Relationship between Glutathione-Dependent Enzymes and the Immunohistochemical Profile of Glial Neoplasms
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Measurement of Tumor Markers
2.3. Preparation of Tissue Homogenates for Biochemical Research
2.4. Analysis of Glutathione Metabolism Parameters
2.5. Analysis of Protein-Protein Interactions
2.6. Statistical Analysis
3. Results
3.1. The Level of Molecular Genetic Markers of Gliomas with Varying Degree of Anaplasia
3.2. Protein-Protein Interactions between Immunohistochemical Markers of Gliomas and Glutathione Metabolism Enzymes
3.3. Metabolic Parameters of Glutathione and Reactive Oxygen Species Production Depending on the Glial Tumor Zone
3.4. Characterization of Glutathione Metabolism as a Function of the Immunohistochemical Profile of Gliomas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corso, C.R.; Acco, A. Glutathione system in animal model of solid tumors: From regulation to therapeutic target. Crit. Rev. Oncol. Hematol. 2018, 128, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.; Sandru, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; WHO Classification of Tumours Editorial Board. Central nervous system tumours. Int. Agency Res. Cancer 2021, 5, 6. [Google Scholar]
- Dimitrov, L.; Hong, C.S.; Yang, C.; Zhuang, Z.; Heiss, J.D. New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int. J. Med. Sci. 2015, 12, 201–213. [Google Scholar] [CrossRef]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef]
- Tamura, R.E.; Vasconcellos, J.F.; Sarkar, D.; Libermann, T.A.; Fisher, P.B.; Zerbini, L.F. GADD45 proteins: Central players in tumorigenesis. Curr. Mol. Med. 2012, 12, 634–651. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Strasser, A.; Kelly, G.L. Tumor-Suppressor Functions of the TP53 Pathwa. Cold Spring Harb. Perspect. Med. 2016, 6, a026062. [Google Scholar] [CrossRef]
- Bullwinkel, J.; Baron-Lühr, B.; Lüdemann, A.; Wohlenberg, C.; Gerdes, J.; Scholzen, T. Ki67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J. Cell. Physiol. 2006, 206, 624–635. [Google Scholar] [CrossRef]
