22, 13384. [Google Scholar]
- Nikolac Perkovic, M.; Videtic Paska, A.; Konjevod, M.; Kouter, K.; Svob Strac, D.; Nedic Erjavec, G.; Pivac, N. Epigenetics of alzheimer’s disease. Biomolecules 2021, 11, 195. [Google Scholar] [CrossRef]
- Frey, H.J.; Mattila, K.M.; Korolainen, M.A.; Pirttilä, T. Problems associated with biological markers of alzheimer’s disease. Neurochem. Res. 2005, 30, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Grünblatt, E.; Zehetmayer, S.; Bartl, J.; Löffler, C.; Wichart, I.; Rainer, M.K.; Jungwirth, S.; Bauer, P.; Danielczyk, W.; Tragl, K.-H.; et al. Genetic risk factors and markers for alzheimer’s disease and/or depression in the vita study. J. Psychiatr. Res. 2009, 43, 298–308. [Google Scholar] [CrossRef]
- Borroni, B.; Costanzi, C.; Padovani, A. Genetic susceptibility to behavioural and psychological symptoms in alzheimer disease. Curr. Alzheimer Res. 2010, 7, 158–164. [Google Scholar] [CrossRef]
- Serretti, A.; Olgiati, P.; De Ronchi, D. Genetics of alzheimer’s disease. A rapidly evolving field. J. Alzheimer’s Dis. 2007, 12, 73–92. [Google Scholar] [CrossRef] [PubMed]
- Russo-Neustadt, A. Brain-derived neurotrophic factor, behavior, and new directions for the treatment of mental disorders. Semin. Clin. Neuropsychiatry 2003, 8, 109–118. [Google Scholar] [CrossRef]
- Wetmore, C.; Ernfors, P.; Persson, H.; Olson, L. Localization of brain-derived neurotrophic factor mrna to neurons in the brain by in situ hybridization. Exp. Neurol. 1990, 109, 141–152. [Google Scholar] [CrossRef]
- Ghosh, A.; Carnahan, J.; Greenberg, M.E. Requirement for bdnf in activity-dependent survival of cortical neurons. Science 1994, 263, 1618–1623. [Google Scholar] [CrossRef]
- Alderson, R.F.; Alterman, A.L.; Barde, Y.A.; Lindsay, R.M. Brain-derived neurotrophic factor increases survival and differentiated functions of rat septal cholinergic neurons in culture. Neuron 1990, 5, 297–306. [Google Scholar] [CrossRef]
- Lindholm, D.; Carroll, P.; Tzimagiorgis, G.; Thoenen, H. Autocrine-paracrine regulation of hippocampal neuron survival by igf-1 and the neurotrophins bdnf, nt-3 and nt-4. Eur. J. Neurosci. 1996, 8, 1452–1460. [Google Scholar] [CrossRef]
- Xiu, M.H.; Hui, L.; Dang, Y.F.; Hou, T.D.; Zhang, C.X.; Zheng, Y.L.; Chen, D.C.; Kosten, T.R.; Zhang, X.Y. Decreased serum bdnf levels in chronic institutionalized schizophrenia on long-term treatment with typical and atypical antipsychotics. Prog. Neuropsychopharmacol. Biol. Psychiatry 2009, 33, 1508–1512. [Google Scholar] [CrossRef]
- Brunoni, A.R.; Lopes, M.; Fregni, F. A systematic review and meta-analysis of clinical studies on major depression and bdnf levels: Implications for the role of neuroplasticity in depression. Int. J. Neuropsychopharmacol. 2008, 11, 1169–1180. [Google Scholar] [CrossRef]
- Tapia-Arancibia, L.; Aliaga, E.; Silhol, M.; Arancibia, S. New insights into brain bdnf function in normal aging and alzheimer disease. Brain Re. Rev. 2008, 59, 201–220. [Google Scholar] [CrossRef]
- Lee, J.; Fukumoto, H.; Orne, J.; Klucken, J.; Raju, S.; Vanderburg, C.R.; Irizarry, M.C.; Hyman, B.T.; Ingelsson, M. Decreased levels of bdnf protein in alzheimer temporal cortex are independent of bdnf polymorphisms. Exp. Neurol. 2005, 194, 91–96. [Google Scholar] [CrossRef]
