Genetic Variations miR-10aA>T, miR-30cA>G, miR-181aT>C, and miR-499bA>G and the Risk of Recurrent Pregnancy Loss in Korean Women
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Antibody Preparation
2.3. Chromosome Analysis
2.4. Genotyping
2.5. Assessment of Plasminogen Activator Inhibitor (PAI-1), Homocysteine, Total Cholesterol, Uric Acid Levels, and Blood Coagulation Status
2.6. Statistical Analyses
2.7. Expression Vector Construction (miR-10aA>T, miR-30cA>G, and miR-181aT>C)
2.8. Quantitative Real-Time PCR (miR-10a, miR-30c, miR-181a Pre- and Mature-Form Primers)
2.9. Prediction of miRNA Binding and Luciferase Reporter Assay
3. Results
3.1. Baseline Characteristics of Recurrent Pregnancy Loss Patients and Control Subjects
3.2. Genotype Frequencies of miRNA Polymorphisms According to the Number of Recurrent Pregnancy Losses
3.3. Adjusted Odds Ratios for Risk of Recurrent Pregnancy Loss Associated with miRNA Polymorphisms Combined with Clinical Factors
3.4. Combination Analysis of miRNA Polymorphisms between Recurrent Pregnancy Loss Patients and Control Subjects
3.5. Allele Combination Analysis of miRNA Polymorphisms in Recurrent Pregnancy Loss Patients and Control Subjects
3.6. Differential Expression of the miR-10aA>T, miR-30cA>G, and miR-499bA>G Polymorphisms
3.7. Differences of Various Clinical Parameters According to miRNA Polymorphisms in RPL Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coulam, C.B.; Coulam, C.B.; Clark, D.A.; Beer, A.E.; Kutteh, W.H.; Kwak, J.; Stephenson, M. Clinical Guidelines Recommendation Committee for Diagnosis and Treatment of Recurrent Spontaneous Abortion. Current clinical options for diagnosis and treatment of recurrent spontaneous abortion. Am. J. Reprod. Immunol. 1997, 38, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Regan, L. Recurrent miscarriage. Lancet 2006, 368, 601–611. [Google Scholar] [CrossRef]
- Sierra, S.; Stephenson, M. Genetics of Recurrent Pregnancy Loss. Semin. Reprod. Med. 2006, 24, 017–024. [Google Scholar] [CrossRef] [PubMed]
- Marjorie, P.P.; Perron, M.P.; Provost, P. Protein interactions and complexes in human microRNA biogenesis and function. Front. Biosci. 2008, 13, 2537–2547. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Cuellar, T.L.; McManus, M.T. MicroRNAs and endocrine biology. J. Endocrinol. 2005, 187, 327–332. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Shiekhattar, R. MicroRNA Biogenesis: Isolation and Characterization of the Microprocessor Complex. MicroRNA Protoc. 2006, 342, 33–48. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short Hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.S.; Biobel, G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 1993, 365, 661–663. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, A.S.; Miller, S.; Haines, N.; Zink, M.C.; Serra, M.J. Comprehensive thermodynamic analysis of 3’ double-nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006, 34, 3338–3344. [Google Scholar] [CrossRef] [PubMed]
