Epigenome-Wide Association Study in Peripheral Tissues Highlights DNA Methylation Profiles Associated with Episodic Memory Performance in Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Samples and Measurements of Episodic Memory
2.2. Episodic Memory Phenotypes
2.3. DNA Extraction and Processing
2.4. DNA Methylation Profiling
2.5. DNA Methylation Data Processing and Quality Control
2.6. Epigenome-Wide Association Analyses to Identify Differentially Methylated Probes
2.7. Calculation of Poly-Epigenetic Scores (PESs) for General Cognitive Abilities and AD
2.8. Epigenetic Age Estimation
2.9. Look-Up of EM-Associated CpGs in an Independent EWAS on AD-Related Phenotypes in Human EC
2.10. Look-Up of EM-Associated CpGs in an Independent Buccal–Brain Correlation Map
2.11. Look-Up of EM-Associated CpGs in a Buccal mQTL Database
2.12. DNAm–mRNA Correlation Analyses
3. Results
3.1. EM Performance Measures in the Analyzed Datasets
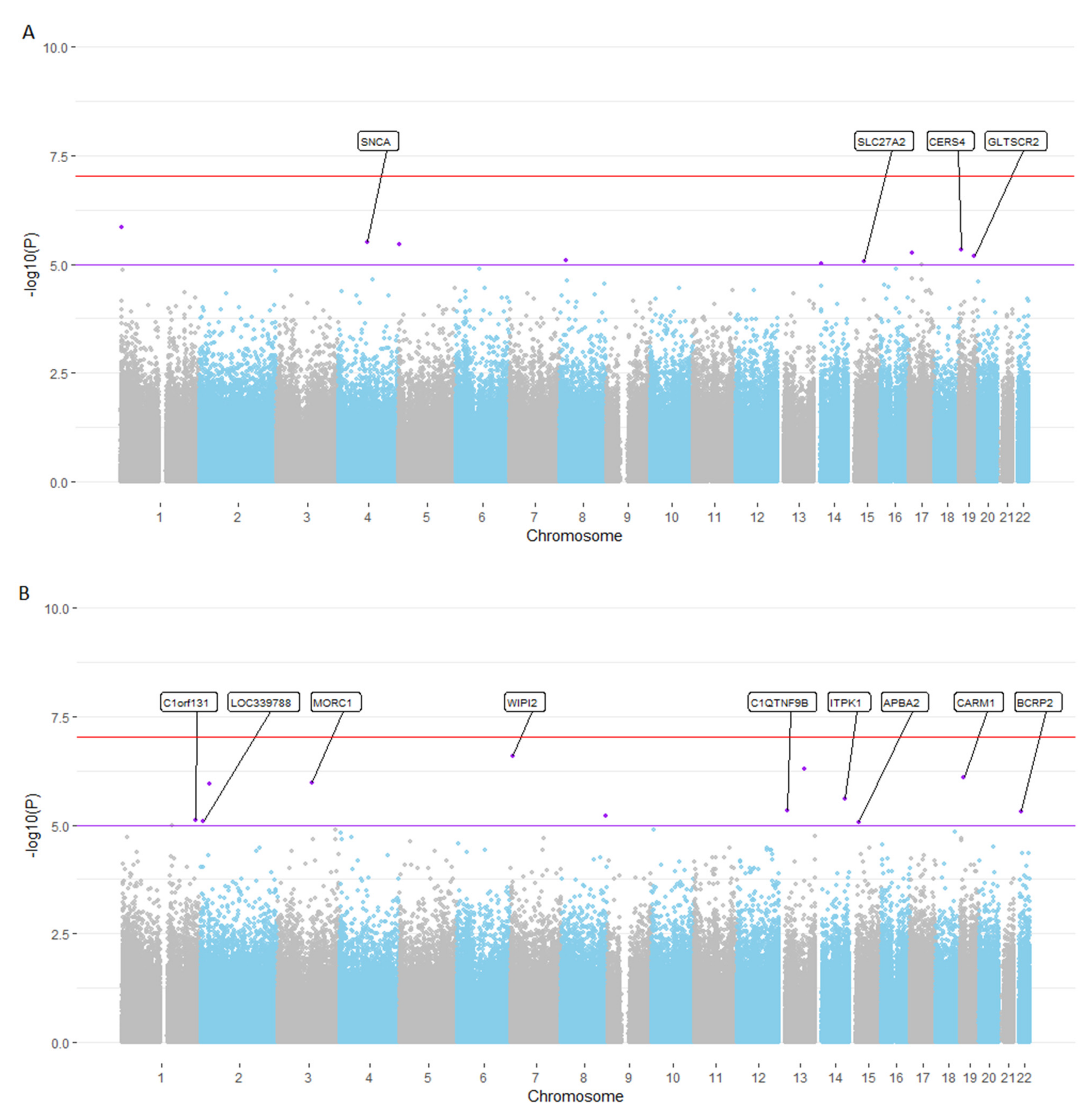
3.2. EWAS Meta-Analyses Highlight Several CpG Loci Showing at Least Genome-Wide Suggestive Association with Cross-Sectional and Longitudinal Changes in EM Performance
3.3. Poly-Epigenetic Score Analysis for Cognitive Abilities and AD Show Only Little Correspondence with Episodic Memory EWAS
3.4. Correspondence of EWAS Results in Blood and Buccal Samples Underscores Tissue Specificity
3.5. Correlation Analysis between DNAm Levels and mRNA Expression in Human Brain Samples Highlights Three Loci
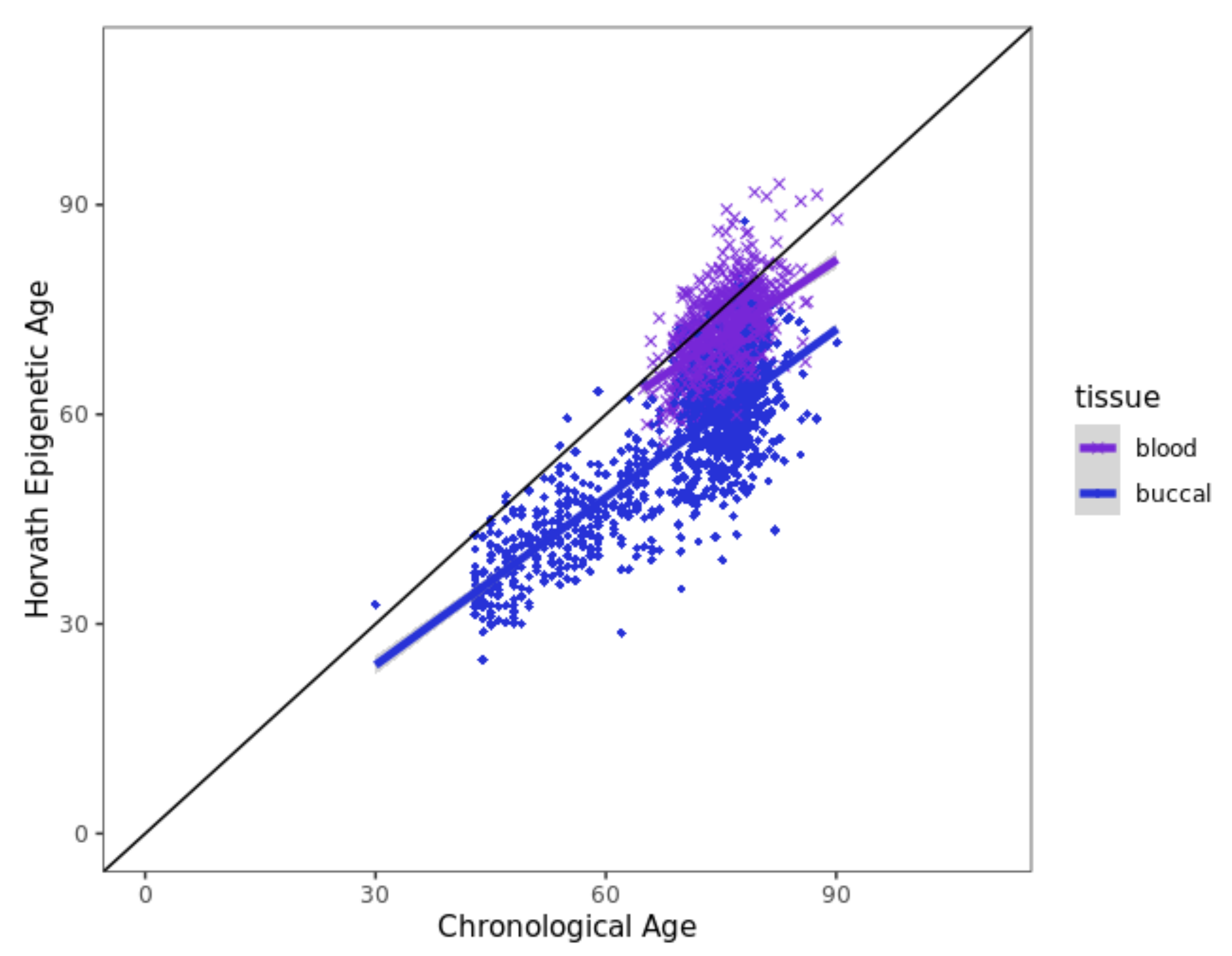
3.6. Horvath Epigenetic Age Acceleration Is Not Associated with EM Performance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dickerson, B.C.; Eichenbaum, H. The Episodic Memory System: Neurocircuitry and Disorders. Neuropsychopharmacology 2010, 35, 86. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, M.A.; Einstein, G.O.; Jacoby, L.L. New Considerations in Aging and Memory: The Glass May Be Half Full. In The Handbook of Aging and Cognition; Psychology Press: New York, NY, USA, 2008; pp. 251–310. [Google Scholar]
- Rugg, M.D.; Morcom, A.M. The Relationship Between Brain Activity, Cognitive Performance, and Aging: The Case of Memory. In Cognitive Neuroscience of Aging: Linking Cognitive and Cerebral Aging; Oxford University Press: Oxford, UK, 2009. [Google Scholar] [CrossRef]
- Friedman, D.; Nessler, D.; Johnson, R. Memory Encoding and Retrieval in the Aging Brain. Clin. EEG Neurosci. 2007, 38, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Sexton, C.E.; Mackay, C.E.; Lonie, J.A.; Bastin, M.E.; Terrière, E.; O’Carroll, R.E.; Ebmeier, K.P. MRI Correlates of Episodic Memory in Alzheimer’s Disease, Mild Cognitive Impairment, and Healthy Aging. Psychiatry Res. Neuroimaging 2010, 184, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Nordahl, C.W.; Ranganath, C.; Yonelinas, A.P.; DeCarli, C.; Reed, B.R.; Jagust, W.J. Different Mechanisms of Episodic Memory Failure in Mild Cognitive Impairment. Neuropsychologia 2005, 43, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Feldman, H.H.; Jacova, C.; DeKosky, S.T.; Barberger-Gateau, P.; Cummings, J.; Delacourte, A.; Galasko, D.; Gauthier, S.; Jicha, G.; et al. Research Criteria for the Diagnosis of Alzheimer’s Disease: Revising the NINCDS-ADRDA Criteria. Lancet Neurol. 