Abstract
Molecularly Imprinted Polymers (MIPs) were synthesised for the selective detection of amoxicillin in aqueous samples. Different functional monomers were tested to determine the optimal composition via batch rebinding experiments. Two different sensor platforms were tested using the same MIP solution; one being bulk synthesized and surface modified Screen Printed Electrodes (SPEs) via drop casting the microparticles onto the electrode surface and the other being UV polymerized directly onto the SPE surface in the form of a thin film. The sensors were used to measure amoxicillin in conjunction with the Heat-Transfer Method (HTM), a low-cost and simple thermal detection method that is based on differences in the thermal resistance at the solid–liquid interface. It was demonstrated that both sensor platforms could detect amoxicillin in the relevant concentration range with Limits of Detection (LOD) of 1.89 ± 1.03 nM and 0.54 ± 0.10 nM for the drop cast and direct polymerisation methods respectively. The sensor platform utilising direct UV polymerisation exhibited an enhanced response for amoxicillin detection, a reduced sensor preparation time and the selectivity of the platform was proven through the addition of nafcillin, a pharmacophore of similar shape and size. The use of MIP-modified SPEs combined with thermal detection provides sensors that can be used for fast and low-cost detection of analytes on-site, which holds great potential for contaminants in environmental aqueous samples. The platform and synthesis methods are generic and by adapting the MIP layer it is possible to expand this sensor platform to a variety of relevant targets.
1. Introduction
The World Health Organisation has stated that antimicrobial resistant (AMR) bacteria pose a fundamental threat to human health due to the overuse and declining effectiveness of antibiotics [1,2]. Antibiotic consumption has increased 65% worldwide between the years 2000 and 2015 [3]. The use of antibiotics in veterinary or clinical practice exerts selective pressure to bacteria, which accelerates the emergence of bacteria with AMR properties due to the accumulation of pharmaceuticals in the food chain and water systems. There are also notable concerns about the discharge of antibiotic waste by pharmaceutical producers, which lead to point-source pollution. Watkinson et al. have shown that even in a first world country such as Australia, there are many water systems that have a presence of beta lactam antibiotics [4]. Recent studies from India, one of the world’s leading pharmaceutical producers, has revealed excessively high concentrations of antibiotic compounds in drinking water that exceed the maximum regulatory limits (MRLs). This has a serious impact on vulnerable populations living near manufacturing facilities, with an estimate of 58,000 new-borns dying in India in these regions from multi-drug resistant infections [5,6,7]. The risk of these infections is considerably higher in developing countries, which has been attributed to the lack of optimal wastewater treatment facilities and high costs associated with vigilant monitoring of pharmaceutical waste [8]. Furthermore, while there are strict European regulations regarding MRL of pharmaceuticals in food products of animal origin, regulations for waste and surface water are yet to be implemented with currently only environmental quality standards set for a number of micropollutants [9]. Antibiotic residues that are most often found in aquatic environments belong to the classes of beta-lactams, fluoroquinolones, macrolides, sulfonamides and tetracyclines [10]. Current commercial screening tests for antibiotics in food and environmental samples either require a long response time (several hours), or are not able to selectively detect antibiotic compounds and solely provide semi-quantitative information. Thus, there is a strong need for low-cost, robust and easy to use sensors that can be used on site to determine trace levels of antibiotics in aquatic systems. Many electrochemical sensors display promise in this respect [11,12,13]. Pellegrini et al. [14] discuss the usability of electrochemical sensors for the determination of antibiotics and promote their use for on-site testing due to their analysis time (~2 h) and small sample volume (~100 µL). These factors combined with the lack of any pre-treatment steps required and a high level of precision give a strong indication of the appropriateness of this type of analysis for the desired purpose. Previous reports in literature have shown the ability of synthetic polymeric recognition elements, namely Molecularly Imprinted Polymers (MIPs), to improve the affinity and selectivity of electrochemical sensors [15]. Yang et al. [16] have demonstrated limits of detection in the nanomolar regime in buffered solutions and in spiked food samples can be achieved with MIP layers grown onto multiwalled carbon nanotubes. Zeinali et al. [17], use magnetic MIPs modified onto carbon paste electrodes combined with cyclic voltammetry to detect amoxicillin with a limit of detection of 0.26 nM. Betlem et al. [18] have shown the effective use for the Heat-Transfer Method (HTM) as an appropriate analysis technique for small molecules such as caffeine.