- Theresia, E.; Malueka, R.G.; Pranacipta, S.; Kameswari, B.; Dananjoyo, K.; Asmedi, A.; Wicaksono, A.S.; Hartanto, R.A.; Dwianingsih, E.K. Association between Ki67 Labeling index and Histopathological Grading of Glioma in Indonesian Population. Asian Pac. J. Cancer Prev. 2020, 21, 1063–1068. [Google Scholar] [CrossRef]
- Yoda, R.A.; Marxen, T.; Longo, L.; Ene, C.; Wirsching, H.G.; Keene, C.D.; Holland, E.C.; Cimino, P.J. Mitotic Index Thresholds Do Not Predict Clinical Outcome for IDH-Mutant Astrocytoma. J. Neuropathol. Exp. Neurol. 2019, 78, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Shiota, S.; Collier, J.; Foote, R.S.; Mitra, S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc. Nat. Acad. Sci. USA 1990, 87, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Leung, S.C.Y.; Nielsen, T.O.; Zabaglo, L.; Arun, I.; Badve, S.S.; Bane, A.L.; Bartlett, J.M.S.; Borgquist, S.; Chang, M.C.; Dodson, A.; et al. Analytical validation of a standardized scoring protocol for Ki67: Phase 3 of an international multicenter collaboration. NPJ Breast Cancer 2016, 2, 16014. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.A.G.; Bangso, J.A.; Lindahl, K.H.; Dahlrot, R.H.; Hjelmborg, J.B.; Hansen, S.; Kristensen, B.W. Evaluation of the proliferation marker Ki67 in gliomas: Interobserver variability and digital quantification. Diagn. Pathol. 2018, 3, 8. [Google Scholar] [CrossRef]
- Younis, S.G.; Khedr, R.A.G.; El-Shorbagy, S.H. Immunohistochemical analysis of O6-methylguanine-DNA methyltransferase (MGMT) protein expression as prognostic marker in glioblastoma patients treated with radiation therapy with concomitant and adjuvant Temozolomide. J. Egypt. Natl. Cancer Inst. 2016, 28, 23–30. [Google Scholar] [CrossRef][Green Version]
- Sánchez-Pérez, Y.; Soto-Reyes, E.; García-Cuellar, C.M.; Cacho-Díaz, B.; Santamaría, A.; Rangel-López, E. Role of Epigenetics and Oxidative Stress in Gliomagenesis. CNS Neurol. Disord.-Drug Targets 2017, 16, 1090–1098. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Sosa, V.; Moliné, T.; Somoza, R.; Paciucci, R.; Kondoh, H.; Leonart, M.E. Oxidative stress and cancer: An overview. Ageing Res. Rev. 2013, 12, 376–390. [Google Scholar] [CrossRef]