2007, 6, 734–746. [Google Scholar] [CrossRef]
- Davies, G.; Marioni, R.E.; Liewald, D.C.; Hill, W.D.; Hagenaars, S.P.; Harris, S.E.; Ritchie, S.J.; Luciano, M.; Fawns-Ritchie, C.; Lyall, D.; et al. Genome-Wide Association Study of Cognitive Functions and Educational Attainment in UK Biobank (N = 112 151). Mol. Psychiatry 2016, 21, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.; Armstrong, N.; Bis, J.C.; Bressler, J.; Chouraki, V.; Giddaluru, S.; Hofer, E.; Ibrahim-Verbaas, C.A.; Kirin, M.; Lahti, J.; et al. Genetic Contributions to Variation in General Cognitive Function: A Meta-Analysis of Genome-Wide Association Studies in the CHARGE Consortium (N = 53 949). Mol. Psychiatry 2015, 20, 183–192. [Google Scholar] [CrossRef]
- Milnik, A.; Heck, A.; Vogler, C.; Heinze, H.J.; de Quervain, D.J.F.; Papassotiropoulos, A. Association of KIBRA with Episodic and Working Memory: A Meta-Analysis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012, 159B, 958–969. [Google Scholar] [CrossRef]
- Papassotiropoulos, A.; Stefanova, E.; Vogler, C.; Gschwind, L.; Ackermann, S.; Spalek, K.; Rasch, B.; Heck, A.; Aerni, A.; Hanser, E.; et al. A Genome-Wide Survey and Functional Brain Imaging Study Identify CTNNBL1 as a Memory-Related Gene. Mol. Psychiatry 2011, 18, 255–263. [Google Scholar] [CrossRef]
- Papassotiropoulos, A.; Stephan, D.A.; Huentelman, M.J.; Hoerndli, F.J.; Craig, D.W.; Pearson, J.V.; Huynh, K.D.; Brunner, F.; Corneveaux, J.; Osborne, D.; et al. Common Kibra Alleles Are Associated with Human Memory Performance. Science 2006, 314, 475–478. [Google Scholar] [CrossRef]
- Liu, T.; Li, S.-C.; Papenberg, G.; Schröder, J.; Roehr, J.T.; Nietfeld, W.; Lindenberger, U.; Bertram, L. No Association between CTNNBL1 and Episodic Memory Performance. Transl. Psychiatry 2014, 4, e454. [Google Scholar] [CrossRef]
- Marioni, R.E.; McRae, A.F.; Bressler, J.; Colicino, E.; Hannon, E.; Li, S.; Prada, D.; Smith, J.A.; Trevisi, L.; Tsai, P.-C.; et al. Meta-Analysis of Epigenome-Wide Association Studies of Cognitive Abilities. Mol. Psychiatry 2018, 23, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- McCartney, D.L.; Hillary, R.F.; Conole, E.L.S.; Banos, D.T.; Gadd, D.A.; Walker, R.M.; Nangle, C.; Flaig, R.; Campbell, A.; Murray, A.D.; et al. Blood-Based Epigenome-Wide Analyses of Cognitive Abilities. Genome Biol. 2022, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Fransquet, P.D.; Lacaze, P.; Saffery, R.; Phung, J.; Parker, E.; Shah, R.; Murray, A.; Woods, R.L.; Ryan, J. Blood DNA Methylation Signatures to Detect Dementia Prior to Overt Clinical Symptoms. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2020, 12, e12056. [Google Scholar] [CrossRef]
