Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Project Approval
2.2. Original Study Approval and Sample Collection
2.3. Original Sample Processing and Storage
2.4. Current Study Protocol
2.5. Growth Factor Administration
2.6. Viability and Proliferation Assays
2.7. RNA Isolation
2.8. cDNA Synthesis
2.9. microRNA Processing
2.10. qPCR Screening
2.11. Statistical Analysis
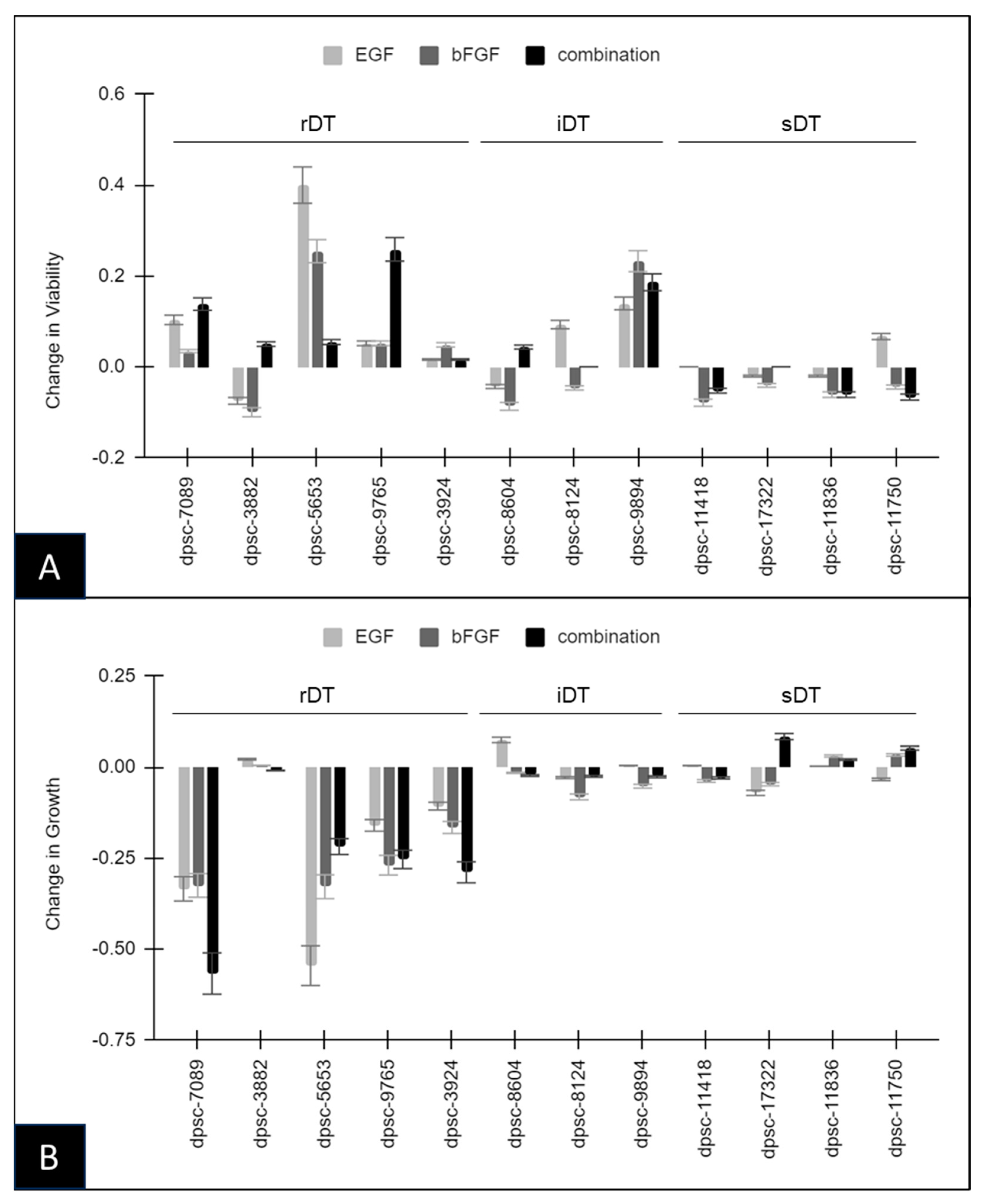
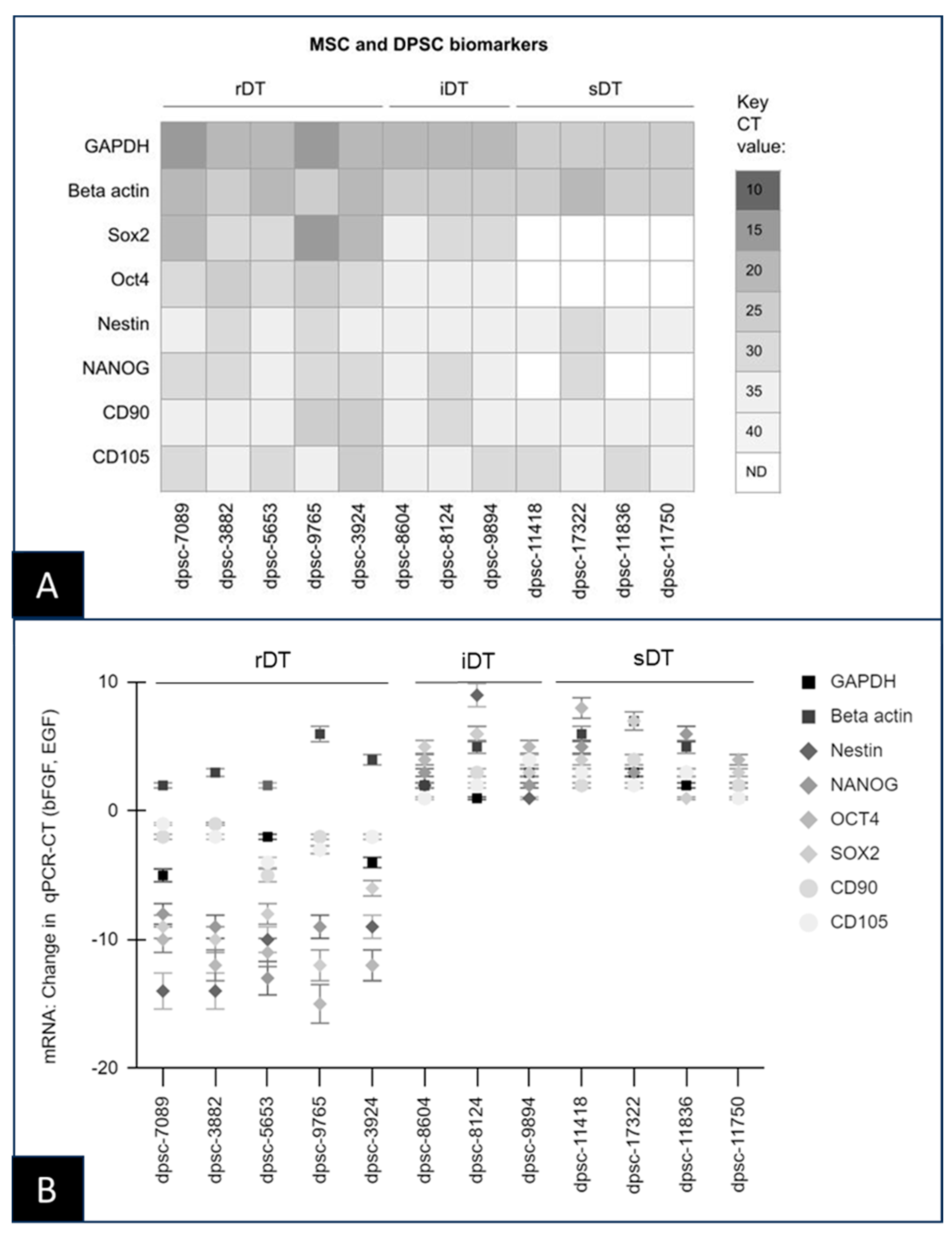
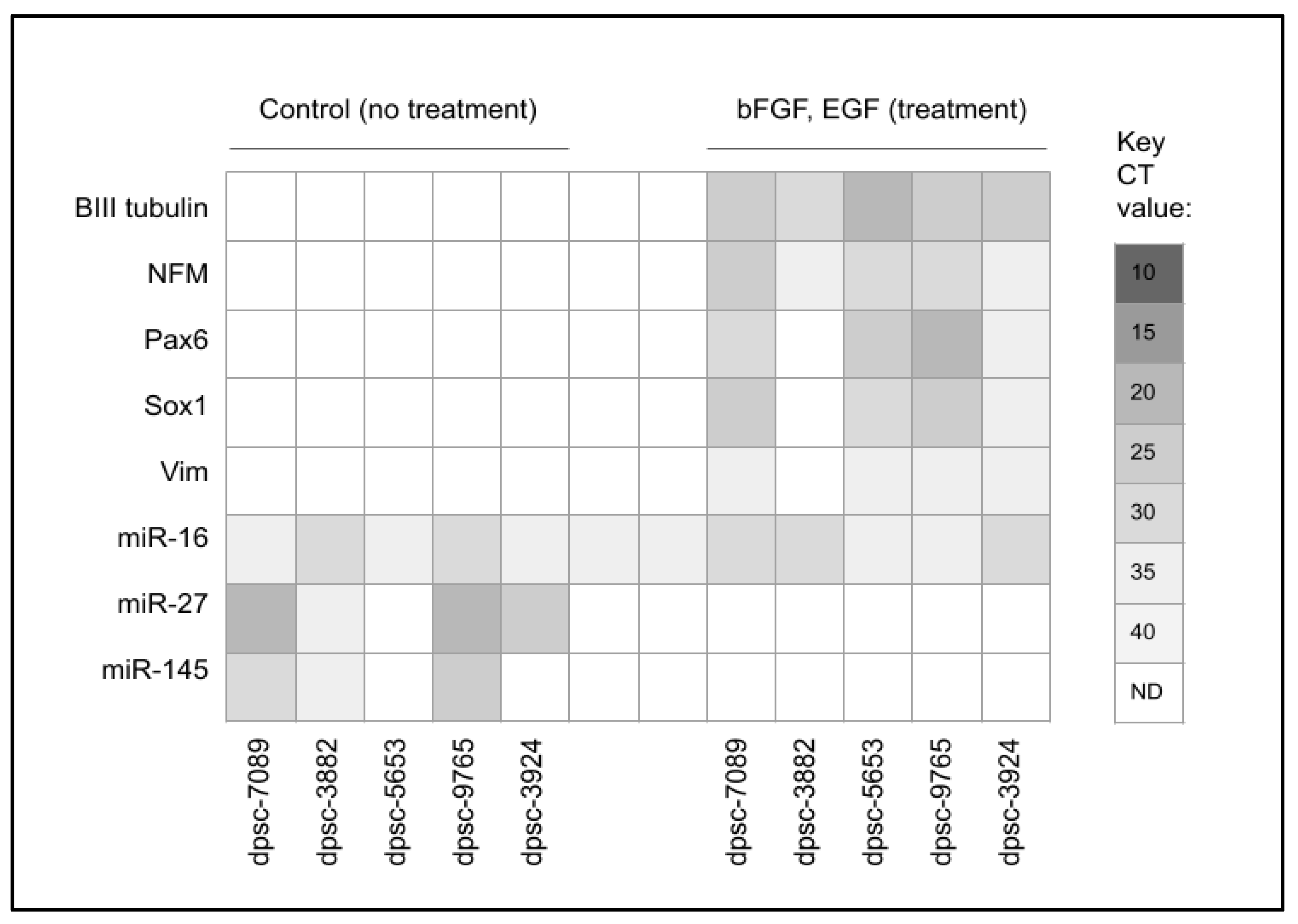
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabaña-Muñoz, M.E.; Fernández, M.J.P.; Parmigiani-Cabaña, J.M.; Parmigiani-Izquierdo, J.M.; Merino, J.J. Adult Mesenchymal Stem Cells from Oral Cavity and Surrounding Areas: Types and Biomedical Applications. Pharmaceutics 2023, 15, 2109. [Google Scholar] [CrossRef] [PubMed]
- Thalakiriyawa, D.S.; Dissanayaka, W.L. Advances in Regenerative Dentistry Approaches: An Update. Int. Dent. J. 2023; Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ostrovidov, S.; Ramalingam, M.; Bae, H.; Orive, G.; Fujie, T.; Shi, X.; Kaji, H. Bioprinting and biomaterials for dental alveolar tissue regeneration. Front. Bioeng. Biotechnol. 2023, 11, 991821. [Google Scholar] [CrossRef] [PubMed]
- Morsczeck, C. Dental stem cells for tooth regeneration: How far have we come and where next? Expert Opin. Biol. Ther. 2023, 23, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Piglionico, S.S.; Pons, C.; Romieu, O.; Cuisinier, F.; Levallois, B.; Panayotov, I.V. In vitro, ex vivo, and in vivo models for dental pulp regeneration. J. Mater. Sci. Mater. Med. 2023, 34, 15. [Google Scholar] [CrossRef] [PubMed]
- EzEldeen, M.; Moroni, L.; Nejad, Z.M.; Jacobs, R.; Mota, C. Biofabrication of engineered dento-alveolar tissue. Mater. Sci. Eng. C 2023, 148, 213371. [Google Scholar] [CrossRef]
- Daltoé, F.P.; Mendonça, P.P.; Mantesso, A.; Deboni, M.C.Z. Can SHED or DPSCs be used to repair/regenerate non-dental tissues? A systematic review of in vivo studies. Braz. Oral Res. 2014, 28, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Mao, J.; Liu, Y. Pulp stem cells derived from human permanent and deciduous teeth: Biological characteristics and therapeutic applications. Stem Cells Transl. Med. 2020, 9, 445–464. [Google Scholar] [CrossRef]
- Namjoynik, A.; Islam, A.; Islam, M. Evaluating the efficacy of human dental pulp stem cells and scaffold combination for bone regeneration in animal models: A systematic review and meta-analysis. Stem Cell Res. Ther. 2023, 14, 132. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, Y.-H.; Zhao, Y.-Q.; Gao, Z.-R.; Ouyang, Z.-Y.; Ye, Q.; Liu, Q.; Chen, Y.; Tan, L.; Zhang, S.-H.; et al. Oral cavity-derived stem cells and preclinical models of jaw-bone defects for bone tissue engineering. Stem Cell Res. Ther. 2023, 14, 39. [Google Scholar] [CrossRef]
