MicroRNAs Associated with Disability Progression and Clinical Activity in Multiple Sclerosis Patients Treated with Glatiramer Acetate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. MicroRNAs Selection and Analysis
2.3. Statistics
3. Results
4. Discussion
Limitations Section
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [PubMed]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Massachi, I.; Manickavel, S.; Singh, S.; Rao, N.P.; Hasan, S.; Mc Curdy, D.K.; Sharma, S.; Wong, D.; Hahn, B.H.; et al. The role of miRNA in inflammation and autoimmunity. Autoimmun. Rev. 2013, 12, 1160–1165. [Google Scholar] [CrossRef] [PubMed]
- Duffy, C.P.; McCoy, C.E. The Role of MicroRNAs in Repair Processes in Multiple Sclerosis. Cells 2020, 9, 1711. [Google Scholar] [CrossRef]
- Martinelli-Boneschi, F.; Fenoglio, C.; Brambilla, P.; Sorosina, M.; Giacalone, G.; Esposito, F.; Serpente, M.; Cantoni, C.; Ridolfi, E.; Rodegher, M.; et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci. Lett. 2012, 508, 4–8. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain J. Neurol. 2009, 132, 3342–3352. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.-P.; Ruprecht, K.; Meese, E. Multiple Sclerosis: MicroRNA Expression Profiles Accurately Differentiate Patients with Relapsing-Remitting Disease from Healthy Controls. PLoS ONE 2009, 4, e7440. [Google Scholar] [CrossRef]
- Otaegui, D.; Baranzini, S.E.; Armañanzas, R.; Calvo, B.; Muñoz-Culla, M.; Khankhanian, P.; Inza, I.; Lozano, J.A.; Castillo-Triviño, T.; Asensio, A.; et al. Differential Micro RNA Expression in PBMC from Multiple Sclerosis Patients. PLoS ONE 2009, 4, e6309. [Google Scholar] [CrossRef]
- Mancuso, R.; Hernis, A.; Agostini, S.; Rovaris, M.; Caputo, D.; Clerici, M. MicroRNA-572 expression in multiple sclerosis patients with different patterns of clinical progression. J. Transl. Med. 2015, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Orlandi, E.; Rossetti, G.; Turatti, M.; Calabrese, M.; Gomez Lira, M.; Gajofatto, A. Upregulated serum miR-128-3p in progressive and relapse-free multiple sclerosis patients. Acta Neurol. Scand. 2020, 142, 511–516. [Google Scholar]
- Hemond, C.C.; Healy, B.C.; Tauhid, S.; Mazzola, M.A.; Quintana, F.J.; Gandhi, R.; Weiner, H.L.; Bakshi, R. MRI phenotypes in MS: Longitudinal changes and miRNA signatures. Neurol. Neuroimmunol. Neuroinflammation 2019, 6, e530. [Google Scholar]
