Development of a New Screen-Printed Transducer for the Electrochemical Detection of Thiram
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Intrumentation
2.2. Methods
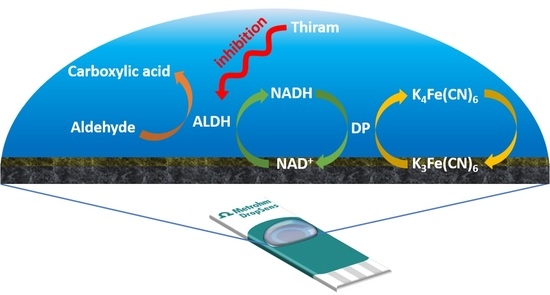
2.2.1. Fabrication of NAD+/K3[Fe(CN)6]/Carbon Transducer
2.2.2. Electrochemical Detection of Thiram
2.3. Determination of Enzyme Activities and Michaelis Constants
2.3.1. Aldehyde Dehydrogenase (ALDH) Activity
2.3.2. Diaphorase (DP) Activity
2.3.3. Michaelis Constants
3. Results
3.1. Optimization of Experimental Conditions
3.2. Detection of Thiram Fungicide with NAD+/K3[Fe(CN)]6/Carbon Transducer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pouchieu, C.; Piel, C.; Carles, C.; Gruber, A.; Helmer, C.; Tual, S.; Marcotullio, E.; Lebailly, P.; Baldi, I. Pesticide use in agriculture and Parkinson’s disease in the AGRICAN cohort study. Int. J. Epidemiol. 2018, 47, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Soleo, L.; Defazio, G.; Scarselli, R.; Zefferino, R.; Livrea, P.; Foà, V. Toxicity of fungicides containing ethylene-bis-dithiocarbamate in serumless dissociated mesencephalic-striatal primary coculture. Arch. Toxicol. 1996, 70, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, A.G.; Rhodes, S.L.; Lulla, A.; Murphy, N.P.; Lam, H.A.; O’Donnell, K.C.; Barnhill, L.; Casida, J.E.; Cockburn, M.; Sagasti, A.; et al. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc. Natl. Acad. Sci. USA 2013, 110, 636–641. [Google Scholar] [CrossRef] [Green Version]
- Woo, Y.T. Carcinogenicity, mutagenicity and teratogenicity of carbamates, thiocarbamates and related compounds: An overview of structure- activity relationships and environmental concerns. J. Environ. Sci. Heal. Part C 1983, C1, 97–133. [Google Scholar] [CrossRef]
- Vettorazzi, G.; Almeida, W.F.; Burin, G.J.; Jaeger, R.B.; Puga, F.R.; Rahde, A.F.; Reyes, F.G.; Schvartsman, S. International safety assessment of pesticides: Dithiocarbamate pesticides, ETU, and PTU-A review and update. Teratog. Carcinog. Mutagen. 1996, 15, 313–337. [Google Scholar] [CrossRef]
- Sharma, V.K.; Aulakh, J.S.; Malik, A.K. Thiram: Degradation, applications and analytical methods. J. Environ. Monit. 2003, 5, 717–723. [Google Scholar] [CrossRef]
- EU Pesticides Database. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32016R0001 (accessed on 21 October 2021).
- Keppel, G.E. Collaborative Study of the Determination of Dithiocarbamate Residues by a Modified Carbon Disulfide Evolution Method. J. Assoc. Off. Anal. Chem. 1971, 54, 528–532. [Google Scholar] [CrossRef]
- Caldas, E.D.; Conceição, M.H.; Miranda, M.C.C.; De Souza, L.C.K.R.; Lima, J.F. Determination of dithiocarbamate fungicide residues in food by a spectrophotometric method using a vertical disulfide reaction system. J. Agric. Food Chem. 2001, 49, 4521–4525. [Google Scholar] [CrossRef]
- Al-Alam, J.; Bom, L.; Chbani, A.; Fajloun, Z.; Millet, M. Analysis of Dithiocarbamate Fungicides in Vegetable Matrices Using HPLC-UV Followed by Atomic Absorption Spectrometry. J. Chromatogr. Sci. 2017, 55, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Lehotay, J.; Kisová, D. HPLC Study of Mancozeb Degradation on Leaves. J. Liq. Chromatogr. 1993, 16, 1015–1022. [Google Scholar] [CrossRef]
- Woodrow, J.E.; Seiber, J.N.; Fitzell, D. Analytical Method for the Dithiocarbamate Fungicides Ziram and Mancozeb in Air: Preliminary Field Results. J. Agric. Food Chem. 1995, 43, 1524–1529. [Google Scholar] [CrossRef]
- Gustafsson, K.H.; Thompson, R.A. High-Pressure Liquid Chromatographic Determination of Fungicidal Dithiocarbamates. J. Agric. Food Chem. 1981, 29, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Azab, H.A.; Duerkop, A.; Mogahed, E.M. Fluorescence and Electrochemical Sensing of Pesticides Methomyl, Aldicarb and Prometryne by the Luminescent Europium-3-Carboxycoumarin Probe. J. Fluoresc. 2012, 22, 659–676. [Google Scholar] [CrossRef]
- Nsibande, S.A.; Forbes, P.B.C. Fluorescence detection of pesticides using quantum dot materials—A review. Anal. Chim. Acta 2016, 945, 9–22. [Google Scholar] [CrossRef]