- Peng, S.; Wuu, J.; Mufson, E.J.; Fahnestock, M. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of alzheimer’s disease. J. Neurochem. 2005, 93, 1412–1421. [Google Scholar] [CrossRef]
- Du, Y.; Wu, H.-T.; Qin, X.-Y.; Cao, C.; Liu, Y.; Cao, Z.-Z.; Cheng, Y. Postmortem brain, cerebrospinal fluid, and blood neurotrophic factor levels in alzheimer’s disease: A systematic review and meta-analysis. J. Mol. Neurosci. 2018, 65, 289–300. [Google Scholar] [CrossRef]
- Ng, T.K.S.; Ho, C.S.H.; Tam, W.W.S.; Kua, E.H.; Ho, R.C.-M. Decreased serum brain-derived neurotrophic factor (bdnf) levels in patients with alzheimer’s disease (ad): A systematic review and meta-analysis. Int. J. Mol. Sci. 2019, 20, 257. [Google Scholar] [CrossRef]
- Wang, Z.-H.; Xiang, J.; Liu, X.; Yu, S.P.; Manfredsson, F.P.; Sandoval, I.M.; Wu, S.; Wang, J.-Z.; Ye, K. Deficiency in bdnf/trkb neurotrophic activity stimulates δ-secretase by upregulating c/ebpβ in alzheimer’s disease. Cell Rep. 2019, 28, 655–669. [Google Scholar] [CrossRef]
- Jiao, S.S.; Shen, L.L.; Zhu, C.; Bu, X.L.; Liu, Y.H.; Liu, C.H.; Yao, X.Q.; Zhang, L.L.; Zhou, H.D.; Walker, D.G.; et al. Brain-derived neurotrophic factor protects against tau-related neurodegeneration of alzheimer’s disease. Transl. Psychiatry 2016, 6, e907. [Google Scholar] [CrossRef]
- Weinstein, G.; Beiser, A.S.; Choi, S.H.; Preis, S.R.; Chen, T.C.; Vorgas, D.; Au, R.; Pikula, A.; Wolf, P.A.; DeStefano, A.L.; et al. Serum brain-derived neurotrophic factor and the risk for dementia: The framingham heart study. JAMA Neurol. 2014, 71, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Männistö, P.T.; Kaakkola, S. Catechol-o-methyltransferase (comt): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective comt inhibitors. Pharmacol. Rev. 1999, 51, 593–628. [Google Scholar] [PubMed]
- Tunbridge, E.M.; Bannerman, D.M.; Sharp, T.; Harrison, P.J. Catechol-o-methyltransferase inhibition improves set-shifting performance and elevates stimulated dopamine release in the rat prefrontal cortex. J. Neurosci. 2004, 24, 5331–5335. [Google Scholar] [CrossRef] [PubMed]
- Craddock, N.; Owen, M.J.; O’Donovan, M.C. The catechol-o-methyl transferase (comt) gene as a candidate for psychiatric phenotypes: Evidence and lessons. Mol. Psychiatry 2006, 11, 446–458. [Google Scholar] [CrossRef]
- Goldman, D.; Weinberger, D.R.; Malhotra, A.K.; Goldberg, T.E. The role of comt val158met in cognition. Biol. Psychiatry 2009, 65, e1–e2. [Google Scholar] [CrossRef] [PubMed]
- Mier, D.; Kirsch, P.; Meyer-Lindenberg, A. Neural substrates of pleiotropic action of genetic variation in comt: A meta-analysis. Mol. Psychiatry 2010, 15, 918–927. [Google Scholar] [CrossRef]
- Barnett, J.H.; Scoriels, L.; Munafò, M.R. Meta-analysis of the cognitive effects of the catechol-o-methyltransferase gene val158/108met polymorphism. Biol. Psychiatry 2008, 64, 137–144. [Google Scholar] [CrossRef]
- Lachman, H.M.; Papolos, D.F.; Saito, T.; Yu, Y.M.; Szumlanski, C.L.; Weinshilboum, R.M. Human catechol-o-methyltransferase pharmacogenetics: Description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 1996, 6, 243–250. [Google Scholar] [CrossRef]
- Perkovic, M.N.; Strac, D.S.; Tudor, L.; Konjevod, M.; Erjavec, G.N.; Pivac, N. Catechol-o-methyltransferase, cognition and alzheimer’s disease. Curr. Alzheimer Res. 2018, 15, 408–419. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar]