- Regev, K.; Healy, B.C.; Khalid, F.; Paul, A.; Chu, R.; Tauhid, S.; Tummala, S.; Diaz-Cruz, C.; Raheja, R.; Mazzola, M.A.; et al. Association Between Serum MicroRNAs and Magnetic Resonance Imaging Measures of Multiple Sclerosis Severity. JAMA Neurol. 2017, 74, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, R.; Healy, B.; Gholipour, T.; Egorova, S.; Musallam, A.; Hussain, M.S.; Nejad, P.; Patel, B.; Hei, H.; Khoury, S.; et al. Circulating MicroRNAs as biomarkers for disease staging in multiple sclerosis. Ann. Neurol. 2013, 73, 729–740. [Google Scholar] [CrossRef]
- Niwald, M.; Migdalska-Sęk, M.; Brzeziańska-Lasota, E.; Miller, E. Evaluation of Selected MicroRNAs Expression in Remission Phase of Multiple Sclerosis and Their Potential Link to Cognition, Depression, and Disability. J. Mol. Neurosci. 2017, 63, 275–282. [Google Scholar] [PubMed]
- Dominguez-Mozo, M.I.; Casanova, I.; De Torres, L.; Aladro-Benito, Y.; Perez-Perez, S.; Garcia-Martínez, A.; Gomez, P.; Abellan, S.; De Antonio, E.; Lopez-De-Silanes, C.; et al. microRNA Expression and Its Association with Disability and Brain Atrophy in Multiple Sclerosis Patients Treated with Glatiramer Acetate. Front. Immunol. 2022, 13, 904683. [Google Scholar] [PubMed]
- Martin, N.A.; Hyrlov, K.H.; Elkjaer, M.L.; Thygesen, E.K.; Wlodarczyk, A.; Elbaek, K.J.; Aboo, C.; Okarmus, J.; Benedikz, E.; Reynolds, R.; et al. Absence of miRNA-146a Differentially Alters Microglia Function and Proteome. Front. Immunol. 2020, 11, 1110. [Google Scholar] [PubMed]
- Quintana, E.; Ortega, F.J.; Robles-Cedeño, R.; Villar, M.L.; Buxó, M.; Mercader, J.M.; Alvarez-Cermeño, J.C.; Pueyo, N.; Perkal, H.; Fernández-Real, J.M.; et al. miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult. Scler. J. 2017, 23, 1716–1726. [Google Scholar] [CrossRef]
- Giuliani, A.; Lattanzi, S.; Ramini, D.; Graciotti, L.; Danni, M.C.; Procopio, A.D.; Silvestrini, M.; Olivieri, F.; Sabbatinelli, J. Potential prognostic value of circulating in-flamma-miR-146a-5p and miR-125a-5p in relapsing-remitting multiple sclerosis. Mult. Scler. Relat. Disord. 2021, 54, 103126. [Google Scholar] [PubMed]
- Saridas, F.; Unlu, H.T.; Cecener, G.; Egeli, U.; Takanlou, M.S.; Takanlou, L.S.; Tunca, B.; Zarifoglu, M.; Turan, O.F.; Taskapilioglu, O. The expression and prognostic value of miR-146a and miR-155 in Turkish patients with multiple sclerosis. Neurol. Res. 2021, 44, 217–223. [Google Scholar] [CrossRef]