- Mattei, V.; Martellucci, S.; Pulcini, F.; Santilli, F.; Sorice, M.; Monache, S.D. Regenerative Potential of DPSCs and Revascularization: Direct, Paracrine or Autocrine Effect? Stem Cell Rev. Rep. 2021, 17, 1635–1646. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Zou, T.; Qi, Y.; Yi, B.; Dissanayaka, W.L.; Zhang, C. DPSCs treated by TGF-β1 regulate angiogenic sprouting of three-dimensionally co-cultured HUVECs and DPSCs through VEGF-Ang-Tie2 signaling. Stem Cell Res. Ther. 2021, 12, 281. [Google Scholar] [CrossRef] [PubMed]
- Victor, A.K.; Reiter, L.T. Dental pulp stem cells for the study of neurogenetic disorders. Hum. Mol. Genet. 2017, 26, R166–R171. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, S.; Venugopal, C.; Parveen, S.; Shobha, K.; Rai, K.S.; Kutty, B.M.; Dhanushkodi, A. Remarkable migration propensity of dental pulp stem cells towards neurodegenerative milieu: An in vitro analysis. NeuroToxicology 2020, 81, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.E.-M.B.; Murakami, M.; Hirose, Y.; Nakashima, M. Therapeutic Potential of Dental Pulp Stem Cell Secretome for Alzheimer’s Disease Treatment: An In Vitro Study. Stem Cells Int. 2016, 2016, 8102478. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Shobha, K.; Sundeep, M.; Pinnelli, V.B.; Parveen, S.; Dhanushkodi, A. Neural Basis of Dental Pulp Stem Cells and its Potential Application in Parkinson’s Disease. CNS Neurol. Disord. Drug Targets 2022, 21, 62–76. [Google Scholar] [CrossRef]
- Goorha, S.; Victor, A.K.; Reiter, L.T. Culturing and Neuronal Differentiation of Human Dental Pulp Stem Cells. Curr. Protoc. 2022, 2, e600. [Google Scholar] [CrossRef]
- Rafiee, F.; Pourteymourfard-Tabrizi, Z.; Mahmoudian-Sani, M.-R.; Mehri-Ghahfarrokhi, A.; Soltani, A.; Hashemzadeh-Chaleshtori, M.; Jami, M.-S. Differentiation of dental pulp stem cells into neuron-like cells. Int. J. Neurosci. 2020, 130, 107–116. [Google Scholar] [CrossRef]
- Madanagopal, T.T.; Franco-Obregón, A.; Rosa, V. Comparative study of xeno-free induction protocols for neural differentiation of human dental pulp stem cells in vitro. Arch. Oral Biol. 2020, 109, 104572. [Google Scholar] [CrossRef]
- Al-Maswary, A.A.; O’reilly, M.; Holmes, A.P.; Walmsley, A.D.; Cooper, P.R.; Scheven, B.A. Exploring the neurogenic differentiation of human dental pulp stem cells. PLoS ONE 2022, 17, e0277134. [Google Scholar] [CrossRef]
- Srikawnawan, W.; Songsaad, A.; Gonmanee, T.; Thonabulsombat, C.; Phruksaniyom, C.; White, K.L.; Ruangsawasdi, N. Rho kinase inhibitor induced human dental pulp stem cells to differentiate into neurons. Life Sci. 2022, 300, 120566. [Google Scholar] [CrossRef]
- Soheilifar, M.H.; Nobari, S.; Hakimi, M.; Adel, B.; Masoudi-Khoram, N.; Reyhani, E.; Neghab, H.K. Current concepts of microRNA-mediated regulatory mechanisms in human pulp tissue-derived stem cells: A snapshot in the regenerative dentistry. Cell Tissue Res. 2023, 393, 229–251. [Google Scholar] [CrossRef] [PubMed]
- Kearney, M.; Duncan, H.F. Micro-RNA Profiling in Dental Pulp Cell Cultures. Methods Mol. Biol. 2023, 2588, 353–367. [Google Scholar]