- Song, F.J.; Chen, K.X. Single-nucleotide polymorphisms among microRNA: Big effects on cancer. Chin. J. Cancer 2011, 30, 381–391. [Google Scholar] [CrossRef]
- Imbar, T.; Eisenberg, I. Regulatory role of microRNAs in ovarian function. Fertil. Steril. 2014, 101, 1524–1530. [Google Scholar] [CrossRef]
- Iwai, N.; Naraba, H. Polymorphisms in human pre-miRNAs. Biochem. Biophys. Res. Commun. 2005, 331, 1439–1444. [Google Scholar] [CrossRef]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar] [CrossRef]
- He, X.-Y.; Chen, J.-X.; Zhang, Z.; Li, C.-L.; Peng, Q.; Peng, H.-M. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J. Cancer Res. Clin. Oncol. 2010, 136, 1023–1028. [Google Scholar] [CrossRef]
- Toloubeydokhti, T.; Bukulmez, O.; Chegini, N. Potential Regulatory Functions of MicroRNAs in the Ovary. Semin. Reprod. Med. 2008, 26, 469–478. [Google Scholar] [CrossRef]
- Medeiros, L.A.; Dennis, L.M.; Gill, M.E.; Houbaviy, H.; Markoulaki, S.; Fu, D.; White, A.C.; Kirak, O.; Sharp, P.A.; Page, D.C.; et al. Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA 2011, 108, 14163–14168. [Google Scholar] [CrossRef] [Green Version]
- Butz, H.; Likó, I.; Czirják, S.; Igaz, P.; Korbonits, M.; Rácz, K.; Patócs, A. MicroRNA profile indicates downregulation of the TGF pathway in sporadic non-functioning pituitary adenomas. Pituitary 2011, 14, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Dien, G.T.; Smith, R.A.; Haupt, L.M.; Griffiths, L.R.; Nguyen, H.T. Genetic polymorphisms inmiRNAs targeting the estrogen receptor and their effect on breast cancer risk. Meta Gene 2014, 2, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, X.; Taylor, H. MicroRNA and gynecological reproductive diseases. Fertil. Steril. 2014, 101, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I. MicroRNA Control of TGF-β Signaling. Int. J. Mol. Sci. 2018, 28, 19. [Google Scholar] [CrossRef] [PubMed]
- Knight, P.G.; Glister, C. TGF-beta superfamily members and ovarian follicle development. Reproduction 2006, 132, 191–206. [Google Scholar] [CrossRef]
- Dong, K.; Xu, Y.; Yang, Q.; Shi, J.; Jiang, J.; Chen, Y.; Song, C.; Wang, K. Associations of functional microRNA binding site polymorphisms in IL23/Th17 inflammatory pathway genes with gastric cancer risk. Mediat. Inflamm. 2017, 6974696. [Google Scholar] [CrossRef]
- Li, C.; Zhu, H.; Bai, W.; Su, L.-L.; Liu, J.-Q.; Cai, W.-X.; Zhao, B.; Gao, J.-X.; Han, S.-C.; Li, J.; et al. MiR-10a and miR-181c regulate collagen type I generation in hypertrophic scars by targeting PAI-1 and uPA. FEBS Lett. 2015, 589, 380–389. [Google Scholar] [CrossRef]
- Ryu, C.S.; Sakong, J.H.; Ahn, E.H.; Kim, J.O.; Ko, D.; Kim, J.H.; Lee, W.S.; Kim, N.K. Association study of the three functional polymorphisms (TAS2R46G>A, OR4C16G>A, and OR4X1A>T) with recurrent pregnancy loss. Genes Genom. 2019, 41, 61–70. [Google Scholar] [CrossRef]