- Li, Y.; Du, C.; Wang, W.; Ma, G.; Cui, L.; Zhou, H.; Tao, H.; Yao, L.; Zhao, B.; Li, K. Genetic Association of MiR-146a with Multiple Sclerosis Susceptibility in the Chinese Population. Cell. Physiol. Biochem. 2015, 35, 281–291. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.G.; Lu, M.; Wang, X.; Shang, X.; Elias, S.B.; Chopp, M. MiR-146a promotes remyelination in a cuprizone model of de-myelinating injury. Neuroscience 2017, 348, 252–263. [Google Scholar] [PubMed]
- Quinn, E.M.; Wang, J.H.; O’callaghan, G.; Redmond, H.P. MicroRNA-146a Is Upregulated by and Negatively Regulates TLR2 Signaling. PLoS ONE 2013, 8, e62232. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Esaba, R.; Sorensen, D.L.; Booth, S.A. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef]
- Waschbisch, A.; Atiya, M.; Linker, R.A.; Potapov, S.; Schwab, S.; Derfuss, T. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS ONE 2011, 6, e24604. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.G.; Lu, M.; Zhang, Y.; Shang, X.; Chopp, M. MiR-146a promotes oligodendrocyte progenitor cell differentiation and enhances remyelination in a model of experimental autoimmune encephalomyelitis. Neurobiol. Dis. 2019, 125, 154–162. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [PubMed]
- Shirani, F.; Baghi, M.; Rostamian Delavar, M.; Shoaraye Nejati, A.; Eshaghiyan, A.; Nasr-Esfahani, M.H.; Peymani, M.; Ghaedi, K. Upregulation of miR-9 and miR-193b over human Th17 cell differentiation. Mol. Genet. Genom. Med. 2020, 8, e1538. [Google Scholar]
- Majd, M.; Hosseini, A.; Ghaedi, K.; Kiani-Esfahani, A.; Tanhaei, S.; Shiralian-Esfahani, H.; Rahnamaee, S.Y.; Mowla, S.J.; Nasr-Esfahani, M.H. MiR-9-5p and miR-106a-5p dysregulated in CD4+ T-cells of multiple sclerosis patients and targeted essential factors of T helper17/regulatory T-cells differentiation. Iran J. Basic Med. Sci. 2018, 21, 277–283. [Google Scholar] [PubMed]
- Yao, H.; Ma, R.; Yang, L.; Hu, G.; Chen, X.; Duan, M.; Kook, Y.; Niu, F.; Liao, K.; Fu, M.; et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat. Commun. 2014, 5, 4386. [Google Scholar] [CrossRef]
- Yue, P.; Jing, L.; Zhao, X.; Zhu, H.; Teng, J. Down-regulation of taurine-up-regulated gene 1 attenuates inflammation by sponging miR-9-5p via targeting NF-κB1/p50 in multiple sclerosis. Life Sci. 2019, 233, 116731. [Google Scholar] [PubMed]