- Ma, X.; Liu, H.; Zheng, Y.; Dai, Y.; Lingling, E.; Zhang, R.; Zhang, S. Genome-Wide Screening of Differentially Expressed Genes and their Potential Associations with Aging Dental Pulp Stem Cells. Comb. Chem. High Throughput Screen. 2023, 26, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu, S.; Dao, J.; Gan, Z.; Zeng, X. Differential expression of lncRNA/miRNA/mRNA and their related functional networks during the osteogenic/odontogenic differentiation of dental pulp stem cells. J. Cell. Physiol. 2020, 235, 3350–3361. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Chu, M.; Zheng, K.; He, P.; Xiao, J. miR-153-3p inhibited osteogenic differentiation of human DPSCs through CBFβ signaling. In Vitro Cell. Dev. Biol. Anim. 2022, 58, 316–324. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, N.; Li, L.; Ge, L.; Jia, H.; Fan, Z. miR-140-3p enhanced the osteo/odontogenic differentiation of DPSCs via inhibiting KMT5B under hypoxia condition. Int. J. Oral Sci. 2021, 13, 41. [Google Scholar] [CrossRef]
- Yang, C.; Xu, X.; Lin, P.; Luo, B.; Luo, S.; Huang, H.; Zhu, J.; Huang, M.; Peng, S.; Wu, Q.; et al. Overexpression of long noncoding RNA MCM3AP-AS1 promotes osteogenic differentiation of dental pulp stem cells via miR-143-3p/IGFBP5 axis. Hum. Cell 2022, 35, 150–162. [Google Scholar] [CrossRef]
- Xie, L.; Guan, Z.; Zhang, M.; Lyu, S.; Thuaksuban, N.; Kamolmattayakul, S.; Nuntanaranont, T. Exosomal circLPAR1 Promoted Osteogenic Differentiation of Homotypic Dental Pulp Stem Cells by Competitively Binding to hsa-miR-31. Biomed. Res. Int. 2020, 2020, 6319395. [Google Scholar] [CrossRef]
- Wu, Y.; Lian, K.; Sun, C. LncRNA LEF1-AS1 promotes osteogenic differentiation of dental pulp stem cells via sponging miR-24-3p. Mol. Cell. Biochem. 2020, 475, 161–169. [Google Scholar] [CrossRef]
- Ji, F.; Zhu, L.; Pan, J.; Shen, Z.; Yang, Z.; Wang, J.; Bai, X.; Lin, Y.; Tao, J. hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front. Cell Dev. Biol. 2020, 8, 470. [Google Scholar] [CrossRef]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. miR-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, R.; Wei, G.; Dai, S.; Zhang, X.; Yang, W.; Li, X.; Bai, C. Long Non-coding RNA Maternally Expressed 3 Increases the Expression of Neuron-Specific Genes by Targeting miR-128-3p in All-Trans Retinoic Acid-Induced Neurogenic Differentiation from Amniotic Epithelial Cells. Front. Cell Dev. Biol. 2019, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Pons-Espinal, M.; Gasperini, C.; Marzi, M.J.; Braccia, C.; Armirotti, A.; Pötzsch, A.; Walker, T.L.; Fabel, K.; Nicassio, F.; Kempermann, G.; et al. MiR-135a-5p Is Critical for Exercise-Induced Adult Neurogenesis. Stem Cell Rep. 2019, 12, 1298–1312. [Google Scholar] [CrossRef]
- Ding, K.; Lai, Z.; Yang, G.; Zeng, L. MiR-140-5p targets Prox1 to regulate the proliferation and differentiation of neural stem cells through the ERK/MAPK signaling pathway. Ann. Transl. Med. 2021, 9, 671. [Google Scholar] [CrossRef]