- Arevalo-Rodriguez, I.; Smailagic, N.; Figuls, M.R.I.; Ciapponi, A.; Sanchez-Perez, E.; Giannakou, A.; Pedraza, O.L.; Bonfill Cosp, X.; Cullum, S. Mini-mental state examination (mmse) for the detection of alzheimer’s disease and other dementias in people with mild cognitive impairment (mci). Cochrane Database Syst. Rev. 2015, 2015, CD010783. [Google Scholar] [CrossRef]
- Boban, M.; Malojčić, B.; Mimica, N.; Vuković, S.; Zrilić, I.; Hof, P.R.; Simić, G. The reliability and validity of the mini-mental state examination in the elderly croatian population. Dement. Geriatr. Cogn. Disord. 2012, 33, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Agrell, B.; Dehlin, O. The clock-drawing test. Age Ageing 1998, 27, 399–403. [Google Scholar] [CrossRef]
- Shulman, K.I. Clock-drawing: Is it the ideal cognitive screening test? Int. J. Geriatr. Psychiatry 2000, 15, 548–561. [Google Scholar] [CrossRef] [PubMed]
- World Medical, A. World medical association declaration of helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at ucsc. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Illumina. PCR Amplicon, P.C.-u., and Index PCR: 16S Metagenomic Sequencing Library Preparation. 2013. Available online: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 25 August 2020).
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data [Online]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 August 2020).
- Krueger, F. Trimgalore. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 25 August 2020).
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- R Development Core Team; R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org (accessed on 25 August 2020).
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. Methylkit: A comprehensive r package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Park, Y.; Figueroa, M.E.; Rozek, L.S.; Sartor, M.A. Methylsig: A whole genome DNA methylation analysis pipeline. Bioinformatics 2014, 30, 2414–2422. [Google Scholar] [CrossRef]
- Pruunsild, P.; Kazantseva, A.; Aid, T.; Palm, K.; Timmusk, T. Dissecting the human bdnf locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics 2007, 90, 397–406. [Google Scholar] [CrossRef]
- Fujimura, H.; Altar, C.A.; Chen, R.; Nakamura, T.; Nakahashi, T.; Kambayashi, J.-i.; Sun, B.; Tandon, N.N. Brain-derived neurotrophic factor is stored in human platelets and released by agonist stimulation. Thromb. Haemost. 2002, 87, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Bocchio-Chiavetto, L.; Zanardini, R.; Milanesi, E.; Placentino, A.; Gennarelli, M. Reduced peripheral brain-derived neurotrophic factor mrna levels are normalized by antidepressant treatment. Int. J Neuropsychopharmacol. 2010, 13, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Gielen, A.; Khademi, M.; Muhallab, S.; Olsson, T.; Piehl, F. Increased brain-derived neurotrophic factor expression in white blood cells of relapsing-remitting multiple sclerosis patients. Scand. J. Immunol. 2003, 57, 493–497. [Google Scholar] [CrossRef]
- Nakahashi, T.; Fujimura, H.; Altar, C.A.; Li, J.; Kambayashi, J.; Tandon, N.N.; Sun, B. Vascular endothelial cells synthesize and secrete brain-derived neurotrophic factor. FEBS Lett. 2000, 470, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Banks, W.A.; Kastin, A.J. Permeability of the blood-brain barrier to neurotrophins. Brain Res. 1998, 788, 87–94. [Google Scholar] [CrossRef]