- Singh, J.; Deshpande, M.; Suhail, H.; Rattan, R.; Giri, S. Targeted Stage-Specific Inflammatory microRNA Profiling in Urine during Disease Progression in Experimental Autoimmune Encephalomyelitis: Markers of Disease Progression and Drug Response. J. Neuroimmune Pharmacol. 2016, 11, 84–97. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA miR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, C.; Edwards, L.J.; de Vries, H.E.; Sharrack, B.; Male, D.K.; Romero, I.A. MiR-126 and miR-126* regulate shear-resistant firm leukocyte adhesion to human brain endothelium. Sci. Rep. 2017, 7, 45284. [Google Scholar] [CrossRef] [PubMed]
- Mattes, J.; Collison, A.; Plank, M.; Phipps, S.; Foster, P.S. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA 2009, 106, 18704–18709. [Google Scholar]
- Mancuso, R.; Agostini, S.; Hernis, A.; Caputo, D.; Galimberti, D.; Scarpini, E.; Clerici, M. Alterations of the miR-126-3p/POU2AF1/Spi-B Axis and JCPyV Reactivation in Multiple Sclerosis Patients Receiving Natalizumab. Front Neurol. 2022, 13, 819911. [Google Scholar] [PubMed]
- Cox, M.B.; Cairns, M.J.; Gandhi, K.S.; Carroll, A.P.; Moscovis, S.; Stewart, G.J.; Broadley, S.; Scott, R.J.; Booth, D.R.; Lechner-Scott, J.; et al. MicroRNAs miR-17 and miR-20a Inhibit T Cell Activation Genes and Are Under-Expressed in MS Whole Blood. PLoS ONE 2010, 5, e12132. [Google Scholar] [CrossRef]
- Baulina, N.; Kulakova, O.; Kiselev, I.; Osmak, G.; Popova, E.; Boyko, A.; Favorova, O. Immune-related miRNA expression patterns in pe-ripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J Neuroimmunol. 2018, 317, 67–76. [Google Scholar]
- Meira, M.; Sievers, C.; Hoffmann, F.; Derfuss, T.; Kuhle, J.; Kappos, L.; Lindberg, R.L. MiR-126: A novel route for natalizumab action? Mult. Scler. J. 2014, 20, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Desi, N.; Teh, V.; Tong, Q.Y.; Lim, C.Y.; Tabatabaeian, H.; Chew, X.H.; Sanchez-Mejias, A.; Chan, J.J.; Zhang, B.; Pitcheshwar, P.; et al. MiR-138 is a potent regulator of the heterogenous MYC transcript population in cancers. Oncogene 2021, 41, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.-H.; Wang, D.-D.; Chen, D.; Liu, S.-W.; Wang, Z.; Yan, D.-L.; Dong, S.-C.; Feng, J.-F. MiR-138: A promising therapeutic target for cancer. Tumor Biol. J. Int. Soc. Oncodevelopmental. Biol. Med. 2017, 39, 1010428317697575. [Google Scholar] [CrossRef]