- Galadari, S.; Rahman, A.; Pallichankandy, S.; Thayyullathil, F. Reactive oxygen species and cancer paradox: To promote or to suppress? Free Radic. Bio. Med. 2017, 104, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Et Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Quintana-Cabrera, R.; Bolaños, J.P. Glutathione and γ-glutamylcysteine in hydrogen peroxide detoxification. Methods Enzymol. 2013, 527, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Dawane, J.S.; Pandit, V.A. Understanding redox homeostasis and its role in cancer. J. Clin. Diagn. Res. JCDR 2012, 6, 1796–1802. [Google Scholar] [CrossRef]
- Wu, B.; Dong, D. Human cytosolic glutathione transferases: Structure, function, and drug discovery. Trends Pharmacol. Sci. 2012, 33, 656–668. [Google Scholar] [CrossRef]
- Tikhanovich, I.; Cox, J.; Weinman, S.A. Forkhead box class O transcription factors in liver function and disease. J. Gastroenterol. Hepatol. 2013, 28, 125–131. [Google Scholar] [CrossRef]
- Klaus, A.; Zorman, S.; Berthier, A.; Polge, C.; Ramirez, S.; Michelland, S.; Sève, M.; Vertommen, D.; Rider, M.; Lentze, N.; et al. Glutathione S-transferases interact with AMP-activated protein kinase: Evidence for S-glutathionylation and activation in vitro. PLoS ONE 2013, 8, e62497. [Google Scholar] [CrossRef]
- Dong, S.; Sha, H.; Xu, X.; Hu, T.; Lou, R.; Li, H.; Wu, J.; Dan, C.; Feng, J. Glutathione S-transferase π: A potential role in antitumor therapy. Drug Des. Res. Pract. Med. J. 2021, 8, 12–22. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Et Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Di Ilio, C.; Federici, L. Glutathione transferases: Substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Gu, X.; Alvarado, A.G.; Du, Y.; Luo, S.; Ahn, E.H.; Kang, S.S.; Ji, B.; Liu, X.; Mao, H.; et al. Discovery of a dual inhibitor of NQO1 and GSTP1 for treating glioblastoma. J. Hematol. Oncol. 