- Jauhari, A.; Singh, T.; Yadav, S. Expression of miR-145 and Its Target Proteins Are Regulated by miR-29b in Differentiated Neurons. Mol. Neurobiol. 2018, 55, 8978–8990. [Google Scholar] [CrossRef] [PubMed]
- Khalil, W.; Tiraihi, T.; Soleimani, M.; Baheiraei, N.; Zibara, K. Conversion of Neural Stem Cells into Functional Neuron-Like Cells by MicroRNA-218: Differential Expression of Functionality Genes. Neurotox. Res. 2020, 38, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Tsan, Y.-C.; Morell, M.H.; O’Shea, K.S. miR-410 controls adult SVZ neurogenesis by targeting neurogenic genes. Stem Cell Res. 2016, 17, 238–247. [Google Scholar] [CrossRef]
- Lott, K.; Collier, P.; Ringor, M.; Howard, K.M.; Kingsley, K. Administration of Epidermal Growth Factor (EGF) and Basic Fibroblast Growth Factor (bFGF) to Induce Neural Differentiation of Dental Pulp Stem Cells (DPSC) Isolates. Biomedicines 2023, 11, 255. [Google Scholar] [CrossRef]
- Lott, K.; Collier, P.; Ringor, M.; Taylor, A.; Truman, B.; Howard, K.M.; Kingsley, K. Assessment and Analysis of Dental Pulp Stem Cells (DPSCs) Biomarkers and Viability Following Cryopreservation Reveals Novel Association with miR-218 Expression. EC Dent. Sci. 2022, 21, 115–128. [Google Scholar]
- Viss, C.; Banning, G.; Swanbeck, S.; Kingsley, K. Differential Viability in Alpha-MEM Culturing Media May Predict Alternative Media Responsiveness in Dental Pulp Stem Cell (DPSC). J. Adv. Biol. Biotechnol. 2022, 25, 1–13. [Google Scholar] [CrossRef]
- Zheng, K.; Feng, G.; Zhang, J.; Xing, J.; Huang, D.; Lian, M.; Zhang, W.; Wu, W.; Hu, Y.; Lu, X.; et al. Basic fibroblast growth factor promotes human dental pulp stem cells cultured in 3D porous chitosan scaffolds to neural differentiation. Int. J. Neurosci. 2021, 131, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Monache, S.D.; Pulcini, F.; Santilli, F.; Martellucci, S.; Santacroce, C.; Fabrizi, J.; Angelucci, A.; Sorice, M.; Mattei, V. Hypoxia Induces DPSC Differentiation versus a Neurogenic Phenotype by the Paracrine Mechanism. Biomedicines 2022, 10, 1056. [Google Scholar] [CrossRef]
- Gaus, S.; Li, H.; Li, S.; Wang, Q.; Kottek, T.; Hahnel, S.; Liu, X.; Deng, Y.; Ziebolz, D.; Haak, R.; et al. Shared Genetic and Epigenetic Mechanisms between the Osteogenic Differentiation of Dental Pulp Stem Cells and Bone Marrow Stem Cells. BioMed Res. Int. 2021, 2021, 6697810. [Google Scholar] [CrossRef] [PubMed]
- Konar, E.; Khatami, S.R.; Pezeshki, S.P.; Shafiei, M.; Hajjari, M.R. The effect of PRP and hyperosmolarity simultaneous use on expression profile alteration of miRNAs associated with cartilage differentiation in human adipose tissue-derived mesenchymal stem cells. Gene 2023, 859, 147188. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-D.; Goto, S.; Hsu, L.-W.; Lin, T.-Y.; Nakano, T.; Lai, C.-Y.; Chang, Y.-C.; Weng, W.-T.; Kuo, Y.-R.; Wang, C.-C.; et al. Identification of miR-27b as a novel signature from the mRNA profiles of adipose-derived mesenchymal stem cells involved in the tolerogenic response. PLoS ONE 2013, 8, e60492. [Google Scholar] [CrossRef] [PubMed]