- Walker, R.M.; Bermingham, M.L.; Vaher, K.; Morris, S.W.; Clarke, T.K.; Bretherick, A.D.; Zeng, Y.; Amador, C.; Rawlik, K.; Pandya, K.; et al. Epigenome-Wide Analyses Identify DNA Methylation Signatures of Dementia Risk. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2020, 12, e12078. [Google Scholar] [CrossRef]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Ritchie, S.J.; Muniz-Terrera, G.; Harris, S.E.; Gibson, J.; Redmond, P.; Cox, S.R.; Pattie, A.; et al. The Epigenetic Clock Is Correlated with Physical and Cognitive Fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 2015, 44, 1388–1396. [Google Scholar] [CrossRef]
- Zheng, Y.; Habes, M.; Gonzales, M.; Pomponio, R.; Nasrallah, I.; Khan, S.; Vaughan, D.E.; Davatzikos, C.; Seshadri, S.; Launer, L.; et al. Mid-Life Epigenetic Age, Neuroimaging Brain Age, and Cognitive Function: Coronary Artery Risk Development in Young Adults (CARDIA) Study. Aging 2022, 14, 1691–1712. [Google Scholar] [CrossRef]
- Beydoun, M.A.; Shaked, D.; Tajuddin, S.M.; Weiss, J.; Evans, M.K.; Zonderman, A.B. Accelerated Epigenetic Age and Cognitive Decline among Urban-Dwelling Adults. Neurology 2020, 94, e613–e625. [Google Scholar] [CrossRef]
- Maddock, J.; Castillo-Fernandez, J.; Wong, A.; Cooper, R.; Richards, M.; Ong, K.K.; Ploubidis, G.B.; Goodman, A.; Kuh, D.; Bell, J.T.; et al. DNA Methylation Age and Physical and Cognitive Aging. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2020, 75, 504–511. [Google Scholar] [CrossRef]
- Vaccarino, V.; Huang, M.; Wang, Z.; Hui, Q.; Shah, A.J.; Goldberg, J.; Smith, N.; Kaseer, B.; Murrah, N.; Levantsevych, O.M.; et al. Epigenetic Age Acceleration and Cognitive Decline: A Twin Study. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2021, 76, 1854–1863. [Google Scholar] [CrossRef]
- Sommerer, Y.; Dobricic, V.; Schilling, M.; Ohlei, O.; Sabet, S.S.; Wesse, T.; Fuß, J.; Franzenburg, S.; Franke, A.; Parkkinen, L.; et al. Entorhinal Cortex EWAS Meta-Analysis Highlights Four Novel Loci Showing Differential Methylation in Alzheimer’s Disease. bioRxiv 2021. [Google Scholar] [CrossRef]
- Walhovd, K.B.; Fjell, A.M.; Westerhausen, R.; Nyberg, L.; Ebmeier, K.P.; Lindenberger, U.; Bartrés-Faz, D.; Baaré, W.F.C.; Siebner, H.R.; Henson, R.; et al. Healthy Minds 0–100 Years: Optimising the Use of European Brain Imaging Cohorts (“Lifebrain”). Eur. Psychiatry 2018, 50, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L.; Böckenhoff, A.; Demuth, I.; Düzel, S.; Eckardt, R.; Li, S.C.; Lindenberger, U.; Pawelec, G.; Siedler, T.; Wagner, G.G.; et al. Cohort Profile: The Berlin Aging Study II (BASE-II). Int. J. Epidemiol. 