- Pan, W.; Banks, W.A.; Fasold, M.B.; Bluth, J.; Kastin, A.J. Transport of brain-derived neurotrophic factor across the blood-brain barrier. Neuropharmacology 1998, 37, 1553–1561. [Google Scholar] [CrossRef]
- Klein, A.B.; Williamson, R.; Santini, M.A.; Clemmensen, C.; Ettrup, A.; Rios, M.; Knudsen, G.M.; Aznar, S. Blood bdnf concentrations reflect brain-tissue bdnf levels across species. Int. J. Neuropsychopharmacol. 2011, 14, 347–353. [Google Scholar] [CrossRef]
- Karege, F.; Schwald, M.; Cisse, M. Postnatal developmental profile of brain-derived neurotrophic factor in rat brain and platelets. Neurosci. Lett. 2002, 328, 261–264. [Google Scholar] [CrossRef]
- Murer, M.G.; Yan, Q.; Raisman-Vozari, R. Brain-derived neurotrophic factor in the control human brain, and in alzheimer’s disease and parkinson’s disease. Prog. Neurobiol. 2001, 63, 71–124. [Google Scholar] [CrossRef]
- Phillips, H.S.; Hains, J.M.; Armanini, M.; Laramee, G.R.; Johnson, S.A.; Winslow, J.W. Bdnf mrna is decreased in the hippocampus of individuals with alzheimer’s disease. Neuron 1991, 7, 695–702. [Google Scholar] [CrossRef]
- Garzon, D.; Yu, G.; Fahnestock, M. A new brain-derived neurotrophic factor transcript and decrease in brain-derived neurotrophic factor transcripts 1, 2 and 3 in alzheimer’s disease parietal cortex. J. Neurochem. 2002, 82, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, S.D.; Malek-Ahmadi, M.H.; Alldred, M.J.; Chen, Y.; Chen, K.; Chao, M.V.; Counts, S.E.; Mufson, E.J. Brain-derived neurotrophic factor (bdnf) and trkb hippocampal gene expression are putative predictors of neuritic plaque and neurofibrillary tangle pathology. Neurobiol. Dis. 2019, 132, 104540. [Google Scholar] [CrossRef] [PubMed]
- Connor, B.; Young, D.; Yan, Q.; Faull, R.L.; Synek, B.; Dragunow, M. Brain-derived neurotrophic factor is reduced in alzheimer’s disease. Brain Res. Mol. Brain Res. 1997, 49, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Narisawa-Saito, M.; Wakabayashi, K.; Tsuji, S.; Takahashi, H.; Nawa, H. Regional specificity of alterations in ngf, bdnf and nt-3 levels in alzheimer’s disease. Neuroreport 1996, 7, 2925–2928. [Google Scholar] [CrossRef]
- Siegel, G.J.; Chauhan, N.B. Neurotrophic factors in alzheimer’s and parkinson’s disease brain. Brain Res. Brain Res. Rev. 2000, 33, 199–227. [Google Scholar] [CrossRef]
- Sampaio, T.B.; Savall, A.S.; Gutierrez, M.E.Z.; Pinton, S. Neurotrophic factors in alzheimer’s and parkinson’s diseases: Implications for pathogenesis and therapy. Neural Regen. Res. 2017, 12, 549–557. [Google Scholar] [CrossRef]
- Durany, N.; Michel, T.; Kurt, J.; Cruz-Sánchez, F.F.; Cervás-Navarro, J.; Riederer, P. Brain-derived neurotrophic factor and neurotrophin-3 levels in alzheimer’s disease brains. Int. J. Dev. Neurosci. 2000, 18, 807–813. [Google Scholar] [CrossRef]
- Angelucci, F.; Spalletta, G.; di Iulio, F.; Ciaramella, A.; Salani, F.; Colantoni, L.; Varsi, A.E.; Gianni, W.; Sancesario, G.; Caltagirone, C.; et al. Alzheimer’s disease (ad) and mild cognitive impairment (mci) patients are characterized by increased bdnf serum levels. Curr. Alzheimer Res. 2010, 7, 15–20. [Google Scholar] [CrossRef]