- Cai, K.; Xiang, Z.; Zhang, J.; Zhou, S.; Feng, Y.; Geng, Z. Determination of eight tobacco alkaloids in flue-cured tobacco samples by gas chromatography with nitrogen chemiluminescence detection (NCD). Anal. Methods 2012, 4, 2095–2100. [Google Scholar] [CrossRef]
- Gámiz-Gracia, L.; García-Campaña, A.M.; Soto-Chinchilla, J.J.; Huertas-Pérez, J.F.; González-Casado, A. Analysis of pesticides by chemiluminescence detection in the liquid phase. Trends Anal. Chem. 2005, 24, 927–942. [Google Scholar] [CrossRef]
- Marty, J.L.; Mionetto, N.; Noguer, T.; Ortega, F.; Roux, C. Enzyme sensors for the detection of pesticides. Biosens. Bioelectron. 1993, 8, 273–280. [Google Scholar] [CrossRef]
- Noguer, T.; Marty, J.L. High sensitive bienzymic sensor for the detection of dithiocarbamate fungicides. Anal. Chim. Acta 1997, 347, 63–70. [Google Scholar] [CrossRef]
- Ibáñez, D.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Detection of dithiocarbamate, chloronicotinyl and organophosphate pesticides by electrochemical activation of SERS features of screen-printed electrodes. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2021, 248, 119174–119180. [Google Scholar] [CrossRef] [PubMed]
- Saute, B.; Premasiri, R.; Ziegler, L.; Narayanan, R. Gold nanorods as surface enhanced Raman spectroscopy substrates for sensitive and selective detection of ultra-low levels of dithiocarbamate pesticides. Analyst 2012, 137, 5082–5087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, Y.; Liu, P.; Tao, X.; Pei, J.; Li, H.; Du, Y. Determination of Pesticides by Surface-enhanced Raman Spectroscopy on Gold Nanoparticle Modified Polymethacrylate. Anal. Lett. 2016, 49, 2268–2278. [Google Scholar] [CrossRef]
- Sharma, V.K.; Aulakh, J.S.; Malik, A.K. Fourth derivative spectrophotometric determination of fungicide thiram (tetramethyldithiocarbamate) using sodium molybdate and its application. Talanta 2005, 65, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Filipe, O.M.S.; Vidal, M.M.; Duarte, A.C.; Santos, E.B.H. A solid-phase extraction procedure for the clean-up of thiram from aqueous solutions containing high concentrations of humic substances. Talanta 2007, 72, 1235–1238. [Google Scholar] [CrossRef]
- Filipe, O.M.S.; Vidal, M.M.; Duarte, A.C.; Santos, E.B.H. Influence of fulvic acids and copper ions on thiram determination in water. J. Agric. Food Chem. 2008, 56, 7347–7354. [Google Scholar] [CrossRef] [PubMed]
- Fernández, C.; Reviejo, A.J.; Polo, L.M.; Pingarrón, J.M. HPLC-Electrochemical detection with graphite-poly(tetrafluoroethylene) electrode. Determination of the fungicides thiram and disulfiram. Talanta 1996, 43, 1341–1348. [Google Scholar] [CrossRef]
- Waseem, A.; Yaqoob, M.; Nabi, A. Determination of thiram in natural waters using flow-injection with cerium(IV)-quinine chemiluminescence system. Luminescence 2010, 25, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Waseem, A.; Yaqoob, M.; Nabi, A. Photodegradation and flow-injection determination of dithiocarbamate fungicides in natural water with chemiluminescence detection. Anal. Sci. 2009, 25, 395–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girotti, S.; Maiolini, E.; Ghini, S.; Ferri, E.; Fini, F.; Nodet, P.; Eremin, S. Quantification of thiram in honeybees: Development of a chemiluminescent ELISA. Anal. Lett. 2008, 41, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Malik, A.K.; Faubel, W. Capillary electrophoretic determination of tetramethylthiuram disulphide (Thiram). Anal. Lett. 2000, 33, 2055–2064. [Google Scholar] [CrossRef]
- Substrates, S.R.S. Direct Detection of Toxic Contaminants in Minimally Processed Food Products Using Dendritic Surface-Enhanced Raman Scattering Substrates. Sensors 2018, 18, 2726. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, B.; Zhu, G.; Zhou, X. Synthesis of silver nanowires as a SERS substrate for the detection of pesticide thiram. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 133, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Saute, B.; Narayanan, R. Solution-based direct readout surface enhanced Raman spectroscopic (SERS) detection of ultra-low levels of thiram with dogbone shaped gold nanoparticles. Analyst 2011, 136, 527–532. [Google Scholar] [CrossRef]