2014, 43, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Gerstorf, D.; Bertram, L.; Lindenberger, U.; Pawelec, G.; Demuth, I.; Steinhagen-Thiessen, E.; Wagner, G.G. Editorial. Gerontology 2016, 62, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Demuth, I.; Banszerus, V.; Drewelies, J.; Düzel, S.; Seeland, U.; Spira, D.; Tse, E.; Braun, J.; Steinhagen-Thiessen, E.; Bertram, L.; et al. Cohort Profile: Follow-up of a Berlin Aging Study II (BASE-II) Subsample as Part of the GendAge Study. BMJ Open 2021, 11, e045576. [Google Scholar] [CrossRef]
- Cattaneo, G.; Bartrés-Faz, D.; Morris, T.P.; Sánchez, J.S.; Macià, D.; Tarrero, C.; Tormos, J.M.; Pascual-Leone, A. The Barcelona Brain Health Initiative: A Cohort Study to Define and Promote Determinants of Brain Health. Front. Aging Neurosci. 2018, 10, 321. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Gorrie-Stone, T.J.; Smart, M.C.; Saffari, A.; Malki, K.; Hannon, E.; Burrage, J.; Mill, J.; Kumari, M.; Schalkwyk, L.C. Bigmelon: Tools for Analysing Large DNA Methylation Datasets. Bioinformatics 2019, 35, 981–986. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Breeze, C.E.; Zheng, S.C.; Beck, S. A Comparison of Reference-Based Algorithms for Correcting Cell-Type Heterogeneity in Epigenome-Wide Association Studies. BMC Bioinform. 2017, 18, 105. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Silva, T.C.; Young, J.I.; Gomez, L.; Schmidt, M.A.; Hamilton-Nelson, K.L.; Kunkle, B.W.; Chen, X.; Martin, E.R.; Wang, L. Epigenome-Wide Meta-Analysis of DNA Methylation Differences in Prefrontal Cortex Implicates the Immune Processes in Alzheimer’s Disease. Nat. Commun. 2020, 11, 6114. [Google Scholar] [CrossRef] [PubMed]
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Babei, M.S.; Sharp, G.; Hatcher, C.; Vega-Salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A Database of Epigenome-Wide Association Studies. OSF Prepr. 2021, 7, 41. [Google Scholar] [CrossRef]
- Lunnon, K.; Smith, R.; Hannon, E.; De Jager, P.L.; Srivastava, G.; Volta, M.; Troakes, C.; Al-Sarraj, S.; Burrage, J.; Macdonald, R.; et al. Methylomic Profiling Implicates Cortical Deregulation of ANK1 in Alzheimer’s Disease. Nat. Neurosci. 2014, 17, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.G.; Pishva, E.; Shireby, G.; Smith, A.R.; Roubroeks, J.A.Y.; Hannon, E.; Wheildon, G.; Mastroeni, D.; Gasparoni, G.; Riemenschneider, M.; et al. A Meta-Analysis of Epigenome-Wide Association Studies in Alzheimer’s Disease Highlights Novel Differentially Methylated Loci across Cortex. Nat. Commun. 2021, 12, 3517. [Google Scholar] [CrossRef]