- Wang, H.; Wu, S.; Wu, J.; Sun, S.; Wu, S.; Bao, W. Association analysis of the SNP (rs345476947) in the FUT2 gene with the production and reproductive traits in pigs. Genes Genom. 2018, 40, 199–206. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-Dimensionality Reduction Reveals High-Order Interactions among Estrogen-Metabolism Genes in Sporadic Breast Cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Teague, E.M.C.O.; Print, C.; Hull, M.L. The role of microRNAs in endometriosis and associated reproductive conditions. Hum. Reprod. Updat. 2010, 16, 142–165. [Google Scholar] [CrossRef] [PubMed]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Moszyńska, A.; Gebert, M.; Collawn, J.F.; Bartoszewski, R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017, 7, 170019. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Agodi, A. The Role of miRNAs as Biomarkers for Pregnancy Outcomes: A Comprehensive Review. J. Genom. 2017, 2017, 8067972. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Peng, Y.; Hu, X.; Xu, J.; Zhu, S.; Yu, Z.; Han, S. miR-30c regulates proliferation, apoptosis and differentiation via the Shh signal-ing pathway in P19 cells. Exp. Mol. Med. 2016, 29, e248. [Google Scholar] [CrossRef]
- Bider, D.; Dulitzky, M.; Goldenberg, M.; Lipitz, S.; Mashiach, S. Intraumbilical vein injection of prostaglandin F2α in retained placenta. Eur. J. Obstet. Gynecol. Reprod. Biol. 1996, 64, 59–61. [Google Scholar] [CrossRef]
- Wolff, M.V.; Thaler, C.J.; Strowitzki, T.; Broome, J.; Stolz, W.; Tabibzadeh, S. Regulated expression of cytokines in human endometrium throughout the menstrual cycle: Dysregulation in habitual abortion. Mol. Hum. Reprod. 2000, 6, 627–634. [Google Scholar] [CrossRef]
- Labied, S.; Blacher, S.; Carmeliet, P.; Noël, A.; Frankenne, F.; Foidart, J.-M.; Munaut, C. Transient reduction of placental angiogenesis in PAI-1-deficient mice. Physiol. Genom. 2011, 43, 188–198. [Google Scholar] [CrossRef]
- Fay, W.P.; Parker, A.C.; Condrey, L.R.; Shapiro, A.D. Human plasminogen activator inhibitor-1 (PAI-1) deficiency: Characterization of a large kindred with a null mutation in the PAI-1 gene. Blood 1997, 90, 204–208. [Google Scholar] [CrossRef] [Green Version]
- Gris, J.-C.; Ripart-Neveu, S.; Maugard, C.; Tailland, M.L.; Brun, S.; Courtieu, C.; Biron, C.; Hoffet, M.; Hédon, B.; Marès, P. Respective evaluation of the prevalence of haemostasis abnormalities in unexplained primary early recurrent miscarriages. The Nimes Obstetricians and Haematologists (NOHA) Study. Thromb. Haemost. 1997, 77, 1096–1103. [Google Scholar] [PubMed]