The current study was built on from this work and tailored the detection system to a beta lactam antibiotic. We used amoxicillin in this manuscript as a case study and compared the sensors characteristics of microparticle modified Screen-Printed Electrodes (SPEs) to those functionalised with a thin film. The structures of amoxicillin and nafcillin, another beta lactam antibiotic used in this study to test the selectivity of the sensors, are shown in Figure 1.
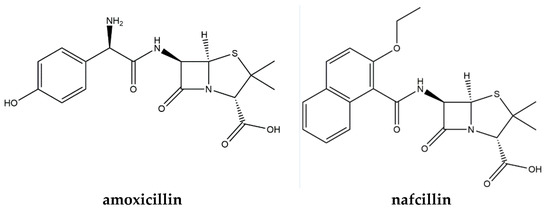
Figure 1.
Structures of amoxicillin and nafcillin.
SPEs are often used in commercial sensors due to their high reproducibility, low-cost and inherent suitability for mass-production [19]. However, integration of polymer structures into SPEs is complicated due to their low resistance to traditional organic solvents such as chloroform and toluene [20].
In this paper, we explore two different electrode modification strategies and compare and contrast towards the analytical detection of amoxicillin. The SPEs are modified with MIPs in two manners; (i) surface modification via dropcasting of MIP microparticles suspended into ethanol directly onto the sensor surface along with a thin layer of polypyrrole to hold the MIPs in place; (ii) directly depositing of MIP thin films onto SPEs using UV polymerization. The disadvantage of working with MIP microparticles is the two-step (indirect) functionalisation process and the fact that the resulting electrodes are insulating and not compatible with traditional electrochemical detection. Recent work in our group has demonstrated that thermal analysis, in particular the Heat-Transfer Method (HTM), can provide an interesting alternative analysis technique that is easy to employ and provides rapid detection [21].
2. Experimental
2.1. Equipment and Reagents
Dimethyl Sulfoxide (DMSO) was sourced from TCI (Oxford, UK). Pyrrole, amoxicillin, nafcillin, azobisisobutyronitrile (AIBN), trimethylolpropane trimethacrylate (TRIM), methacrylic acid (MAA), acrylamide (AA) and 2-vinylpyridine were sourced from Sigma Aldrich (Gillingham, UK). Ethanol was sourced from Fisher Scientific (Loughborough, UK). The orbital shaker used was a Stuart mini orbital shaker SSM1 (Staffordshire, UK). All experiments were carried out in Newcastle University and Manchester Metropolitan University. Experiments were carried out at 20 ± 1 °C unless otherwise noted. UV/Vis analysis was carried out on an Agilent 8453 UV-Vis spectrophotometer (Santa Clara, CA, USA). Phosphate Buffered Saline (PBS) tablets were purchased from Sigma (Gillingham, UK) and used to maintain pH level (pH = 7.4) and the ionic strength of solutions used throughout the experimental portion of the study.
2.2. MIP and NIP Microparticle Syntheses
MIPs for amoxicillin were synthesised with varying compositions, which are listed in Table 1. The general functionalisation procedure includes mixing of the template (0.35 mmol) with the monomer (1.4 mmol) in 3.3 mL of DMSO. Subsequently, the crosslinker monomer TRIM (2.8 mmol) was added followed by the initiator AIBN (0.22 mmol). The mixture was sonicated and degassed with N2. Polymerization was initiated by exposing the samples to a UV lamp (200 mW, Polytec UV LC-S, Karlsbad, Germany) for 10–15 min. The obtained polymer block was washed with water and subsequently ground to obtain a fine powder. The template was removed by continuous Soxhlet extraction with a mixture of methanol and water (50/50) until UV-Vis analysis no longer showed traces of amoxicillin in the extract. The powders were washed with water and dried overnight under vacuum. Non-Imprinted Polymers (NIPs) were synthesized in the same manner but without the addition of the template.