- Sommerer, Y.; Ohlei, O.; Dobricic, V.; Oakley, D.H.; Wesse, T.; Sabet, S.S.; Demuth, I.; Franke, A.; Hyman, B.T.; Lill, C.M.; et al. A Correlation Map of Genome-Wide DNA Methylation Patterns between Paired Human Brain and Buccal Samples. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix EQTL: Ultra Fast EQTL Analysis via Large Matrix Operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT Improves Functional Interpretation of Cis-Regulatory Regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA Methylation Studies: Statistical Insights from the Illumina EPIC Array. BMC Genom. 2019, 20, 366. [Google Scholar] [CrossRef]
- Gasparoni, G.; Bultmann, S.; Lutsik, P.; Kraus, T.F.J.; Sordon, S.; Vlcek, J.; Dietinger, V.; Steinmaurer, M.; Haider, M.; Mulholland, C.B.; et al. DNA Methylation Analysis on Purified Neurons and Glia Dissects Age and Alzheimer’s Disease-Specific Changes in the Human Cortex. Epigenetics Chromatin 2018, 11, 41. [Google Scholar] [CrossRef]
- Smith, R.G.; Hannon, E.; De Jager, P.L.; Chibnik, L.; Lott, S.J.; Condliffe, D.; Smith, A.R.; Haroutunian, V.; Troakes, C.; Al-Sarraj, S.; et al. Elevated DNA Methylation across a 48-Kb Region Spanning the HOXA Gene Cluster Is Associated with Alzheimer’s Disease Neuropathology. Alzheimers Dement. 2018, 14, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Narayan, P.; Grant, M.J.; Langfelder, P.; Wang, N.; Kwak, S.; Wilkinson, H.; Chen, R.Z.; Chen, J.; Simon Bawden, C.; et al. DNA Methylation Study of Huntington’s Disease and Motor Progression in Patients and in Animal Models. Nat. Commun. 2020, 11, 4529. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, J.A.; Sears, R.L.; Gao, F.; Klein, E.D.; Karydas, A.; Geschwind, M.D.; Rosen, H.J.; Boxer, A.L.; Guo, W.; et al. An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy. PLoS Genet. 2014, 10, e1004211. [Google Scholar] [CrossRef] [PubMed]
- Vallerga, C.L.; Zhang, F.; Fowdar, J.; McRae, A.F.; Qi, T.; Nabais, M.F.; Zhang, Q.; Kassam, I.; Henders, A.K.; Wallace, L.; et al. Analysis of DNA Methylation Associates the Cystine-Glutamate Antiporter SLC7A11 with Risk of Parkinson’s Disease. Nat. Commun. 2020, 11, 1238. [Google Scholar] [CrossRef]
- Vetter, V.M.; Kalies, C.H.; Sommerer, Y.; Spira, D.; Drewelies, J.; Regitz-Zagrosek, V.; Bertram, L.; Gerstorf, D.; Demuth, I. Relationship Between 5 Epigenetic Clocks, Telomere Length, and Functional Capacity Assessed in Older Adults: Cross-Sectional and Longitudinal Analyses. J. Gerontol. Ser. A 2022, 77, 1724–1733. [Google Scholar] [CrossRef]
- Lawn, R.B.; Anderson, E.L.; Suderman, M.; Simpkin, A.J.; Gaunt, T.R.; Teschendorff, A.E.; Widschwendter, M.; Hardy, R.; Kuh, D.; Relton, C.L.; et al. Psychosocial Adversity and Socioeconomic Position during Childhood and Epigenetic Age: Analysis of Two Prospective Cohort Studies. Hum. Mol. Genet. 2018, 27, 1301–1308. [Google Scholar] [CrossRef]
- Logan, T.; Bendor, J.; Toupin, C.; Thorn, K.; Edwards, R.H. α-Synuclein Promotes Dilation of the Exocytotic Fusion Pore. Nat. Neurosci. 2017, 20, 681–689. [Google Scholar] [CrossRef]