- Rasoolnezhad, M.; Safaralizadeh, R.; Hosseinpourfeizi, M.A.; Banan-Khojasteh, S.M.; Baradaran, B. MiRNA-138–5p: A strong tumor suppressor targeting PD-L-1 inhibits proliferation and motility of breast cancer cells and induces apoptosis. Eur. J. Pharmacol. 2021, 896, 173933. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Hu, J.; Zhan, F.; Luo, D.; Hua, F.; Xu, G. MicroRNA-138-5p Regulates Hippocampal Neuroinflammation and Cognitive Impairment by NLRP3/Caspase-1 Signaling Pathway in Rats. J. Inflamm. Res. 2021, 14, 1125–1143. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, J.M.K.; Anderson, R.C.; McDermott, K.W. MicroRNA: Key regulators of oligodendrocyte development and pathobiol-ogy. Int. J. Biochem. Cell Biol. 2015, 65, 134–138. [Google Scholar] [CrossRef]
- Varma-Doyle, A.V.; Lukiw, W.J.; Zhao, Y.; Lovera, J.; Devier, D. A hypothesis-generating scoping review of miRs identified in both multiple sclerosis and dementia, their protein targets, and miR signaling pathways. J. Neurol. Sci. 2021, 420, 117202. [Google Scholar] [CrossRef]
- Chen, J.; Qin, R. MicroRNA-138-5p regulates the development of spinal cord injury by targeting SIRT1. Mol. Med. Rep. 2020, 22, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tan, L.; Lu, Y.; Peng, J.; Zhu, Y.; Zhang, Y.; Sun, Z. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015, 589, 726–729. [Google Scholar] [CrossRef]
- Weiss, K.; Treiber, T.; Meister, G.; Schratt, G. The nuclear matrix protein Matr3 regulates processing of the synaptic mi-croRNA-138-5p. Neurobiol. Learn Mem. 2019, 159, 36–45. [Google Scholar] [CrossRef]
- Mazdeh, M.; Kordestani, H.; Komaki, A.; Eftekharian, M.; Arsang-Jang, S.; Branicki, W.; Taheri, M.; Ghafouri-Fard, S. Expression Profile of Selected MicroRNAs in the Peripheral Blood of Multiple Sclerosis Patients: A Multivariate Statistical Analysis with ROC Curve to Find New Biomarkers for Fingolimod. J. Mol. Neurosci. 2019, 68, 153–161. [Google Scholar]
- Mousavi, S.R.; Tahmasebivand, M.; Khorrami, M.; Ayromlou, H.; Khalili, S.K.; Khorvash, F.; Rikhtegar, R.; Khademi, B.; Bahmanpour, Z.; Emamalizadeh, B. Connection of miR-185 and miR-320a Expression Levels With Response to Interferon-Beta in Multiple Sclerosis Patients. Mult. Scler. Relat. Disord. 2020, 44, 102264. [Google Scholar] [CrossRef] [PubMed]