- Laske, C.; Stransky, E.; Leyhe, T.; Eschweiler, G.W.; Wittorf, A.; Richartz, E.; Bartels, M.; Buchkremer, G.; Schott, K. Stage-dependent bdnf serum concentrations in alzheimer’s disease. J. Neural Transm. 2006, 113, 1217–1224. [Google Scholar] [CrossRef]
- Laske, C.; Stransky, E.; Leyhe, T.; Eschweiler, G.W.; Maetzler, W.; Wittorf, A.; Soekadar, S.; Richartz, E.; Koehler, N.; Bartels, M.; et al. Bdnf serum and csf concentrations in alzheimer’s disease, normal pressure hydrocephalus and healthy controls. J. Psychiatr. Res. 2007, 41, 387–394. [Google Scholar] [CrossRef]
- Salat, D.H.; Kaye, J.A.; Janowsky, J.S. Selective preservation and degeneration within the prefrontal cortex in aging and alzheimer disease. Arch. Neurol. 2001, 58, 1403–1408. [Google Scholar] [CrossRef] [PubMed]
- Tenhunen, J.; Salminen, M.; Jalanko, A.; Ukkonen, S.; Ulmanen, I. Structure of the rat catechol-o-methyltransferase gene: Separate promoters are used to produce mrnas for soluble and membrane-bound forms of the enzyme. DNA Cell Biol. 1993, 12, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Weickert, C.S.; Beltaifa, S.; Kolachana, B.; Chen, J.; Hyde, T.M.; Herman, M.M.; Weinberger, D.R.; Kleinman, J.E. Catechol o-methyltransferase (comt) mrna expression in the dorsolateral prefrontal cortex of patients with schizophrenia. Neuropsychopharmacology 2003, 28, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Lundström, K.; Tenhunen, J.; Tilgmann, C.; Karhunen, T.; Panula, P.; Ulmanen, I. Cloning, expression and structure of catechol-o-methyltransferase. Biochim. Biophys. Acta 1995, 1251, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Serretti, A.; Olgiati, P. Catechol-o-methyltransferase and alzheimer’s disease: A review of biological and genetic findings. CNS Neurol. Disord. Drug Targets 2012, 11, 299–305. [Google Scholar] [CrossRef]
- Forero, D.A.; Benítez, B.; Arboleda, G.; Yunis, J.J.; Pardo, R.; Arboleda, H. Analysis of functional polymorphisms in three synaptic plasticity-related genes (bdnf, comt and uchl1) in alzheimer’s disease in colombia. Neurosci. Res. 2006, 55, 334–341. [Google Scholar] [CrossRef]
- Shibata, N.; Nagata, T.; Tagai, K.; Shinagawa, S.; Ohnuma, T.; Kawai, E.; Kasanuki, K.; Shimazaki, H.; Toda, A.; Tagata, Y.; et al. Association between the catechol-o-methyltransferase polymorphism val158met and alzheimer’s disease in a japanese population. Int. J. Geriatr. Psychiatry 2015, 30, 927–933. [Google Scholar] [CrossRef]
- Lanni, C.; Garbin, G.; Lisa, A.; Biundo, F.; Ranzenigo, A.; Sinforiani, E.; Cuzzoni, G.; Govoni, S.; Ranzani, G.N.; Racchi, M. Influence of comt val158met polymorphism on alzheimer’s disease and mild cognitive impairment in italian patients. J. Alzheimers Dis. 2012, 32, 919–926. [Google Scholar] [CrossRef]
- Chang, C.-C.; Tsai, S.-J.; Chen, N.-C.; Huang, C.-W.; Hsu, S.-W.; Chang, Y.-T.; Liu, M.-E.; Chang, W.-N.; Tsai, W.-C.; Lee, C.-C. Catechol-o-methyltransferase val158met polymorphism on striatum structural covariance networks in alzheimer’s disease. Mol. Neurobiol. 2018, 55, 4637–4649. [Google Scholar] [CrossRef]
- Thornton, V.; Warden, D.; Talbot, C.; Mastana, S.S.; Bandelow, S.; Hogervorst, E. Modification of estrogen’s association with alzheimer’s disease risk by genetic polymorphisms. Brain Res. 2011, 1379, 213–223. [Google Scholar] [CrossRef]