- Fernández, C.; Reviejo, A.J.; Pingarrón, J.M. Development of graphite-poly(tetrafluoroethylene) composite electrodes Voltammetric determination of the herbicides thiram and disulfiram. Anal. Chim. Acta 1995, 305, 192–199. [Google Scholar] [CrossRef]
- Hernández-Olmos, M.A.; Agüí, L.; Yáñez-Sedeño, P.; Pingarrón, J.M. Analytical voltammetry in low-permitivity organic solvents using disk and cylindrical microelectrodes. Determination of thiram in ethyl acetate. Electrochim. Acta 2000, 46, 289–296. [Google Scholar] [CrossRef]
- Law Al, A.T.; Adeloju, S.B. Progress and recent advances in phosphate sensors: A review. Talanta 2013, 114, 191–203. [Google Scholar] [CrossRef]
- Fernández-Fernández, M.; Sanromán, M.Á.; Moldes, D. Recent developments and applications of immobilized laccase. Biotechnol. Adv. 2013, 31, 1808–1825. [Google Scholar] [CrossRef]
- Arduini, F.; Micheli, L.; Moscone, D.; Palleschi, G.; Piermarini, S.; Ricci, F.; Volpe, G. Electrochemical biosensors based on nanomodified screen-printed electrodes: Recent applications in clinical analysis. TrAC-Trends Anal. Chem. 2016, 79, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Avramescu, A.; Andreescu, S.; Noguer, T.; Bala, C.; Andreescu, D.; Marty, J.L. Biosensors designed for environmental and food quality control based on screen-printed graphite electrodes with different configurations. Anal. Bioanal. Chem. 2002, 374, 25–32. [Google Scholar] [CrossRef]
- Neves, M.M.P.S.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Streptavidin functionalized nickel nanowires: A new ferromagnetic platform for biotinylated-based assays. Talanta 2015, 144, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.M.P.S.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Screen-Printed Electrochemical 96-Well Plate: A High-Throughput Platform for Multiple Analytical Applications. Electroanalysis 2014, 26, 2764–2772. [Google Scholar] [CrossRef]
- Noguer, T.; Marty, J.L. An amperometric bienzyme electrode for acetaldehyde detection. Enzyme Microb. Technol. 1995, 17, 453–456. [Google Scholar] [CrossRef]
- Honda, K.; Hara, N.; Cheng, M.; Nakamura, A.; Mandai, K.; Okano, K.; Ohtake, H. In vitro metabolic engineering for the salvage synthesis of NAD+. Metab. Eng. 2016, 35, 114–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachisuka, S.; Sato, T.; Atomi, H. Metabolism Dealing with Thermal Degradation of NAD+ in the Hyperthermophilic Archaeon Thermococcus kodakarensis. J. Bacteriol. 2017, 199, e00162-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Detection Technique | LOD (ppm) | Reference |
|---|---|---|
| Spectrophotometry | 0.3 | [23] |
| Spectrophotometry | 0.33 | [24] |
| HPLC-UV | 0.088 | [25] |
| HPLC-EC | 0.14 | [26] |
| FI-CL | 0.0075 | [27] |
| FI-CL | 0.005 | [28] |
| CL-ELISA | 0.009 | [29] |
| Electrophoresis | 0.5 | [30] |
| SERS | 0.115 | [31] |
| SERS | 0.024 | [32] |
| SERS | 0.0024 | [33] |
| EC | 0.013 | [34] |
| EC | 0.103 | [35] |
| EC | 0.09 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibáñez, D.; Izquierdo-Bote, D.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Development of a New Screen-Printed Transducer for the Electrochemical Detection of Thiram. Chemosensors 2021, 9, 303. https://doi.org/10.3390/chemosensors9110303
Ibáñez D, Izquierdo-Bote D, González-García MB, Hernández-Santos D, Fanjul-Bolado P. Development of a New Screen-Printed Transducer for the Electrochemical Detection of Thiram. Chemosensors. 2021; 9(11):303. https://doi.org/10.3390/chemosensors9110303
Chicago/Turabian StyleIbáñez, David, Daniel Izquierdo-Bote, María Begoña González-García, David Hernández-Santos, and Pablo Fanjul-Bolado. 2021. "Development of a New Screen-Printed Transducer for the Electrochemical Detection of Thiram" Chemosensors 9, no. 11: 303. https://doi.org/10.3390/chemosensors9110303
APA StyleIbáñez, D., Izquierdo-Bote, D., González-García, M. B., Hernández-Santos, D., & Fanjul-Bolado, P. (2021). Development of a New Screen-Printed Transducer for the Electrochemical Detection of Thiram. Chemosensors, 9(11), 303. https://doi.org/10.3390/chemosensors9110303