- Kara, N.; Wei, C.; Commanday, A.C.; Patton, J.G. miR-27 regulates chondrogenesis by suppressing focal adhesion kinase during pharyngeal arch development. Dev. Biol. 2017, 429, 321–334. [Google Scholar] [CrossRef]
- Li, C.-J.; Hong, T.; Tung, Y.-T.; Yen, Y.-P.; Hsu, H.-C.; Lu, Y.-L.; Chang, M.; Nie, Q.; Chen, J.-A. MicroRNA filters Hox temporal transcription noise to confer boundary formation in the spinal cord. Nat. Commun. 2017, 8, 14685. [Google Scholar] [CrossRef]
- Verbus, E.A.; Kenyon, J.D.; Sergeeva, O.; Awadallah, A.; Yuan, L.; Welter, J.F.; Caplan, A.I.; Schluchter, M.D.; Khalil, A.M.; Lee, Z. Expression of miR-145-5p During Chondrogenesis of Mesenchymal Stem Cells. J. Stem Cell Res. 2017, 1, 1–10. [Google Scholar] [CrossRef]
- Kenyon, J.D.; Sergeeva, O.; Somoza, R.A.; Li, M.; Caplan, A.I.; Khalil, A.M.; Lee, Z. Analysis of -5p and -3p Strands of miR-145 and miR-140 During Mesenchymal Stem Cell Chondrogenic Differentiation. Tissue Eng. Part A 2019, 25, 80–90. [Google Scholar] [CrossRef]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Yamahara, K.; Homma, K.; Suzuki, S.; Fujii, S.; Morizane, R.; Monkawa, T.; Matsuzaki, Y.; Kangawa, K.; Itoh, H. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis 2011, 219, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, C. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014, 23, 2851–2861. [Google Scholar] [CrossRef] [PubMed]
- Gunaratne, P.H. Embryonic stem cell microRNAs: Defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cell Res Ther. 2009, 4, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Morgado, A.L.; Rodrigues, C.M.; Solá, S. MicroRNA-145 Regulates Neural Stem Cell Differentiation Through the Sox2-Lin28/let-7 Signaling Pathway. Stem Cells 2016, 34, 1386–1395. [Google Scholar] [CrossRef]
- Cao, L.-L.; Zhang, Y.-J.; Wang, J.-W.; Tian, F.; Wang, C.-F. Studies on microRNA regulation of multidirectional differentiation of dental pulp stem cells: A narrative review. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 1816–1824. [Google Scholar] [CrossRef]
- Åkerblom, M.; Petri, R.; Sachdeva, R.; Klussendorf, T.; Mattsson, B.; Gentner, B.; Jakobsson, J. microRNA-125 distinguishes developmentally generated and adult-born olfactory bulb interneurons. Development 2014, 141, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Gräfe, A.; Seiler, A.; Schumacher, S.; Nitsch, R.; Wulczyn, F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005, 21, 1469–1477. [Google Scholar] [CrossRef]
- Santos, M.C.T.; Tegge, A.N.; Correa, B.R.; Mahesula, S.; Kohnke, L.Q.; Qiao, M.; Ferreira, M.A.R.; Kokovay, E.; Penalva, L.O.F. miR-124, -128, and -137 Orchestrate Neural Differentiation by Acting on Overlapping Gene Sets Containing a Highly Connected Transcription Factor Network. Stem Cells 2016, 34, 220–232. [Google Scholar] [CrossRef]