- Gennatas, E.D.; Cholfin, J.A.; Zhou, J.; Crawford, R.K.; Sasaki, D.A.; Karydas, A.; Boxer, A.L.; Bonasera, S.J.; Rankin, K.P.; Gorno-Tempini, M.L.; et al. Comt val158met genotype influences neurodegeneration within dopamine-innervated brain structures. Neurology 2012, 78, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, Y.-C.; Xu, H.-D.; Liu, X.; Zhu, J.; Zhang, F.; Wang, D.; Wang, Y.; Jin, C. Lack of association between comt polymorphism rs4680 and risk of alzheimer’s disease in asians: Evidence from a meta-analysis. Psychiatry Res. 2015, 228, 979–981. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Song, G.G. Comt val158met and pparγ pro12ala polymorphisms and susceptibility to alzheimer’s disease: A meta-analysis. Neurol. Sci. 2014, 35, 643–651. [Google Scholar] [CrossRef]
- Ni, P.; Liu, M.; Wang, D.; Tian, Y.; Zhao, L.; Wei, J.; Yu, X.; Qi, X.; Li, X.; Yu, H.; et al. Association analysis between catechol-o-methyltransferase expression and cognitive function in patients with schizophrenia, bipolar disorder, or major depression. Neuropsychiatr. Dis. Treat. 2021, 17, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. Cpg islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Wang, Y.; Ji, H.; Dai, D.; Xu, X.; Jiang, D.; Hong, Q.; Ye, H.; Zhang, X.; Zhou, X.; et al. Elevation of peripheral bdnf promoter methylation links to the risk of alzheimer’s disease. PLoS ONE 2014, 9, e110773. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Xu, Y.; Liu, Z.; Liu, W.; Jiang, L.; Zhang, R.; Cui, D.; Zhang, Q.; Xu, S. Elevation of peripheral bdnf promoter methylation predicts conversion from amnestic mild cognitive impairment to alzheimer’s disease: A 5-year longitudinal study. J Alz.Heimers Dis. 2017, 56, 391–401. [Google Scholar] [CrossRef]
- Nagata, T.; Kobayashi, N.; Ishii, J.; Shinagawa, S.; Nakayama, R.; Shibata, N.; Kuerban, B.; Ohnuma, T.; Kondo, K.; Arai, H.; et al. Association between DNA methylation of the bdnf promoter region and clinical presentation in alzheimer’s disease. Dement. Geriatr. Cogn. Dis. Extra 2015, 5, 64–73. [Google Scholar] [CrossRef]
- Carboni, L.; Lattanzio, F.; Candeletti, S.; Porcellini, E.; Raschi, E.; Licastro, F.; Romualdi, P. Peripheral leukocyte expression of the potential biomarker proteins bdnf, sirt1, and psen1 is not regulated by promoter methylation in alzheimer’s disease patients. Neurosci. Lett 2015, 605, 44–48. [Google Scholar] [CrossRef]
- Angeloni, A.; Bogdanovic, O. Sequence determinants, function, and evolution of cpg islands. Biochem. Soc. Trans. 2021, 49, 1109–1119. [Google Scholar] [CrossRef]
| Amplicon | Position (Reference Genome Build hg19, Strand) | Target Amplicon Length (bp) * | Number of Interrogated CpGs |
|---|---|---|---|
| COMT_1 | chr22:19951071-19951343 | 273 | 14 |
| COMT_2 | chr22:19929042-19929349 | 308 | 36 |
| COMT_4 | chr22:19950002-19950320 | 319 | 13 |
| BDNF_1 | chr11:27744260-27744605 (-) | 346 | 22 |
| BDNF_2 | chr11:27743702-27743960 (-) | 259 | 10 |
| BDNF_3 | chr11:27743454-27743762 (-) | 309 | 20 |
| BDNF_4 | chr11:27741988-27742250 (-) | 263 | 13 |
| BDNF_5 | chr11:27740916-27741131 (-) | 216 | 16 |
| BDNF_6 | chr11:27740607-27740901 (-) | 295 | 30 |
| BDNF_7 | chr11:27721638-27721854 (-) | 217 | 19 |