- Huang, C.C.; Chiu, T.Y.; Lee, T.Y.; Hsieh, H.J.; Lin, C.C.; Kao, L.S. Soluble α-Synuclein Facilitates Priming and Fusion by Releasing Ca 2+ from the Thapsigargin-Sensitive Ca 2+ Pool in PC12 Cells. J. Cell Sci. 2018, 131, jcs.213017. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. Alpha-Synuclein Promotes SNARE-Complex Assembly in Vivo and in Vitro. Science 2010, 329, 1663–1667. [Google Scholar] [CrossRef]
- Butler, B.; Saha, K.; Rana, T.; Becker, J.P.; Sambo, D.; Davari, P.; Goodwin, J.S.; Khoshbouei, H. Dopamine Transporter Activity Is Modulated by α-Synuclein. J. Biol. Chem. 2015, 290, 29542–29554. [Google Scholar] [CrossRef]
- Zarranz, J.J.; Alegre, J.; Gómez-Esteban, J.C.; Lezcano, E.; Ros, R.; Ampuero, I.; Vidal, L.; Hoenicka, J.; Rodriguez, O.; Atarés, B.; et al. The New Mutation, E46K, of Alpha-Synuclein Causes Parkinson and Lewy Body Dementia. Ann. Neurol. 2004, 55, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Lill, C.M.; Roehr, J.T.; McQueen, M.B.; Kavvoura, F.K.; Bagade, S.; Schjeide, B.M.M.; Schjeide, L.M.; Meissner, E.; Zauft, U.; Allen, N.C.; et al. Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson’s Disease Genetics: The PDGene Database. PLoS Genet. 2012, 8, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Heilbron, K.; Vallerga, C.L.; Bandres-Ciga, S.; von Coelln, R.; Pihlstrøm, L.; Simón-Sánchez, J.; Schulte, C.; Sharma, M.; Krohn, L.; et al. Parkinson’s Disease Age at Onset Genome-Wide Association Study: Defining Heritability, Genetic Loci, and α-Synuclein Mechanisms. Mov. Disord. 2019, 34, 866–875. [Google Scholar] [CrossRef]
- Guerreiro, R.; Ross, O.A.; Kun-Rodrigues, C.; Hernandez, D.G.; Orme, T.; Eicher, J.D.; Shepherd, C.E.; Parkkinen, L.; Darwent, L.; Heckman, M.G.; et al. Investigating the Genetic Architecture of Dementia with Lewy Bodies: A Two-Stage Genome-Wide Association Study. Lancet. Neurol. 2018, 17, 64–74. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Ren, Y.; Zhang, Y.; Feng, L. A53T α-Synuclein Induces Neurogenesis Impairment and Cognitive Dysfunction in Line M83 Transgenic Mice and Reduces the Proliferation of Embryonic Neural Stem Cells. Brain Res. Bull. 2022, 182, 118–129. [Google Scholar] [CrossRef]
- Gu, Q.P.; Sun, Y.; Ream, L.W.; Whanger, P.D. Selenoprotein W Accumulates Primarily in Primate Skeletal Muscle, Heart, Brain and Tongue. Mol. Cell. Biochem. 2000, 204, 49–56. [Google Scholar] [CrossRef]
- Situ, J.; Huang, X.; Zuo, M.; Huang, Y.; Ren, B.; Liu, Q. Comparative Proteomic Analysis Reveals the Effect of Selenoprotein W Deficiency on Oligodendrogenesis in Fear Memory. Antioxidants 2022, 11, 999. [Google Scholar] [CrossRef]
- Homo Sapiens Inositol-Tetrakisphosphate 1-Kinase (ITPK1), Transcript v—Nucleotide—NCBI. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NM_001142594 (accessed on 25 February 2022).