- Franzoi, A.E.d.A.; Machado, F.S.d.M.; Junior, W.L.G.d.M.; Bandeira, I.P.; Brandão, W.N.; Gonçalves, M.V.M. Altered expression of microRNAs and B lymphocytes during Natalizumab therapy in multiple sclerosis. Heliyon 2021, 7, e07263. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebivand, M.; Mousavi, S.R.; Khorrami, M.; Ayromlou, H.; Rikhtegar, R.; Saberi, L.; Khademi, B.; Bahmanpour, Z.; Emamalizadeh, B. miR-504 Expression Level is Increased in Multiple Sclerosis Patients Responder to Interferon-Beta. J. Neuroimmunol. 2020, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Mameli, G.; Arru, G.; Caggiu, E.; Niegowska, M.; Leoni, S.; Madeddu, G.; Babudieri, S.; Sechi, G.P.; Sechi, L.A. Natalizumab Therapy Modulates miR-155, miR-26a and Proinflammatory Cytokine Expression in MS Patients. PLoS ONE 2016, 11, e0157153. [Google Scholar] [CrossRef]
- Mazdeh, M.; Kordestani, H.; Komaki, A.; Eftekharian, M.M.; Arsang-Jang, S.; Branicki, W.; Taheri, M.; Ghafouri-Fard, S. Assessment of Expression Profile of microRNAs in Multiple Sclerosis Patients Treated with Fingolimod. J. Mol. Neurosci. 2020, 70, 1274–1281. [Google Scholar] [CrossRef]
- Ntranos, A.; Ntranos, V.; Bonnefil, V.; Liu, J.; Kim-Schulze, S.; He, Y.; Zhu, Y.; Brandstadter, R.; Watson, C.T.; Sharp, A.J.; et al. Fumarates Target the Metabolic-Epigenetic Interplay of Brain-Homing T Cells in Multiple Sclerosis. Brain 2019, 142, 647–661. [Google Scholar] [CrossRef]
- Neuhaus, O.; Kieseier, B.C.; Hartung, H.-P. Pharmacokinetics and Pharmacodynamics of the Interferon-Betas, Glatiramer Acetate, and Mitoxantrone in Multiple sclerosis. J. Neurol. Sci. 2007, 259, 27–37. [Google Scholar] [CrossRef] [PubMed]
| microRNas | Clinical Association | MRI Volume |
|---|---|---|
| 9.5p | EDSS | Thalamus |
| 126.3p | SDMT | - |
| 138.5p | - | Pallidum and amygdala |
| 146a.5p | EDSS and SDMT | - |
| 200c.3p | - | Cerebellum and pallidum |
| 223.3p | - | Caudate |
| Sex N (F:M) | Age at MS Onset (Years) Md (ICR) | Age at GA Onset (Years) Md (ICR) | Time with GA at Study Onset (Years) Md (ICR) | Basal EDSS Mean (±SD) |
|---|---|---|---|---|
| 18:8 | 31.9 (25.1–41.9) | 32.8 (26.6–44.9) | 4 (2.1–6.4) | 1.4 (1.7) |
| Clinical Data | 2 Years (n = 25) | 5 Years (n = 24) |
|---|---|---|
| NEDA-3 | 70.8% (n = 17/24) | 56.5% (n = 13/23) |
| Relapse | 8% (n = 2) | 13% (n = 3) |
| 6-month CDP | 12% (n = 3) | 17.4% (n = 4) |
| MRI activity | 8% (n = 2) | 13% (n = 3) |
| Treatment GA | 60% (n = 15) | 20.8% (n = 5) |
| GA failure | 28% (n = 7) | 42.3% (n = 11) |
| GA change (not failure) | 12% (n = 3) | 30.8% (n = 8) |
| miRNA | EDSS 2 Years | |
| β (CI 95%) | p | |
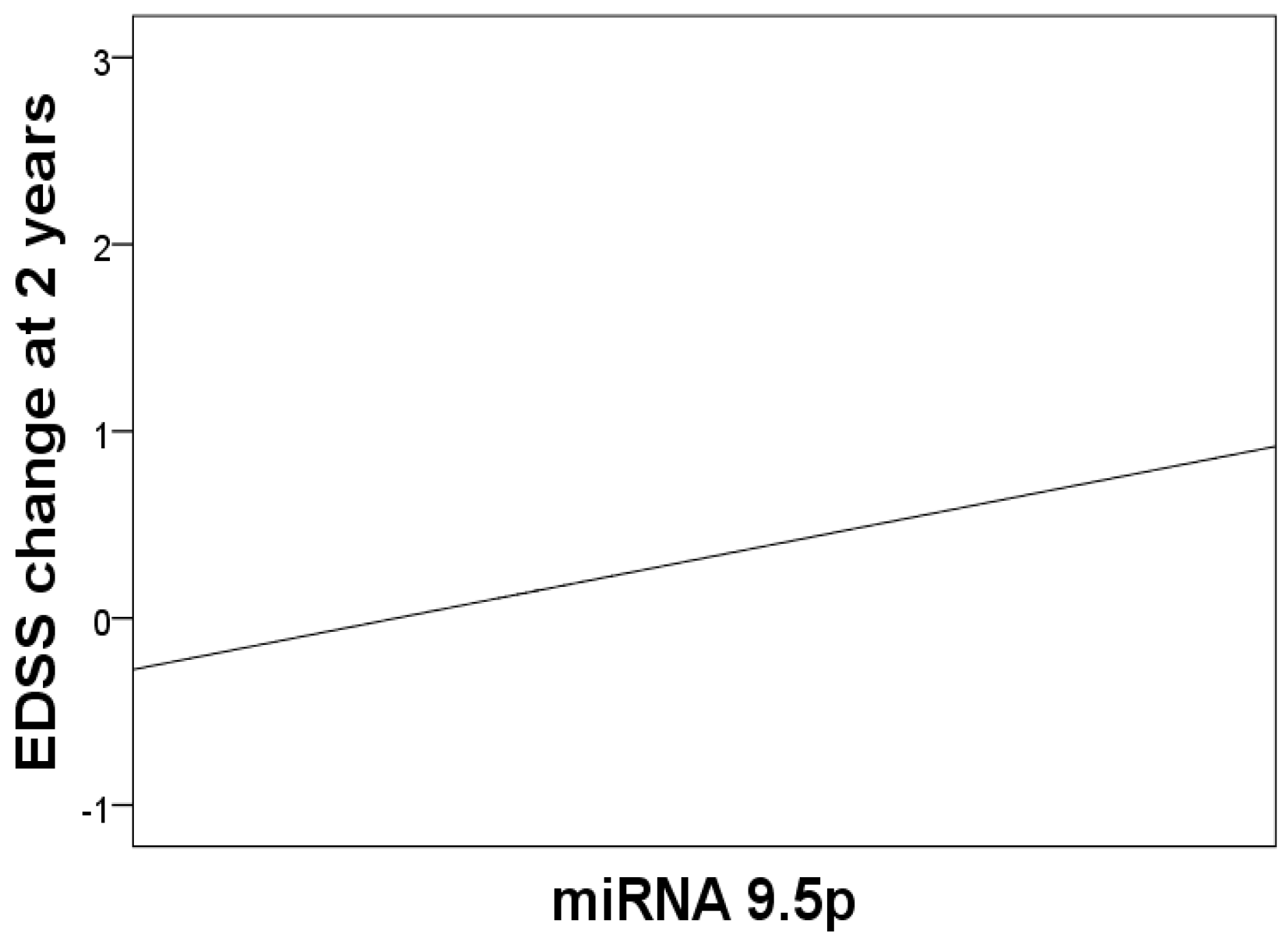
| 9.5p | 0.23 (0.04–0.46) | * 0.047 |
| Excluded variables (β; p) | ||