- Hu, F.; Sun, B.; Xu, P.; Zhu, Y.; Meng, X.-H.; Teng, G.-J.; Xiao, Z.-D. MiR-218 Induces Neuronal Differentiation of ASCs in a Temporally Sequential Manner with Fibroblast Growth Factor by Regulation of the Wnt Signaling Pathway. Sci. Rep. 2017, 7, 39427. [Google Scholar] [CrossRef]
- Hu, F.; Zhang, X.; Liu, H.; Xu, P.; Doulathunnisa; Teng, G.; Xiao, Z. Neuronally differentiated adipose-derived stem cells and aligned PHBV nanofiber nerve scaffolds promote sciatic nerve regeneration. Biochem. Biophys. Res. Commun. 2017, 489, 171–178. [Google Scholar] [CrossRef]
- Raik, S.; Kumar, A.; Rattan, V.; Seth, S.; Kaur, A.; Charyya, S.B. Assessment of Post-thaw Quality of Dental Mesenchymal Stromal Cells after Long-Term Cryopreservation by Uncontrolled Freezing. Appl. Biochem. Biotechnol. 2020, 191, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Conde, M.C.M.; Chisini, L.A.; Grazioli, G.; Francia, A.; De Carvalho, R.V.; Alcázar, J.C.B.; Tarquinio, S.B.C.; Demarco, F.F. Does Cryopreservation Affect the Biological Properties of Stem Cells from Dental Tissues? A Systematic Review. Braz. Dent. J. 2016, 27, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Hollands, P.; Aboyeji, D.; Orcharton, M. Dental pulp stem cells in regenerative medicine. Br. Dent. J. 2018, 224, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Ohshima, H. Biological characteristics of dental pulp stem cells and their potential use in regenerative medicine. J. Oral Biosci. 2022, 64, 26–36. [Google Scholar] [CrossRef]
- Zha, K.; Yang, Y.; Tian, G.; Sun, Z.; Yang, Z.; Li, X.; Sui, X.; Liu, S.; Zhao, J.; Guo, Q. Nerve growth factor (NGF) and NGF receptors in mesenchymal stem/stromal cells: Impact on potential therapies. Stem Cells Transl. Med. 2021, 10, 1008–1020. [Google Scholar] [CrossRef]
- Morys, J.; Borkowska, P.; Zielinska, A.; Kowalski, J. Study of the influence of NGF-β gene overexpression in human mesenchymal stem cells on the expression level of SOX1 and neural pathway genes. Mol. Biol. Rep. 2022, 49, 4435–4441. [Google Scholar] [CrossRef]
- Huat, T.J.; Khan, A.A.; Abdullah, J.M.; Idris, F.M.; Jaafar, H. MicroRNA expression profile of bone marrow mesenchymal stem cell-derived neural progenitor by microarray under the influence of EGF, bFGF and IGF-1. Genom. Data 2015, 5, 201–205. [Google Scholar] [CrossRef]
| Primers | Sequence |
|---|---|
| Positive control primers | |
| Forward primer, Beta actin | 5′-GTG GGG TCC TGT GGT GTG-3′ |
| Reverse primer, Beta actin | 5′-GAA GGG GAC AGG CAG TGA-3′ |
| Forward primer, GAPDH | 5′-ATC TTC CAG GAG CGA GAT CC-3′ |
| Reverse primer, GAPDH | 5′-ACC ACT GAC ACG TTG GCA GT-3′ |
| MSC primers | |
| Forward primer, Nestin | 5′-CGT TGG AAC AGA GGT TGG AG-3′ |
| Reverse primer, Nestin | 5′-TCC TGA AAG CTG AGG GAA G-3′ |
| Forward primer, NANOG | 5′-GCT GAG ATG CCT CAC ACG GAG-3′ |
| Reverse primer, NANOG | 5′-TCT GTT TCT TGA CTG GGA CCT TGT C-3′ |