- Majerus, P.W.; Wilson, D.B.; Zhang, C.; Nicholas, P.J.; Wilson, M.P. Expression of Inositol 1,3,4-Trisphosphate 5/6-Kinase (ITPK1) and Its Role in Neural Tube Defects. Adv. Enzym. Regul. 2010, 50, 365–372. [Google Scholar] [CrossRef][Green Version]
- Yu, L.; Petyuk, V.A.; Gaiteri, C.; Mostafavi, S.; Young-Pearse, T.; Shah, R.C.; Buchman, A.S.; Schneider, J.A.; Piehowski, P.D.; Sontag, R.L.; et al. Targeted Brain Proteomics Uncover Multiple Pathways to Alzheimer’s Dementia. Ann. Neurol. 2018, 84, 78–88. [Google Scholar] [CrossRef]
- Kim, N.; Yu, L.; Dawe, R.; Petyuk, V.A.; Gaiteri, C.; De Jager, P.L.; Schneider, J.A.; Arfanakis, K.; Bennett, D.A. Microstructural Changes in the Brain Mediate the Association of AK4, IGFBP5, HSPB2, and ITPK1 with Cognitive Decline. Neurobiol. Aging 2019, 84, 17–25. [Google Scholar] [CrossRef]
- Biederer, T.; Südhof, T.C. Mints as Adaptors. Direct Binding to Neurexins and Recruitment of Munc18. J. Biol. Chem. 2000, 275, 39803–39806. [Google Scholar] [CrossRef]
- Tomita, S.; Ozaki, T.; Taru, H.; Oguchi, S.; Takeda, S.; Yagi, Y.; Sakiyama, S.; Kirino, Y.; Suzuki, T. Interaction of a Neuron-Specific Protein Containing PDZ Domains with Alzheimer’s Amyloid Precursor Protein. J. Biol. Chem. 1999, 274, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Shiono, M.; Itoh, G.; Takei, N.; Matsushima, T.; Maeda, M.; Taru, H.; Hata, S.; Yamamoto, T.; Saito, Y.; et al. Increased Amyloidogenic Processing of Transgenic Human APP in X11-like Deficient Mouse Brain. Mol. Neurodegener. 2010, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.C.; Ariff, B.B.; Yates, D.M.; Lau, K.F.; Perkinton, M.S.; Rogelj, B.; Stephenson, J.D.; Miller, C.C.J.; Mcloughlin, D.M. X11beta Rescues Memory and Long-Term Potentiation Deficits in Alzheimer’s Disease APPswe Tg2576 Mice. Hum. Mol. Genet. 2009, 18, 4492–4500. [Google Scholar] [CrossRef] [PubMed]
- Starnawska, A.; Tan, Q.; Lenart, A.; McGue, M.; Mors, O.; Børglum, A.D.; Christensen, K.; Nyegaard, M.; Christiansen, L. Blood DNA Methylation Age Is Not Associated with Cognitive Functioning in Middle-Aged Monozygotic Twins. Neurobiol. Aging 2017, 50, 60–63. [Google Scholar] [CrossRef]
| A: Cross-sectional EM performance. | ||||||
|---|---|---|---|---|---|---|
| Analysis | N | % F | Age (mean ± sd) | Age range | DNAm PCs | Genetic PCs |
| BASE-II buccal-1 | 433 | 52 | 76 ± 4 | 65–86 | 12 | 11 |
| BASE-II buccal-2 | 245 | 50 | 76 ± 4 | 66–90 | 9 | 11 |
| BBHI | 341 | 46 | 54 ± 7 | 30–67 | 11 | 4 |
| Meta-analysis | 1019 | 50 | 69 ± 11 | 30–90 | NA | NA |
| BASE-II blood | 800 | 50 | 76 ± 4 | 65–90 | 10 | 13 |
| B: Longitudinal EM performance. | ||||||
| Analysis | N | % F | Age (mean ± sd) | Age range | DNAm PCs | Genetic PCs |
| BASE-II buccal-1 | 403 | 52 | 70 ± 4 | 61–81 | 12 | 11 |
| BASE-II buccal-2 | 223 | 50 | 70 ± 4 | 61–85 | 8 | 7 |
| Meta-analysis | 626 | 51 | 70 ± 4 | 61–85 | NA | NA |
| BASE-II blood | 735 | 50 | 70 ± 4 | 61–85 | 9 | 8 |
| A: Cross-sectional EM performance. | ||||||
|---|---|---|---|---|---|---|
| CpG | Location | Gene annotation * | Effectcross-sect. | pcross-sect. | Effectlongitudinal | plongitudinal |
| cg25311963 | chr1:1546691 | MIB2 | −7.15 | 1.41 × 10−6 | −0.10 | 0.93 |
| cg15402943 | chr4:90659260 | SNCA, TIGD2 | −6.38 | 3.07 × 10−6 | −0.81 | 0.45 |
| cg18370700 | chr5:2212396 | IRX4, IRX2 | 5.38 | 3.48 × 10−6 | 0.98 | 0.28 |