- Kim, Y.K.; Wasser, S.K.; Fujimoto, V.Y.; Klein, N.A.; Moore, D.E.; Soules, M.R. Utility of follicle stimulating hormone (FSH), luteinizing hormone (LH), oestradiol and FSH:LH ratio in predicting reproductive age in normal women. Hum. Reprod. 1997, 12, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Stilley, J.A.W.; Segaloff, D.L. FSH Actions and Pregnancy: Looking Beyond Ovarian FSH Receptors. Endocrinology 2018, 159, 4033–4042. [Google Scholar] [CrossRef] [PubMed]
- Bergen, N.E.; Jaddoe, V.W.; Timmermans, S.; Hofman, A.; Lindemans, J.; Russcher, H.; Raat, H.; Steegers-Theunissen, R.P.; Steegers, E.A. Homocysteine and folate concentrations in early pregnancy and the risk of adverse pregnancy outcomes: The Generation R Study. BJOG 2012, 119, 739–751. [Google Scholar] [CrossRef]


| Characteristics | Controls (n = 281) | RPL Patients (n = 381) | p * |
|---|---|---|---|
| Age (years, mean ± SD) | 33.00 ± 5.73 | 33.03 ± 4.36 | 0.94 |
| BMI (kg/m2, mean ± SD) | 21.58 ± 3.18 | 21.35 ± 4.04 | 0.558 |
| Previous pregnancy losses | None | 3.01 ± 1.50 | |
| Average no. of gestational weeks | 39.28 ± 1.67 | 7.36 ± 1.93 | <0.0001 |
| CD56 NK cells (%, mean ± SD) | None | 18.12 ± 7.98 | |
| Homocysteine (μmol/L, mean ± SD) | None | 6.98 ± 2.10 | |
| Folate (nmol/L, mean ± SD) | None | 14.18 ± 12.01 | |
| Total cholesterol (mg/dL, mean ± SD) | None | 187.73 ± 49.41 | |
| Uric acid (mg/dL, mean ± SD) | 4.19 ± 1.44 | 3.80 ± 0.83 | 0.172 |
| PLT (103/μL, mean ± SD) | 235.17 ± 63.60 | 255.43 ± 59.22 | 0.0007 |
| aPTT (sec, mean ± SD) | 30.77 ± 4.60 | 32.23 ± 4.32 | 0.005 |
| PAI-1 (ng/mL) | None | 10.53 ± 5.72 | |
| BUN (mg/dL) | None | 9.98 ± 2.76 | |
| Creatinine (mg/dL) | None | 0.72 ± 0.12 | |
| FSH (mIU/mL) | 8.11 ± 2.84 | 7.51 ± 10.54 | 0.557 |
| LH (mIU/mL) | 3.32 ± 1.74 | 6.32 ± 12.11 | 0.011 |
| E2 (pg/mL) | 26.00 ± 14.74 | 35.64 ± 29.53 | 0.001 |
| PT (sec, mean ± SD) | 11.53 ± 3.10 | 11.58 ± 0.85 | 0.84 |
| Genotype | Controls (n = 281) | RPL Patients (n = 381) | AOR (95% CI) | p | FDR-p | PL ≥ 3 | AOR (95% CI) | p | FDR-p | PL ≥ 4 | AOR (95% CI) | p | FDR-p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 201) | (n = 81) | ||||||||||||
| miR-10aA>T | |||||||||||||
| AA | 230 (81.9) | 285 (74.8) | 1.000 (reference) | 151 (75.1) | 1.000 (reference) | 60 (74.1) | 1.000 (reference) | ||||||
| AT | 50 (17.8) | 88 (23.1) | 1.420(0.963–2.094) | 0.077 | 0.584 | 44 (21.9) | 1.365 (0.865–2.154) | 0.181 | 0.362 | 19 (23.5) | 1.470 (0.805–2.683) | 0.21 | 0.21 |
| TT | 1 (0.4) | 8 (2.1) | 6.476(0.804–52.176) | 0.079 | 0.237 | 6 (3.0) | 9.484 (1.128–79.759) | 0.038 | 0.274 | 2 (2.5) | 7.931 (0.705–89.228) | 0.094 | 0.156 |
| Dominant (AA vs. AT+TT) | 1.520(1.038–2.227) | 0.032 | 0.709 | 1.524 (0.979–2.372) | 0.062 | 0.124 | 1.595 (0.889–2.862) | 0.117 | 0.156 | ||||
| Recessive (AA+AT vs. TT) | 6.003(0.746–48.285) | 0.092 | 0.276 | 8.847 (1.055–74.186) | 0.045 | 0.631 | 7.206 (0.644–80.641) | 0.109 | 0.156 | ||||
| HWE P | 0.318 | 0.695 | 0.217 | 0.738 | |||||||||
| miR-30cA>G | |||||||||||||