| BDNF_8 | chr11:27722466-27722696 (-) | 231 | 13 |
| BDNF_9 | chr11:27722209-27722487 (-) | 279 | 23 |
| Characteristics | Participants | Mann–Whitney U Test | ||
|---|---|---|---|---|
| MCI | AD | U | p | |
| Age (years) | 71.0 (57.0–87.0) | 79.0 (63.0–89.0) | 5111.5 | <0.001 |
| BMI (kg/m2) | 22.0 (18.4–32.4) | 22.9 (18.5–31.9) | 3355.5 | 0.550 |
| Waist circumference (cm) | 86.0 (71.0–101.0) | 86.0 (72.0–99.0) | 3173.0 | 0.975 |
| Total cholesterol (mmol/L) | 5.7 (3.2–8.8) | 5.6 (3.2–8.8) | 2936.5 | 0.400 |
| HDL cholesterol (mmol/L) | 1.3 (0.7–3.0) | 1.3 (0.7–3.0) | 3012.5 | 0.560 |
| LDL cholesterol (mmol/L) | 3.5 (0.8–5.6) | 3.2 (0.8–5.8) | 3048.5 | 0.647 |
| Triglycerides | 1.8 (0.7–6.7) | 1.7 (0.7–6.7) | 3048.0 | 0.645 |
| Blood glucose (mmol/L) | 5.5 (4.5–11.8) | 5.6 (4.7–11.8) | 3415.0 | 0.423 |
| MMSE score | 27.0 (21.0–28.0) | 13.0 (10.0–24.0) | 27.5 | <0.001 |
| CDT score | 5.0 (1.0–5.0) | 2.0 (1.0–5.0) | 187.0 | <0.001 |
| Gene | Amplicon Name | Number of DMCs/Number of Investigated CpGs | DNAm % ADs | DNAm % MCIs | DNAm % Difference | Mann–Whitney Test | |
|---|---|---|---|---|---|---|---|
| U | p-Value | ||||||
| BDNF | BDNF_1 | 2/22 | 5.095 | 4.549 | −0.546 | 196 | 0.5499 |
| BDNF_2 | 2/10 | 6.635 | 5.912 | −0.723 | 36 | 0.3150 | |
| BDNF_3 | 3/20 | 3.552 | 3.989 | 0.436 | 186 | 0.3963 | |
| BDNF_4 | 0/13 | 11.39 | 11.36 | −0.036 | 81 | 0.8798 | |
| BDNF_5 | 1/16 | 7.019 | 6.803 | −0.216 | 143 | 0.9729 | |
| BDNF_6 | 0/30 | 6.654 | 6.957 | 0.302 | 473 | 0.9221 | |
| BDNF_7 | 2/19 | 1.938 | 1.915 | −0.023 | 167 | 0.7075 | |
| BDNF_8 | 0/13 | 1.242 | 1.238 | −0.004 | 75 | 0.6498 | |
| BDNF_9 | 8/23 | 0.855 | 0.990 | 0.135 | 202 | 0.0778 | |
| COMT | COMT_1 | 0/14 | 96.00 | 96.02 | 0.017 | 111 | 0.9674 |
| COMT_2 | 0/36 | 0.1725 | 0.1730 | 0.001 | 617 | 0.7328 | |
| COMT_4 | 2/13 | 95.99 | 95.78 | −0.210 | 56 | 0.7969 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouter, K.; Nikolac Perkovic, M.; Nedic Erjavec, G.; Milos, T.; Tudor, L.; Uzun, S.; Mimica, N.; Pivac, N.; Videtic Paska, A. Difference in Methylation and Expression of Brain-Derived Neurotrophic Factor in Alzheimer’s Disease and Mild Cognitive Impairment. Biomedicines 2023, 11, 235. https://doi.org/10.3390/biomedicines11020235
Kouter K, Nikolac Perkovic M, Nedic Erjavec G, Milos T, Tudor L, Uzun S, Mimica N, Pivac N, Videtic Paska A. Difference in Methylation and Expression of Brain-Derived Neurotrophic Factor in Alzheimer’s Disease and Mild Cognitive Impairment. Biomedicines. 2023; 11(2):235. https://doi.org/10.3390/biomedicines11020235
Chicago/Turabian StyleKouter, Katarina, Matea Nikolac Perkovic, Gordana Nedic Erjavec, Tina Milos, Lucija Tudor, Suzana Uzun, Ninoslav Mimica, Nela Pivac, and Alja Videtic Paska. 2023. "Difference in Methylation and Expression of Brain-Derived Neurotrophic Factor in Alzheimer’s Disease and Mild Cognitive Impairment" Biomedicines 11, no. 2: 235. https://doi.org/10.3390/biomedicines11020235
APA StyleKouter, K., Nikolac Perkovic, M., Nedic Erjavec, G., Milos, T., Tudor, L., Uzun, S., Mimica, N., Pivac, N., & Videtic Paska, A. (2023). Difference in Methylation and Expression of Brain-Derived Neurotrophic Factor in Alzheimer’s Disease and Mild Cognitive Impairment. Biomedicines, 11(2), 235. https://doi.org/10.3390/biomedicines11020235