| cg12160320 | chr19:8279608 | CERS4, CD320 | −5.76 | 4.74 × 10−6 | −2.04 | 0.04 |
| cg13468767 | chr17:9672024 | DHRS7C, USP43 | −2.69 | 5.60 × 10−6 | −0.53 | 0.27 |
| cg27110655 | chr19:48259098 | GLTSCR2, SEPW1 | 6.90 | 6.39 × 10−6 | 1.39 | 0.26 |
| cg14408927 | chr8:18875728 | PSD3 | 4.57 | 7.96 × 10−6 | 1.90 | 0.02 |
| cg17268483 | chr15:50477521 | SLC27A2, HDC | 13.02 | 8.48 × 10−6 | 5.07 | 0.03 |
| cg02064414 | chr14:23122560 | OXA1L, OR6J1 | 4.24 | 9.69 × 10−6 | 0.97 | 0.21 |
| B: Longitudinal EM performance. | ||||||
| CpG | Location | Gene annotation* | Effectlongitudinal | plongitudinal | Effectcross-sect. | pcross-sect. |
| cg08891989 | chr7:5272846 | WIPI2, SLC29A4 | 8.96 | 2.56 × 10−7 | 5.19 | 0.02 |
| ch.13.1159947F | chr13:78677888 | EDNRB, POU4F1 | −16.43 | 5.05 × 10−7 | −5.90 | 0.16 |
| cg16525470 | chr19:10983625 | CARM1, YIPF2 | −5.50 | 7.75 × 10−7 | −5.03 | 7.22 × 10−4 |
| cg19857541 | chr3:108836800 | MORC1 | 9.13 | 1.05 × 10−6 | 5.93 | 0.01 |
| cg05275832 | chr2:27984686 | SUPT7L, MRPL33 | −4.87 | 1.11 × 10−6 | −2.89 | 0.03 |
| cg14744604 | chr14:93559541 | ITPK1, CHGA | −5.50 | 2.50 × 10−6 | −1.36 | 0.36 |
| cg19531475 | chr13:24472454 | C1QTNF9B | −7.04 | 4.55 × 10−6 | −4.28 | 0.02 |
| cg21390166 | chr22:21455845 | BCRP2, SLC7A4, GGT2 | −4.13 | 4.76 × 10−6 | −1.84 | 0.10 |
| cg18632612 | chr8:140116347 | COL22A1, KCNK9 | 3.70 | 6.21 × 10−6 | 1.24 | 0.25 |
| cg19100344 | chr1:231372247 | C1orf131, GNPAT | 4.00 | 7.50 × 10−6 | 1.48 | 0.21 |
| cg16655166 | chr2:8063199 | LOC339788, ID2 | 5.04 | 7.96 × 10−6 | −0.08 | 0.96 |
| cg27184903 | chr15:29285727 | APBA2, NDNL2 | 6.01 | 8.79 × 10−6 | 6.22 | 3.93 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sommerer, Y.; Dobricic, V.; Schilling, M.; Ohlei, O.; Bartrés-Faz, D.; Cattaneo, G.; Demuth, I.; Düzel, S.; Franzenburg, S.; Fuß, J.; et al. Epigenome-Wide Association Study in Peripheral Tissues Highlights DNA Methylation Profiles Associated with Episodic Memory Performance in Humans. Biomedicines 2022, 10, 2798. https://doi.org/10.3390/biomedicines10112798
Sommerer Y, Dobricic V, Schilling M, Ohlei O, Bartrés-Faz D, Cattaneo G, Demuth I, Düzel S, Franzenburg S, Fuß J, et al. Epigenome-Wide Association Study in Peripheral Tissues Highlights DNA Methylation Profiles Associated with Episodic Memory Performance in Humans. Biomedicines. 2022; 10(11):2798. https://doi.org/10.3390/biomedicines10112798
Chicago/Turabian StyleSommerer, Yasmine, Valerija Dobricic, Marcel Schilling, Olena Ohlei, David Bartrés-Faz, Gabriele Cattaneo, Ilja Demuth, Sandra Düzel, Sören Franzenburg, Janina Fuß, and et al. 2022. "Epigenome-Wide Association Study in Peripheral Tissues Highlights DNA Methylation Profiles Associated with Episodic Memory Performance in Humans" Biomedicines 10, no. 11: 2798. https://doi.org/10.3390/biomedicines10112798
APA StyleSommerer, Y., Dobricic, V., Schilling, M., Ohlei, O., Bartrés-Faz, D., Cattaneo, G., Demuth, I., Düzel, S., Franzenburg, S., Fuß, J., Lindenberger, U., Pascual-Leone, Á., Sabet, S. S., Solé-Padullés, C., Tormos, J. M., Vetter, V. M., Wesse, T., Franke, A., Lill, C. M., & Bertram, L. (2022). Epigenome-Wide Association Study in Peripheral Tissues Highlights DNA Methylation Profiles Associated with Episodic Memory Performance in Humans. Biomedicines, 10(11), 2798. https://doi.org/10.3390/biomedicines10112798