| Forward primer, Oct4 | 5′-TGG AGA AGG AGA AGC TGG AGC AAA A-3′ |
| Reverse primer, Oct4 | 5′-GGC AGA TGG TCG TTT GGC TGA ATA-3′ |
| Forward primer, Sox2 | 5′-ATG GGC TCT GTG GTC AAG TC-3′ |
| Reverse primer, Sox2 | 5′-CCC TCC CAA TTC CCT TGT AT-3′ |
| ISCT control primers | |
| Forward primer, CD45 | 5′-CAT ATT TAT TTT GTC CTT CTC CCA-3′ |
| Reverse primer, CD45 | 5′-GAA AGT TTC CAC GAA CGG-3′ |
| Forward primer, CD90 | 5′-ATG AAC CTG GCC ATC AGC A-3′ |
| Reverse primer, CD90 | 5′-GTG TGC TCA GGC ACC CC-3′ |
| Forward primer, CD105 | 5′-CCA CTA GCC AGG TCT CGA AG-3′ |
| Reverse primer, CD105 | 5′-GAT GCA GGA AGA CAC TGC TG-3′ |
| Neuronal differentiation primers | |
| Forward primer, BIII tubulin | 5′-GGC CAA GGG TCA CTA CAC G-3′ |
| Reverse primer, BIII tubulin | 5′-GCA GTC GCA GTT TTC ACA CTC-3′ |
| Forward primer, NFM | 5′-GCT CGT CAT TTG CGC GAA TAC-3′ |
| Reverse primer, NFM | 5′-TTT CTG TAC GCA GCG ATT TCT AT-3′ |
| Forward primer, Pax6 | 5′-TGG GCA GGT ATT ACG AGA CTG-3′ |
| Reverse primer, Pax6 | 5′-ACT CCC GCT TAT ACT GGG CTA-3′ |
| Forward primer, Sox1 | 5′-CAG TAC AGC CCC ATC TCC AAC-3′ |
| Reverse primer, Sox1 | 5′-GCG GGC AAG TAC ATG CTG A-3′ |
| Forward primer, Vim | 5′-GAC GCC ATC AAC ACC GAG TT-3′ |
| Reverse primer, Vim | 5′-CTT TGT CGT TGG TTA GCT GGT-3′ |
| microRNA primers | |
| Forward primer, miR-16 | 5′-TAG CAG CAC GTA AAT ATT GGC G-3′ |
| Reverse primer, miR-16 | 5′-TGC GTG TCG TGG AGT C-3′ |
| Forward primer, miR-27 | 5′-ATA TGA GAA AAG AGC TTC CCT GTG-3′ |
| Reverse primer, miR-27 | 5′-CAA GGC CAG AGG AGG TGA G-’3′ |
| Forward primer, miR-125 | 5′-GCC CTC CCT GAG ACC TCA A-3′ |
| Reverse primer, miR-125 | 5′-GTG CAG GGT CCG AGG T-3′ |
| Forward primer, miR-128 | 5′-TCT CCT AAA GAG CCC GAA CA-3′ |
| Reverse primer, miR-128 | 5′-TTG CAT TCA TAG CTG CAT CC-3′ |
| Forward primer, miR-135 | 5′-CGA TAT GGC TTT TTA TTC CTA -3′ |
| Reverse primer, miR-135 | 5′-GAG CAG GGT CCG AGG T -3′ |
| Forward primer, miR-140 | 5′-GGG CAG TGG TTT TAC CCT A -3′ |
| Reverse primer, miR-140 | 5′-CAG TGC GTG TCG TGG AGT -3′ |
| Forward primer, miR-145 | 5′-AGA GAA CTC CAG CTG-3′ |
| Reverse primer, miR-145 | 5′-GGC AAC TGT GGG GTG-3′ |
| Forward primer, miR-218 | 5′-TCG GGC TTG TGC TTG ATC T-3′ |
| Reverse primer, miR-218 | 5′-GTG CAG GGT CCG AGT G-3′ |
| Forward primer, miR-410 | 5′-CCG CAC GAT ATA ACA CAG ATG-3′ |
| Reverse primer, miR-410 | 5′-GTG CAG GGT CCG AGG TAT TC-3′ |
| Rapid Doubling Time (rDT) | Intermediate Doubling Time (iDT) | Slow Doubling Time (sDT) | Viability (Baseline) | |
|---|---|---|---|---|
| dpsc-7089 | 1.9 days | 62% | ||
| dpsc-3882 | 1.8 days | 58% | ||
| dpsc-5653 | 1.9 days | 77% | ||
| dpsc-9765 | 2.1 days | 69% | ||
| dpsc-3924 | 2.2 days | 66% | ||
| rDT average | 1.98 days | 66.40% | ||
| dpsc-8604 | 5.1 days | 50% | ||
| dpsc-8124 | 5.4 days | 53% | ||
| dpsc-9894 | 5.2 days | 66% | ||
| iDT average | 5.23 days | 56.30% | ||
| dpsc-11418 | 10.2 days | 63% | ||