2020, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, C.A.; Doyle, S.; Brunner, C.; Dunne, A.; Brint, E.; Wietek, C.; Walch, E.; Wirth, T.; O’Neill, L.A. Bruton’s tyrosine kinase is a Toll/interleukin-1 receptor domain-binding protein that participates in nuclear factor kappaB activation by Toll-like receptor 4. J. Biol. Chem. 2003, 278, 26258–26264. [Google Scholar] [CrossRef] [PubMed]
- Krell, D.; Assoku, M.; Galloway, M.; Mulholland, P.; Tomlinson, I.; Bardella, C. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PLoS ONE 2011, 6, e19868. [Google Scholar] [CrossRef]
- Ohka, F.; Ito, M.; Ranjit, M.; Senga, T.; Motomura, A.; Motomura, K.; Saito, K.; Kato, K.; Kato, Y.; Wakabayashi, T.; et al. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 5911–5920. [Google Scholar] [CrossRef]
- Peeters, T.H.; Lenting, K.; Breukels, V.; van Lith, S.; van den Heuvel, C.; Molenaar, R.; van Rooij, A.; Wevers, R.; Span, P.N.; Heerschap, A.; et al. Isocitrate dehydrogenase 1-mutated cancers are sensitive to the green tea polyphenol epigallocatechin-3-gallate. Cancer Metab. 2019, 7, 4. [Google Scholar] [CrossRef]
- Mustafa, D.A.N.; Swagemakers, S.M.; Buise, L.; Spek, P.J.; Kros, J.M. Metabolic alterations due to IDH1 mutation in glioma: Opening for therapeutic opportunities? Acta Neuropathol. Commun. 2014, 2, 3. [Google Scholar] [CrossRef]
- Zhu, Z.; Du, S.; Du, Y.; Ren, J.; Ying, G.; Yan, Z. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 2018, 144, 93–104. [Google Scholar] [CrossRef]
- Dokic, I.; Hartmann, C.; Herold-Mende, C.; Régnier-Vigouroux, A. Glutathione peroxidase 1 activity dictates the sensitivity of glioblastoma cells to oxidative stress. Glia 2012, 60, 1785–1800. [Google Scholar] [CrossRef]
- Tang, X.; Fu, X.; Liu, Y.; Yu, D.; Cai, S.J.; Yang, C. Blockade of Glutathione Metabolism in IDH1-Mutated Glioma. Mol. Cancer Ther. 2020, 19, 221–230. [Google Scholar] [CrossRef]