| AA | 106 (37.7) | 163 (42.8) | 1.000 (reference) | 130 (64.7) | 1.000 (reference) | 52 (64.2) | 1.000 (reference) | ||||||
| AG | 144 (51.2) | 182 (47.8) | 0.821(0.591–1.139) | 0.237 | 0.237 | 64 (31.8) | 1.044 (0.647–1.686) | 0.86 | 0.994 | 27 (33.3) | 1.170 (0.640–2.139) | 0.611 | 0.994 |
| GG | 32 (11.0) | 36 (9.4) | 0.742(0.432–1.276) | 0.281 | 0.422 | 7 (3.5) | - | 0.994 | 0.994 | 2 (2.5) | - | 0.994 | 0.994 |
| Dominant (AA vs. AG+GG) | 0.810(0.591–1.111) | 0.191 | 0.191 | 1.162 (0.724–1.864) | 0.534 | 0.994 | 1.262 (0.696–2.288) | 0.443 | 0.994 | ||||
| Recessive (AA+AG vs. GG) | 0.842(0.506–1.400) | 0.507 | 0.606 | - | 0.994 | 0.994 | - | 0.994 | 0.994 | ||||
| HWE P | 0.104 | 0.144 | |||||||||||
| miR-181aT>C | |||||||||||||
| TT | 198 (70.5) | 247 (64.8) | 1.000 (reference) | 79 (39.3) | 1.000 (reference) | 32 (39.5) | 1.000 (reference) | ||||||
| TC | 78 (27.8) | 125 (32.8) | 1.286(0.916–1.805) | 0.147 | 0.221 | 104 (51.7) | 1.376 (0.866–2.185) | 0.177 | 0.223 | 42 (51.9) | 1.403 (0.779–2.525) | 0.259 | 0.345 |
| CC | 5 (1.8) | 9 (2.4) | 1.483(0.488–4.509) | 0.487 | 0.487 | 18 (9.0) | 2.094 (0.812–5.399) | 0.126 | 0.223 | 7 (8.6) | 2.035 (0.621–6.665) | 0.241 | 0.345 |
| Dominant (TT vs. TC+CC) | 1.294(0.929–1.803) | 0.128 | 0.191 | 1.448 (0.925–2.269) | 0.106 | 0.223 | 1.462 (0.826–2.585) | 0.192 | 0.345 | ||||
| Recessive (TT+TC vs. CC) | 1.337(0.443–4.037) | 0.606 | 0.606 | 1.764 (0.707–4.400) | 0.223 | 0.223 | 1.664 (0.548–5.057) | 0.369 | 0.369 | ||||
| HWE P | 0.393 | 0.137 | |||||||||||
| miR-499bA>G | |||||||||||||
| AA | 188 (66.9) | 221 (58.0) | 1.000 (reference) | 116 (57.7) | 1.000 (reference) | 46 (56.8) | 1.000 (reference) | ||||||
| AG | 87 (31.0) | 139 (36.5) | 1.361(0.977–1.896) | 0.068 | 0.204 | 77 (38.3) | 2.037 (1.240–3.347) | 0.005 | 0.01 | 34 (42.0) | 2.274 (1.241–4.168) | 0.008 | 0.016 |
| GG | 6 (2.1) | 21 (5.5) | 2.956(1.168–7.482) | 0.022 | 0.066 | 8 (4.0) | 3.890 (0.767–19.730) | 0.101 | 0.135 | 1 (1.2) | 1.970 (0.152–25.590) | 0.604 | 0.805 |
| Dominant (AA vs. AG+GG) | 1.465(1.062–2.020) | 0.02 | 0.06 | 2.136 (1.314–3.472) | 0.002 | 0.008 | 2.259 (1.240–4.114) | 0.008 | 0.016 | ||||
| Recessive (AA+AG vs. GG) | 2.677(1.066–6.725) | 0.036 | 0.108 | 2.998 (0.605–14.857) | 0.179 | 0.179 | 1.361 (0.111–16.739) | 0.81 | 0.81 | ||||
| HWE P | 0.263 | 0.888 | |||||||||||
| Variable | miR-10aAT + TT | miR-30cAG + GG | miR-181aTC + CC | miR-499bAG + GG | ||||
|---|---|---|---|---|---|---|---|---|
| AOR (95% CI) | p | AOR (95% CI) | p | AOR (95% CI) | p | AOR (95%CI) | p | |
| Age (years) | ||||||||
| <33 | 1.476 (0.870–2.505) | 0.149 | 0.583 (0.371–0.918) | 0.02 | 1.677 (1.038–2.709) | 0.035 | 1.329 (0.848–2.083) | 0.216 |
| ≥33 | 1.566 (0.902–2.717) | 0.111 | 1.124 (0.721–1.754) | 0.606 | 0.996 (0.626–1.583) | 0.985 | 1.631 (1.028–2.588) | 0.038 |
| Homocysteine | ||||||||
| <6.97µmol/L | 1.186 (0.127–11.086) | 0.881 | 0.364 (0.040–3.344) | 0.372 | - | - | 3.063 (0.333–28.174) | 0.323 |