| 126.3p | −1.94; 0.554 | |
| 138.5p | 2.45; 0.396 | |
| 146a.5p | −3.21; 0.262 | |
| 200c.3p | −1.03; 0.764 | |
| 223.3p | 1.9; 0.583 | |
| miRNA | EDSS 5 Years | |
| Excluded variables (β; p) | ||
| 9.5p | 1.41; 0.697 | |
| 126.3p | 0.36; 0.305 | |
| 138.5p | −2.92; 0.413 | |
| 146a.5p | 0.41; 0.234 | |
| 200c.3p | 0.36; 0.305 | |
| 223.3p | −2.95; 0.408 | |
| miRNA | Clinical Evolution (2 Years) | |||
|---|---|---|---|---|
| EDSS | Relapse/MRI Activity | CDP | NEDA-3 | |
| (Multivariate Regression Analysis) | (U Mann–Whitney Test) | |||
| 9.5p | + | - | - | - |
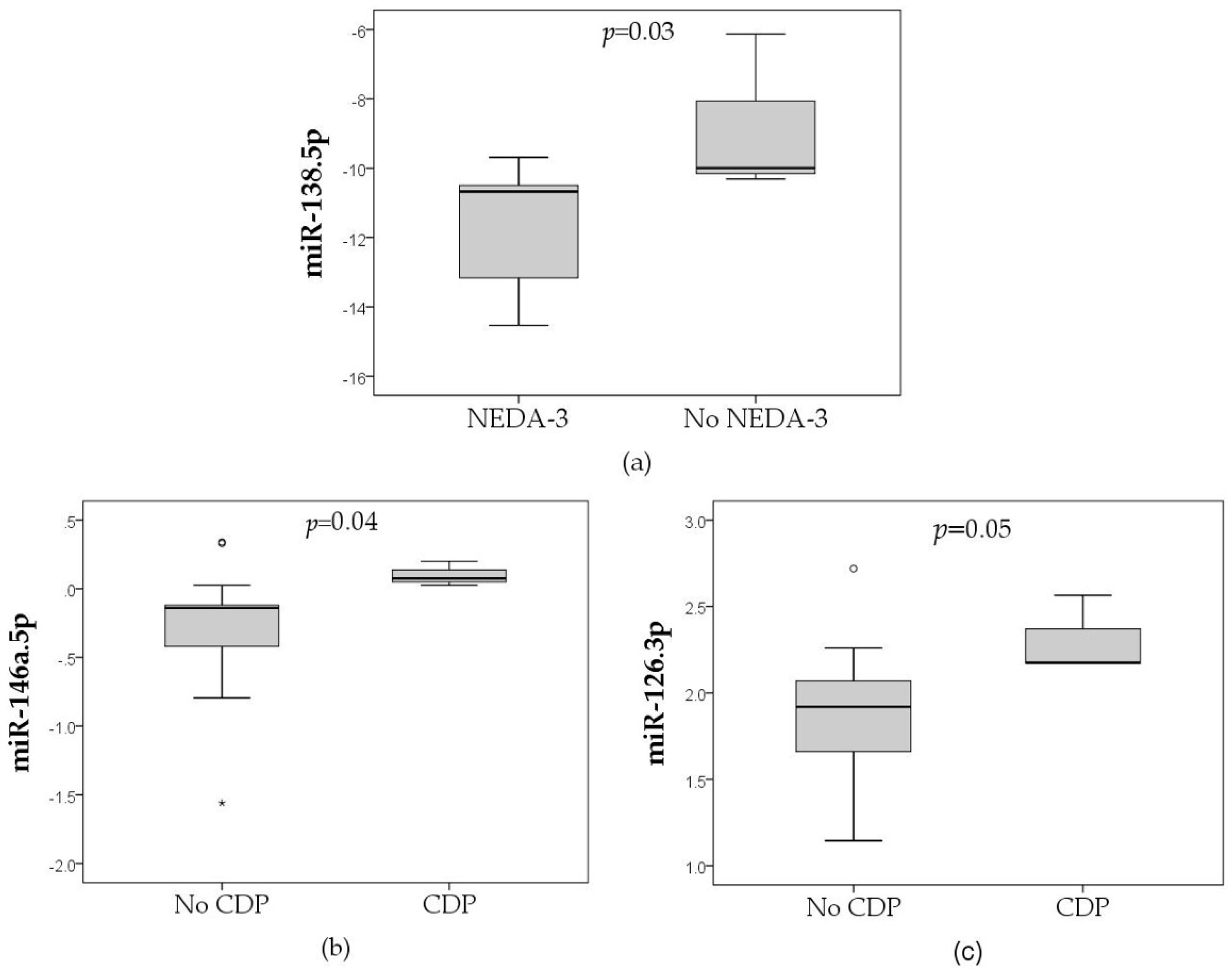
| 126.3p | - | - | + | - |
| 138.5p | - | - | - | + |
| 146a.5p | - | - | + | - |
| 200c.3p | - | - | - | - |
| 223.3p | - | - | - | - |
| Clinical evolution (5 years) | ||||
| No miRNAs associated with any clinical/radiological variable | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casanova, I.; Domínguez-Mozo, M.I.; De Torres, L.; Aladro-Benito, Y.; García-Martínez, Á.; Gómez, P.; Abellán, S.; De Antonio, E.; Álvarez-Lafuente, R. MicroRNAs Associated with Disability Progression and Clinical Activity in Multiple Sclerosis Patients Treated with Glatiramer Acetate. Biomedicines 2023, 11, 2760. https://doi.org/10.3390/biomedicines11102760
Casanova I, Domínguez-Mozo MI, De Torres L, Aladro-Benito Y, García-Martínez Á, Gómez P, Abellán S, De Antonio E, Álvarez-Lafuente R. MicroRNAs Associated with Disability Progression and Clinical Activity in Multiple Sclerosis Patients Treated with Glatiramer Acetate. Biomedicines. 2023; 11(10):2760. https://doi.org/10.3390/biomedicines11102760
Chicago/Turabian StyleCasanova, Ignacio, María I. Domínguez-Mozo, Laura De Torres, Yolanda Aladro-Benito, Ángel García-Martínez, Patricia Gómez, Sara Abellán, Esther De Antonio, and Roberto Álvarez-Lafuente. 2023. "MicroRNAs Associated with Disability Progression and Clinical Activity in Multiple Sclerosis Patients Treated with Glatiramer Acetate" Biomedicines 11, no. 10: 2760. https://doi.org/10.3390/biomedicines11102760
APA StyleCasanova, I., Domínguez-Mozo, M. I., De Torres, L., Aladro-Benito, Y., García-Martínez, Á., Gómez, P., Abellán, S., De Antonio, E., & Álvarez-Lafuente, R. (2023). MicroRNAs Associated with Disability Progression and Clinical Activity in Multiple Sclerosis Patients Treated with Glatiramer Acetate. Biomedicines, 11(10), 2760. https://doi.org/10.3390/biomedicines11102760