- Lv, S.; Luo, H.; Huang, K.; Zhu, X. The Prognostic Role of Glutathione Peroxidase 1 and Immune Infiltrates in Glioma Investigated Using Public Datasets. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e926440. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.W.; Stephenson, L.; Cao, X.; Milas, M.; Pollock, R.; Ali-Osman, F. Identification and functional characterization of the human glutathione S-transferase P1 gene as a novel transcriptional target of the p53 tumor suppressor gene. Mol. Cancer Res. 2008, 6, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Pandith, A.A.; Qasim, I.; Zahoor, W.; Shah, P.; Bhat, A.R.; Sanadhya, D.; Shah, Z.A.; Naikoo, N.A. Concordant association validates MGMT methylation and protein expression as favorable prognostic factors in glioma patients on alkylating chemotherapy (Temozolomide). Sci. Rep. 2018, 8, 6704. [Google Scholar] [CrossRef]
- Dahlrot, R.H.; Larsen, P.; Boldt, H.B.; Kreutzfeldt, M.S.; Hansen, S.; Hjelmborg, J.B.; Kristensen, B.W. Posttreatment Effect of MGMT Methylation Level on Glioblastoma Survival. J. Neuropathol. Exp. Neurol. 2019, 78, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Natsume, A. Overview of DNA methylation in adult diffuse gliomas. Brain Tumor Pathol. 2019, 36, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Hatem, E.; El Banna, N.; Huang, M.E. Multifaceted Roles of Glutathione and Glutathione-Based Systems in Carcinogenesis and Anticancer Drug Resistance. Antioxid. Redox Signal. 2017, 27, 1217–1234. [Google Scholar] [CrossRef]
- Desideri, E.; Ciccarone, F.; Ciriolo, M.R. Targeting Glutathione Metabolism: Partner in Crime in Anticancer Therapy. Nutrients 2019, 11, 1926. [Google Scholar] [CrossRef]
- Ichimura, K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012, 29, 131–139. [Google Scholar] [CrossRef]
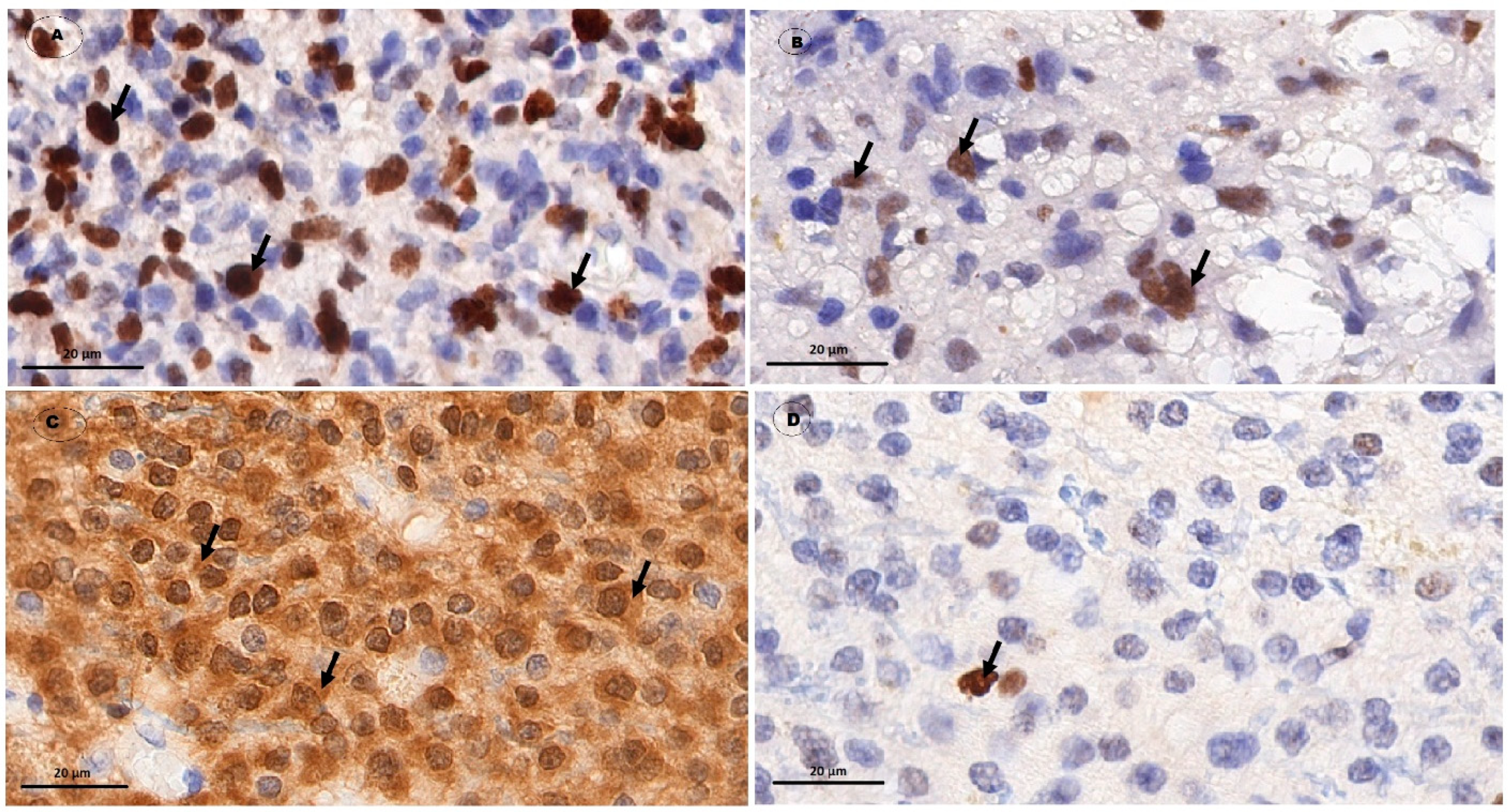





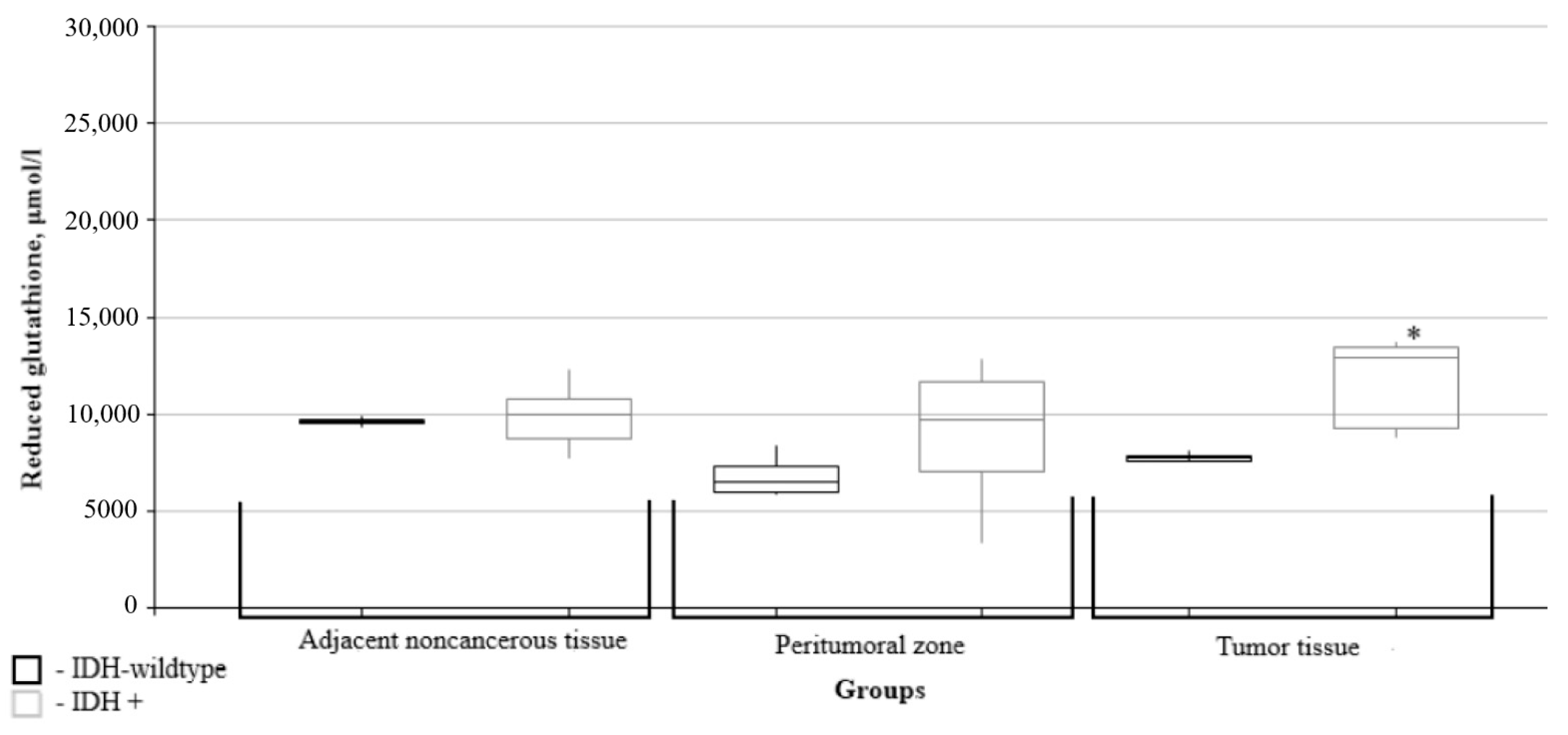



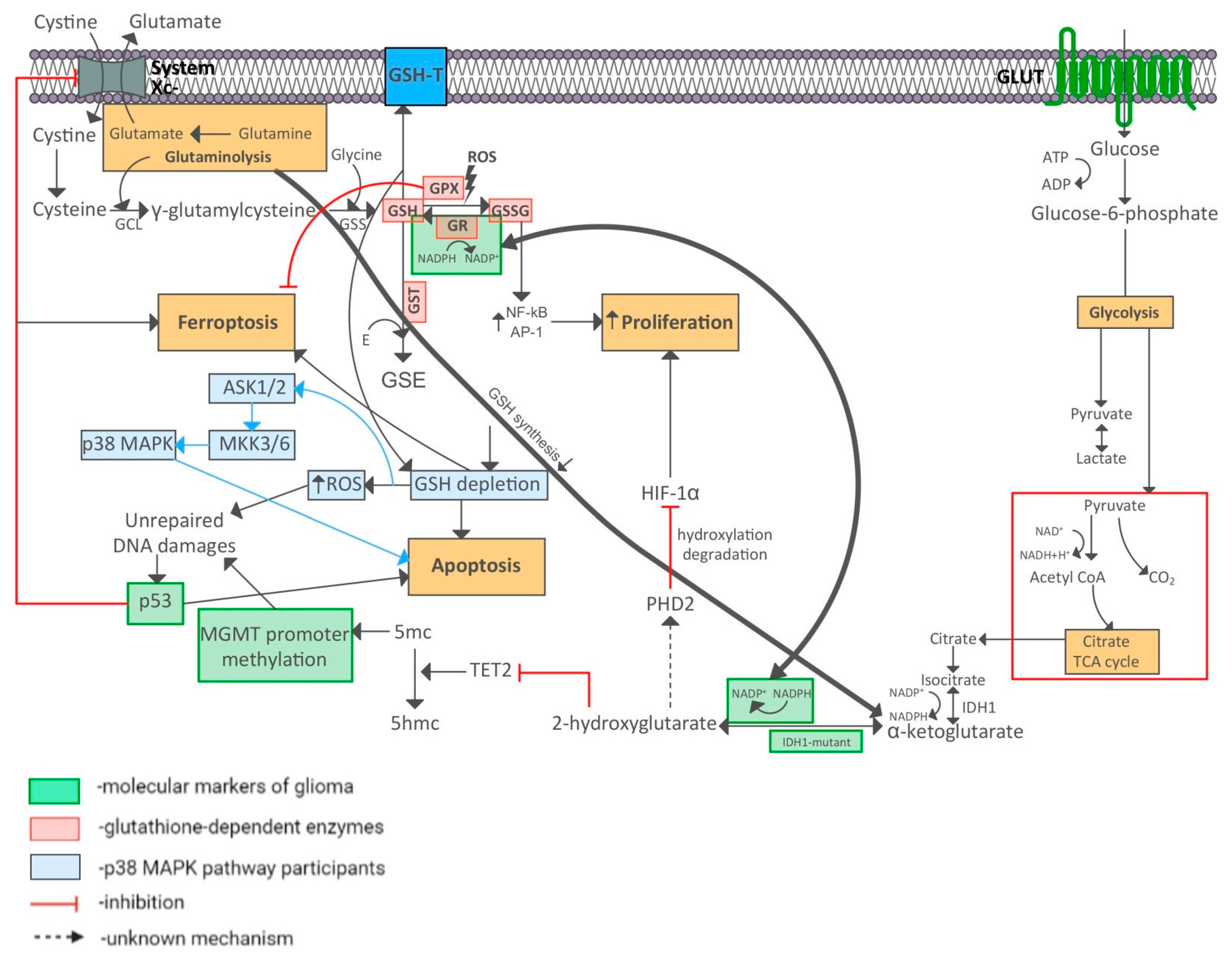
| Grade | I | II | III | IV |
|---|---|---|---|---|
| Number of patients per group | 1 | 6 | 3 | 10 |
| IDH1—% of mutation detection | 0% | 100% | 100% | 30% |