| ≥6.97µmol/L | 1.690 (0.190–15.041) | 0.638 | 0.275 (0.032–2.399) | 0.243 | 0.590 (0.124–2.816) | 0.509 | 0.846 (0.177–4.050) | 0.835 |
| BMI | ||||||||
| <25 kg/m2 | 1.399 (0.928–2.108) | 0.109 | 0.834 (0.593–1.174) | 0.298 | 1.401 (0.979–2.005) | 0.065 | 1.456 (1.029–2.059) | 0.034 |
| ≥25 kg/m2 | 2.840 (1.544–5.223) | 0.001 | 0.949 (0.591–1.524) | 0.829 | 1.194 (0.725–1.967) | 0.485 | 2.284 (1.377–3.789) | 0.001 |
| Platelet | ||||||||
| <255.62 × 103/μL | 1.133 (0.624–2.057) | 0.681 | 1.008 (0.606–1.678) | 0.976 | 1.779 (1.038–3.048) | 0.036 | 1.468 (0.878–2.455) | 0.144 |
| ≥255.62 × 103/μL | 2.019 (0.933–4.370) | 0.075 | 0.539 (0.287–1.011) | 0.054 | 0.820 (0.429–1.569) | 0.55 | 1.256 (0.665–2.369) | 0.483 |
| PT | ||||||||
| ≥11.58 s | 1.476 (0.870–2.505) | 0.149 | 1.845 (0.468–7.277) | 0.382 | 0.557 (0.145–2.139) | 0.394 | 0.368 (0.090–1.514) | 0.166 |
| <11.58 s | 1.566 (0.902–2.717) | 0.111 | 0.699 (0.335–1.458) | 0.339 | 1.031 (0.495–2.151) | 0.935 | 1.023 (0.522–2.006) | 0.947 |
| aPTT | ||||||||
| <32.83 s | 1.476 (0.870–2.505) | 0.149 | 0.364 (0.185–0.717) | 0.004 | 1.714 (0.862–3.409) | 0.125 | 1.409 (0.763–2.604) | 0.273 |
| ≥32.83 s | 1.566 (0.902–2.717) | 0.111 | 0.976 (0.426–2.237) | 0.954 | 1.069 (0.459–2.493) | 0.877 | 0.639 (0.284–1.439) | 0.279 |
| Genotype Combination | Controls (n = 281) | RPL Patients | AOR (95% CI) | pa | FDR-p b |
|---|---|---|---|---|---|
| (n = 381) | |||||
| miR-10aA>T/miR-30cA>G | |||||
| AA/AA | 162 (57.7) | 190 (49.9) | 1.000 (reference) | ||
| AT/AA | 35 (12.5) | 51 (13.4) | 1.243 (0.770–2.008) | 0.374 | 0.499 |
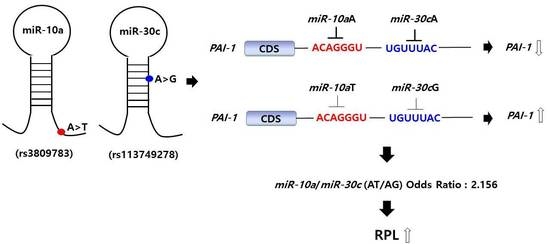
| AT/AG | 14 (5.0) | 35 (9.2) | 2.156 (1.120–4.151) | 0.022 | 0.088 |
| AT/GG | 1 (0.4) | 2 (0.5) | 1.770 (0.159–19.773) | 0.643 | 0.643 |
| TT/AA | 1 (0.4) | 6 (1.6) | 4.958 (0.589–41.748) | 0.141 | 0.282 |
| miR-10aA>T/miR-181aT>C | |||||
| AA/TT | 88 (31.3) | 113 (29.7) | 1.000 (reference) | ||
| AA/TC | 121 (43.1) | 142 (37.3) | 0.915 (0.632–1.324) | 0.637 | 0.812 |
| AA/CC | 21 (7.5) | 30 (7.9) | 1.079 (0.576–2.020) | 0.812 | 0.812 |
| AT/TT | 18 (6.4) | 45 (11.8) | 1.974 (1.065–3.658) | 0.031 | 0.124 |
| AT/TC | 22 (7.8) | 37 (9.7) | 1.272 (0.698–2.317) | 0.433 | 0.812 |
| miR-10aA>T/miR-499A>G | |||||
| AA/AA | 154 (54.8) | 168 (44.1) | 1.000 (reference) | ||
| AA/AG | 71 (25.3) | 102 (26.8) | 1.317 (0.907–1.914) | 0.148 | 0.197 |
| AA/GG | 5 (1.8) | 15 (3.9) | 2.719 (0.964–7.665) | 0.059 | 0.118 |
| AT/AA | 34 (12.1) | 47 (12.3) | 1.264 (0.772–2.069) | 0.351 | 0.351 |
| AT/AG | 15 (5.3) | 36 (9.4) | 2.195 (1.156–4.169) | 0.016 | 0.064 |
| AT/GG | 1 (0.4) | 5 (1.3) | 4.508 (0.520–39.109) | 0.172 | 0.344 |
| TT/AG | 1 (0.4) | 1 (0.3) | 0.922 (0.057–14.874) | 0.954 | 0.954 |
| miR-30cA>G/miR-181aT>C | |||||
| AA/TT | 79 (28.1) | 101 (26.5) | 1.000 (reference) | ||