| dpsc-17322 | 10.6 days | 56% | ||
| dpsc-11836 | 12.1 days | 57% | ||
| dpsc-11750 | 11.9 days | 62% | ||
| sDT average | 11.2 days | 59.50% |
| RNA Concentration | RNA Purity (A260:A280 Ratio) | cDNA Concentration | DNA Purity (A260:A280 Ratio) | |
|---|---|---|---|---|
| dpsc-7089 | 481 ng/µL | 1.88 | 1541 ng/µL | 1.89 |
| dpsc-3882 | 357 ng/µL | 1.67 | 1480 ng/µL | 1.92 |
| dpsc-5653 | 443 ng/µL | 1.82 | 1606 ng/µL | 1.82 |
| dpsc-9765 | 432 ng/µL | 1.73 | 1587 ng/µL | 1.9 |
| dpsc-3924 | 320 ng/µL | 1.82 | 1527 ng/µL | 1.92 |
| rDT average | 406.6 ng/µL | 1.78 | 1548.2 ng/µL | 1.89 |
| dpsc-8604 | 447 ng/µL | 1.84 | 1589 ng/µL | 1.8 |
| dpsc-8124 | 302 ng/µL | 1.66 | 1551 ng/µL | 1.89 |
| dpsc-9894 | 577 ng/µL | 1.85 | 1561 ng/µL | 1.81 |
| iDT average | 442.0 ng/µL | 1.78 | 1567.0 ng/µL | 1.83 |
| dpsc-11418 | 484 ng/µL | 1.77 | 1607 ng/µL | 1.81 |
| dpsc-17322 | 519 ng/µL | 1.78 | 1610 ng/µL | 1.89 |
| dpsc-11836 | 321 ng/µL | 1.86 | 1489 ng/µL | 1.86 |
| dpsc-11750 | 567 ng/µL | 1.85 | 1492 ng/µL | 1.92 |
| sDT average | 472.8 ng/µL | 1.81 | 1549.5 ng/µL | 1.87 |
| ICST Biomarkers | MSC Biomarkers | Neuronal Biomarkers (Post-Treated) | microRNA Expression | |
|---|---|---|---|---|
| rDT dpsc-7089 dpsc-2883 dpsc-5653 dpsc-9765 dpsc-3924 | CD90, CD105 (all rDT) CD45 (no rDT) | NANOG Nestin Oct4 Sox2 (all rDT) | BIII tubulin, NFM (all rDT) Pax, Sox1, Vim (all rDT but dpsc-3882) | miR-27 (all rDT but dpsc-5653) miR-145 (dpsc-7089, dpsc-3882, dpsc-9765) miR-125, miR-128 miR-135 miR-140 miR-218 miR-410 (no rDT) |
| iDT dpsc-8604 dpsc-8124 dpsc-9894 | CD90 (all iDT) CD105 (all iDT) CD45 (no iDT) | NANOG Nestin Oct4 Sox2 (all iDT) | BIII tubulin NFM Pax Sox1 Vim (no iDT) | miR-27 miR-125, miR-128 miR-135 miR-140 miR-145 miR-218 miR-410 (no iDT) |
| sDT dpsc-11418 dpsc-17322 dpsc-11836 dpsc-11750 | CD90 (all sDT) CD105 (all sDT) CD45 (no sDT) | NANOG (dpsc-17322) Nestin (all sDT) Sox2, Oct4 (no sDT) | BIII tubulin NFM Pax Sox1 Vim (no sDT) | miR-27 (all sDT) miR-145 (all sDT) miR-125, miR-128 miR-135 miR-140 miR-218 miR-410 (no sDT) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassett, C.; Triplett, H.; Lott, K.; Howard, K.M.; Kingsley, K. Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli. Biomedicines 2023, 11, 3003. https://doi.org/10.3390/biomedicines11113003
Bassett C, Triplett H, Lott K, Howard KM, Kingsley K. Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli. Biomedicines. 2023; 11(11):3003. https://doi.org/10.3390/biomedicines11113003
Chicago/Turabian StyleBassett, Charlton, Hunter Triplett, Keegan Lott, Katherine M. Howard, and Karl Kingsley. 2023. "Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli" Biomedicines 11, no. 11: 3003. https://doi.org/10.3390/biomedicines11113003
APA StyleBassett, C., Triplett, H., Lott, K., Howard, K. M., & Kingsley, K. (2023). Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli. Biomedicines, 11(11), 3003. https://doi.org/10.3390/biomedicines11113003