| MGMT—% of methylation detection | 100% | 83% | 100% | 30% |
| p53—% of detection | 0% | 66% | 0% | 80% |
| High Ki67 (>10%)—% of detection | 0% | 0% | 100% | 100% |
| Low Ki67 (<10%)—% of detection | 100% | 100% | 0% | 0% |
| Ki67 median | - | 2% | 12% | 37% |
| Ki67 interquartile intervals (25–75) | - | 1.5–3% | - | 25–40% |
| Glutathione Metabolism Parameter | Adjacent Noncancerous Tissues (Median; Quartiles) | Peritumoral Zone | Tumor | |||
|---|---|---|---|---|---|---|
| Median; Quartiles | Mann-Whitney U-Test | Median; Quartiles | Mann-Whitney U-Test | |||
| Oxidized glutathione, μmol/l | Grade I (n = 1) | 12,583.00 | 13,636.00 | 10,532.00 | ||
| Grade II, (n = 6) | 11,514.50 (10,897.465– 12,141.75) | 14,359.00 (13,350.75–15,753.50) | * p = 0.054 | 11,430.50 (9772.00–12,059.25) | p = 0.631 | |
| Grade III, (n = 3) | 12,256.00 (11,191.00– 12,661.04) | 8137.00 (8124.79 9019.50) | * p = 0.049 | 4013.00 (3635.89 4975.50) | * p = 0.050 | |
| Grade IV, (n = 10) | 11,363.33 (11,146.00– 11,853.17) | 9021.00 (8462.97–9837.00) | * p = 0.007 | 6854.65 (6464.75–7006.58) | * p = 0.004 | |
| Reduced glutathione, μmol/l | Grade I (n = 1) | 11,239.00 | 15,210.00 | 1323.00 | ||
| Grade II, (n = 6) | 9342.00 (8783.75–9922.75) | 10,639.50 (7043.55–11,928.00) | p = 0.522 | 14,132.00 (13,294.81–14,675.25) | * p = 0.003 | |
| Grade III, (n = 3) | 10,396.00 (10,033.00–11,347.78) | 6921.00 (5174.08–7725.50) | * p = 0.049 | 7813.00 (7059.00–8641.78) | * p = 0.049 | |
| Grade IV, (n = 10) | 9705.62 (9417.06– 9970.00) | 8004.50 (6478.75–8315.22) | p = 0.109 | 8006.50 (7777.00–8636.50) | * p = 0.054 | |
| Glutathione peroxidase, µg/g of protein | Grade I (n = 1) | 8.25 | 24.71 | 21.30 | ||
| Grade II, (n = 6) | 6.84 (5.89–7.44) | 20.00 (19.25–21.50) | * p = 0.004 | 3.79 (3.52–4.73) | * p = 0.010 | |
| Grade III, (n = 3) | 6.43 (4.33–7.06) | 3.88 (2.89–4.07) | p = 0.275 | 3.78 (2.43–5.12) | p = 0.513 | |
| Grade IV, (n = 10) | 7.72 (7.06–12.89) | 3.22 (2.56–4.37) | * p = 0.039 | 2.63 (1.41–3.78) | * p = 0.0002 | |