| AA/TC | 89 (31.7) | 120 (31.5) | 1.058 (0.707–1.583) | 0.784 | 0.784 |
| AA/CC | 30 (10.7) | 26 (6.8) | 0.692 (0.377–1.268) | 0.233 | 0.466 |
| AG/TT | 25 (8.9) | 58 (15.2) | 1.839 (1.054–3.210) | 0.032 | 0.128 |
| AG/TC | 52 (18.5) | 59 (15.5) | 0.886 (0.551–1.425) | 0.617 | 0.784 |
| miR-30cA>G/miR-499A>G | |||||
| AA/AA | 137 (48.8) | 146 (38.3) | 1.000 (reference) | ||
| AA/AG | 57 (20.3) | 83 (21.8) | 1.355 (0.898–2.044) | 0.147 | 0.196 |
| AA/GG | 4 (1.4) | 18 (4.7) | 4.324 (1.423–13.141) | 0.01 | 0.026 |
| AG/AA | 49 (17.4) | 68 (17.8) | 1.319 (0.852–2.041) | 0.214 | 0.214 |
| AG/AG | 27 (9.6) | 55 (14.4) | 1.921 (1.145–3.224) | 0.013 | 0.026 |
| AG/GG | 2 (0.7) | 2 (0.5) | 0.908 (0.124–6.641) | 0.924 | 0.924 |
| miR-181aT>C/miR-499A>G | |||||
| TT/AA | 72 (25.6) | 98 (25.7) | 1.000 (reference) | ||
| TT/AG | 33 (11.7) | 54 (14.2) | 1.155 (0.677–1.970) | 0.597 | 0.796 |
| TT/GG | 1 (0.4) | 11 (2.9) | 8.320 (1.043–66.384) | 0.046 | 0.184 |
| TC/AA | 95 (33.8) | 107 (28.1) | 0.818 (0.542–1.236) | 0.34 | 0.68 |
| TC/AG | 47 (16.7) | 67 (17.6) | 1.040 (0.642–1.685) | 0.872 | 0.872 |
| Allele Combination | Controls | RPL Patients | OR (95% CI) | pa | FDR-p b |
|---|---|---|---|---|---|
| (n = 281) | (n = 381) | ||||
| miR-10aA>T/miR-181aT>C/miR-30cA>G/miR-499A>G | |||||
| A-T-A-A | 236 (41.9) | 271 (35.6) | 1.000 (reference) | ||
| A-T-A-G | 38 (6.9) | 66 (8.6) | 1.468 (0.953–2.263) | 0.085 | 0.128 |
| A-T-G-A | 128 (22.9) | 139 (18.2) | 0.935 (0.695–1.257) | 0.705 | 0.705 |
| A-T-G-G | 27 (4.9) | 63 (8.2) | 1.952 (1.210–3.149) | 0.006 | 0.036 |
| A-C-A-A | 44 (7.8) | 59 (7.7) | 1.163 (0.759–1.784) | 0.516 | 0.619 |
| A-C-A-G | 9 (1.7) | 26 (3.5) | 2.343 (1.111–4.942) | 0.026 | 0.056 |
| A-C-G-A | 12 (2.3) | 31 (4.1) | 2.136 (1.095–4.165) | 0.028 | 0.056 |
| T-T-A-G | 7 (1.2) | 22 (3.0) | 2.851 (1.202–6.764) | 0.014 | 0.056 |
| T-T-G-A | 20 (3.7) | 10 (1.4) | 0.455 (0.215–0.962) | 0.044 | 0.088 |
| T-T-G-G | 3 (0.7) | 1 (0.2) | 0.434 (0.079–2.391) | 0.425 | 0.425 |
| T-C-A-A | 9 (1.6) | 18 (2.3) | 1.735 (0.765–3.937) | 0.235 | 0.313 |
| T-C-G-A | 0 (0.0) | 6 (0.9) | 13.020 (0.739–229.300) | 0.017 | 0.017 |
| aPTT | Creatinine (mg/dL) | E2 (pg/mL) | FSH (mIU/mL) | Hct | Hcy | LH (mIU/mL) | PT | T. Chol (mg/dL) | |
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD |
| miR-10a A>T | |||||||||
| AA | 31.86 ± 4.43 | 1.87 ± 2.75 | 35.07 ± 25.54 | 7.84 ± 11.93 | 36.28 ± 3.90 | 6.92 ± 2.00 | 5.72 ± 7.14 | 11.61 ± 1.87 | 154.54 ± 83.14 |
| AT | 31.39 ± 4.64 | 2.47 ± 3.36 | 38.07 ± 41.91 | 6.30 ± 3.79 | 36.36 ± 4.20 | 7.31 ± 2.37 | 8.38 ± 21.83 | 11.41 ± 0.89 | 140.93 ± 89.67 |
| TT | 31.47 ± 3.42 | 0.73 ± 0.15 | 31.20 ± 9.11 | 9.94 ± 4.91 | 36.63 ± 6.47 | 5.64 ± 1.10 | 5.52 ± 1.60 | 11.80 ± 0.35 | 251.00 ± 118.01 |
| p | 0.719 | 0.53 | 0.837 | 0.642 | 0.974 | 0.228 | 0.435 | 0.696 | 0.079 |
| miR-30c A>G | |||||||||
| AA | 32.46 ± 4.71 | 1.19 ± 1.94 | 43.02 ± 38.58 | 6.96 ± 4.29 | 36.65 ± 3.73 | 6.84 ± 2.00 | 5.41 ± 3.63 | 11.83 ± 2.21 | 172.56 ± 64.85 |
| AG | 31.92 ± 4.11 | 1.55 ± 2.35 | 30.80 ± 19.30 | 6.98 ± 8.47 | 36.27 ± 4.02 | 7.01 ± 2.16 | 6.46 ± 14.43 | 11.41 ± 1.28 | 167.25 ± 79.17 |
| GG | 27.56 ± 3.59 | 6.26 ± 3.71 | 15.10 ± 10.66 | 33.82 ± 55.85 | 34.14 ± 4.49 | 7.73 ± 1.86 | 20.00 ± 32.76 | 11.68 ± 0.83 | 38.15 ± 76.11 |
| p | 0.001 | 0.001 | 0.015 | 0.001 | 0.013 | 0.199 | 0.062 | 0.135 | 0.001 |
| miR-181a T>C | |||||||||
| TT | 31.51 ± 4.45 | 2.38 ± 3.24 | 33.91 ± 21.97 | 6.42 ± 3.17 | 36.26 ± 4.08 | 6.76 ± 2.01 | 4.81 ± 2.74 | 11.43 ± 1.14 | 136.96 ± 86.18 |
| TC | 32.23 ± 4.48 | 1.17 ± 1.76 | 39.85 ± 41.63 | 9.76 ± 17.78 | 36.45 ± 3.70 | 7.36 ± 1.99 | 9.50 ± 20.64 | 11.91 ± 2.48 | 185.65 ± 76.23 |
| CC | 36.05 ± 3.32 | - | 25.67 ± 5.08 | 6.73 ± 1.80 | 31.35 ± 0.64 | 9.98 ± 4.50 | 4.20 ± 0.71 | 10.20 ± 0.28 | - |
| p | 0.17 | 0.011 | 0.409 | 0.115 | 0.19 | 0.001 | 0.038 | 0.048 | 0.001 |
| miR-499bA>G | |||||||||
| AA | 31.20 ± 4.29 | 2.14 ± 3.00 | 35.86 ± 26.02 | 6.84 ± 7.73 | 36.43 ± 3.85 | 7.00 ± 2.06 | 5.37 ± 4.90 | 11.54 ± 1.72 | 144.26 ± 86.46 |
| AG | 32.62 ± 4.69 | 1.82 ± 2.84 | 34.70 ± 37.33 | 9.32 ± 15.27 | 36.25 ± 4.24 | 7.01 ± 2.15 | 8.62 ± 20.48 | 11.54 ± 1.25 | 164.37 ± 84.75 |
| GG | 32.10 ± 4.18 | 1.67 ± 2.54 | 38.39 ± 23.85 | 5.46 ± 3.78 | 34.60 ± 3.54 | 6.80 ± 1.99 | 4.55 ± 4.01 | 12.17 ± 3.59 | 166.09 ± 82.36 |
| p | 0.026 | 0.66 | 0.94 | 0.251 | 0.18 | 0.942 | 0.198 | 0.453 | 0.223 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, H.-J.; Cho, S.-H.; Park, H.-S.; Kim, J.-H.; Kim, Y.-R.; Lee, W.-S.; Lee, J.-R.; Joo, S.-S.; Ahn, E.-H.; Kim, N.-K. Genetic Variations miR-10aA>T, miR-30cA>G, miR-181aT>C, and miR-499bA>G and the Risk of Recurrent Pregnancy Loss in Korean Women. Biomedicines 2022, 10, 2395. https://doi.org/10.3390/biomedicines10102395
An H-J, Cho S-H, Park H-S, Kim J-H, Kim Y-R, Lee W-S, Lee J-R, Joo S-S, Ahn E-H, Kim N-K. Genetic Variations miR-10aA>T, miR-30cA>G, miR-181aT>C, and miR-499bA>G and the Risk of Recurrent Pregnancy Loss in Korean Women. Biomedicines. 2022; 10(10):2395. https://doi.org/10.3390/biomedicines10102395
Chicago/Turabian StyleAn, Hui-Jeong, Sung-Hwan Cho, Han-Sung Park, Ji-Hyang Kim, Young-Ran Kim, Woo-Sik Lee, Jung-Ryeol Lee, Seong-Soo Joo, Eun-Hee Ahn, and Nam-Keun Kim. 2022. "Genetic Variations miR-10aA>T, miR-30cA>G, miR-181aT>C, and miR-499bA>G and the Risk of Recurrent Pregnancy Loss in Korean Women" Biomedicines 10, no. 10: 2395. https://doi.org/10.3390/biomedicines10102395
APA StyleAn, H.-J., Cho, S.-H., Park, H.-S., Kim, J.-H., Kim, Y.-R., Lee, W.-S., Lee, J.-R., Joo, S.-S., Ahn, E.-H., & Kim, N.-K. (2022). Genetic Variations miR-10aA>T, miR-30cA>G, miR-181aT>C, and miR-499bA>G and the Risk of Recurrent Pregnancy Loss in Korean Women. Biomedicines, 10(10), 2395. https://doi.org/10.3390/biomedicines10102395