| Glutathione reductase, µg/g of protein | Grade I (n = 1) | 2105.00 | 3179.00 | 2769.23 | ||
| Grade II, (n = 6) | 1802.00 (1625.25–1872.17) | 3188.50 (3029.00–3452.25) | * p = 0.007 | 1536.49 (1351.56–1698.41) | p = 0.149 | |
| Grade III, (n = 3) | 1750.28 (1657.14–1810.64 | 973.00 (569.84–987.00) | * p = 0.049 | 1134.00 (895.02–1338.36) | * p = 0.049 | |
| Grade IV, (n = 10) | 1894.00 (1482.49–2252.00) | 994.35 (685.42–1739.20) | p = 0.065 | 907.79 (420.39– 1374.25) | * p = 0.011 | |
| Glutathione transferase, µg/g of protein | Grade I (n = 1) | 28.25 | 78.19 | 67.31 | ||
| Grade II, (n = 6) | 20.07 (16.60–22.08) | 70.70 (66.60 –74.61) | * p = 0.004 | 54.47 (41.82–61.49) | * p = 0.007 | |
| Grade III, (n = 3) | 27.00 (26.02–28.50) | 19.00 (18.52–21.00) | * p = 0.005 | 15.00 (13.48–24.17) | p = 0.513 | |
| Grade IV, (n = 10) | 35.17 (32.89–55.20) | 33.24 (23.21–40.51) | p = 0.064 | 12.09 (7.37–16.32) | * p = 0.001 | |
| Parameter of Glutathione Metabolism | Markers of Gliomas | ||||
|---|---|---|---|---|---|
| IDH | Ki67 | MGMT | p53 | ||
| Oxidized glutathione Rho (p) | Adjacent noncancerous tissue | 0.598 (0.052) * | −0.260 (0.392) | 0.169 (0.689) | −0.173 (0.656) |
| Peritumoral zone | 0.570 (0.086) | −0.534 (0.060) | 0.169 (0.689) | 0.087 (0.825) | |
| Tumor tissue | 0.597 (0.051) * | −0.674 (0.012) * | 0.169 (0.689) | 0.260 (0.500) | |
| Reduced glutathione Rho (p) | Adjacent noncancerous tissue | 0.239 (0.479) | −0.273 (0.391) | 0.056 (0.895) | −0.346 (0.361) |
| Peritumoral zone | 0.359 (0.279) | −0.550 (0.051) * | 0.056 (0.,895) | 0.327 (0.429) | |
| Tumor tissue | 0.837 (0.001) * | −0.765 (0.002) * | 0.169 (0.689) | −0.173 (0.656) | |
| Glutathione peroxidase Rho (p) | Adjacent noncancerous tissue | −0.210 (0.536) | 0.359 (0.252) | −0.510 (0.197) | −0.433 (0.244) |
| Peritumoral zone | 0.570 (0.049) * | −0.616 (0.025) * | 0.056 (0.895) | −0.087 (0.825) | |
| Tumor tissue | −0.179 (0.598) | −0.282 (0.351) | 0.394.(0.334) | 0.519 (0.051) * | |
| Glutathione reductase Rho (p) | Adjacent noncancerous tissue | −0.119 (0.726) | 0.166 (0.588) | −0.056 (0.895) | −0.606 (0.043) * |
| Peritumoral zone | 0.538 (0.048) * | −0.338 (0.283) | −0.507 (0.199) | 0.001 (0.999) | |
| Tumor tissue | −0.179 (0.598) | −0.268 (0.376) | 0.394.(0.334) | −0.086 (0.824) | |
| Glutathione transferase Rho (p) | Adjacent noncancerous tissue | −0.120 (0.726) | 0.663 (0.014) * | −0.158 (0.735) | −0.606 (0.043) * |
| Peritumoral zone | 0.359 (0.279) | −0.531 (0.062) | −0.282 (0.244) | 0.001 (0.999) | |
| Tumor tissue | 0.119 (0.726) | −0.597 (0.031) * | 0.620 (0.012) * | −0.087 (0.825) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obukhova, L.; Kopytova, T.; Murach, E.; Shchelchkova, N.; Kontorshchikova, C.; Medyanik, I.; Orlinskaya, N.; Grishin, A.; Kontorshchikov, M.; Badanina, D. Relationship between Glutathione-Dependent Enzymes and the Immunohistochemical Profile of Glial Neoplasms. Biomedicines 2022, 10, 2393. https://doi.org/10.3390/biomedicines10102393
Obukhova L, Kopytova T, Murach E, Shchelchkova N, Kontorshchikova C, Medyanik I, Orlinskaya N, Grishin A, Kontorshchikov M, Badanina D. Relationship between Glutathione-Dependent Enzymes and the Immunohistochemical Profile of Glial Neoplasms. Biomedicines. 2022; 10(10):2393. https://doi.org/10.3390/biomedicines10102393
Chicago/Turabian StyleObukhova, Larisa, Tatiana Kopytova, Elena Murach, Natalya Shchelchkova, Claudia Kontorshchikova, Igor Medyanik, Natalia Orlinskaya, Artem Grishin, Michael Kontorshchikov, and Dariya Badanina. 2022. "Relationship between Glutathione-Dependent Enzymes and the Immunohistochemical Profile of Glial Neoplasms" Biomedicines 10, no. 10: 2393. https://doi.org/10.3390/biomedicines10102393
APA StyleObukhova, L., Kopytova, T., Murach, E., Shchelchkova, N., Kontorshchikova, C., Medyanik, I., Orlinskaya, N., Grishin, A., Kontorshchikov, M., & Badanina, D. (2022). Relationship between Glutathione-Dependent Enzymes and the Immunohistochemical Profile of Glial Neoplasms. Biomedicines, 10(10), 2393. https://doi.org/10.3390/biomedicines10102393