Table 1.
The composition of the different Molecularly Imprinted Polymers (MIPs), listing the amount of template, functional monomer, crosslinker monomers, initiator and porogen used.
2.3. Batch Rebinding Experiments Evaluated with Optical Detection
Batch rebinding experiments were performed and optical detection was used to evaluate the binding of amoxicillin to the MIP and NIP powders. The antibiotic concentration was determined by measuring the absorbance at λ = 272 nm. In each experiment, 10 mg of MIP or NIP powder was added to 5.0 mL of PBS solutions with amoxicillin concentrations between 0.5–1.0 mM. The resulting suspensions were placed on a rocking table (110 rpm) for 60 min and subsequently filtered. The free concentration of amoxicillin (Cf) in the filtrate was determined by comparing the absorbance to that of a pre-determined calibration curve. Subsequently, the amount of bound template (Cb) was calculated by subtracting the initial concentration added to the solution (Ci) by the free template concentration (Cf), . The molecule of template bound per gram (Sb) was obtained by multiplying Cb by the volume in litres and then dividing this by the amount of polymer used in the experiment (10 mg). The Imprint Factor (IF) was determined by dividing Sb for a given MIP by the Sb for the corresponding NIP at a Cf value of 0.5 mM [22].
2.4. Thin Film Polymerization
A pre-polymerization mixture of MIP 2 was produced as described in Section 2.2. A total of 0.3 µL of the solution was applied onto the working electrode of the SPE and subsequently exposed to UV light (365 nm) for 1 min. A cover slip was placed over the droplet and it was exposed to UV light for a further 1 min to complete the polymerization. The functionalised SPE was then washed in methanol and the template extracted in deionized water (40 mL) using an orbital shaker (160 rpm) overnight and stored in a fresh deionized water solution until use. A NIP was produced using an identical protocol without the presence of the target amoxicillin.
2.5. Electrochemical Deposition of Polypyrrole
Electrochemical experiments were performed using a standard three electrode set up. Graphite Screen-Printed macroelectrodes (SPE, Manchester Metropolitan University, UK) [23,24] were utilised as the working electrode alongside a nickel/chromium wire auxiliary electrode and silver silver chloride (Ag AgCl) reference electrode (BASi, West Lafayette, IN, USA). These were controlled by an Ivium Compactstat (Eindhoven, The Netherlands) connected to a Desktop PC (Dell, UK). The SPEs were fabricated in-house using a stencil design to achieve a 3.1 mm diameter working electrode using a graphite ink (Product Ink: C2000802P2; Gwent Electronic Materials Ltd., Pontypool, UK) and were printed using a DEK 248 screen printer machine (DEK, Weymouth, UK) onto a polyester flexible film (Autostat, Milan, Italy, 250 micron thickness). The layer was cured in a fan oven at 60 °C for 30 min and finally, a dielectric paste (Product Code: D2070423D5; Gwent Electronic Materials Ltd., Pontypool, UK) was then printed onto the polyester substrate to cover the connections. The SPEs were then cured for an additional 30 min at 60 °C before use [25,26].
All electrochemical measurements were carried out at 20 ± 1 °C and using deionised water of resistivity ≥18.2 MΩ cm. Prior to all electrochemical measurements the solutions were thoroughly degassed with highly pure nitrogen for 15 min.
MIP microparticles were dispersed in ethanol (1 mg/mL), dropcast (30 µL) onto the SPE surface and allowed to dry. Following this, a thin layer of polypyrrole was applied to the SPE surface to hold the MIPs in place. A droplet (30 µL) of pyrrole (0.1 M) in PBS was applied to the electrode surface. Electropolymerization was performed through chronoamperometry, using the three electrode system described above and applying a potential of +0.8 V for 30 s. The electrodes were thoroughly rinsed with deionised water and the presence of MIPs on the surface confirmed by Scanning Electron Microscopy (SEM) (Figure S1A).
2.6. Scanning Electron Microscopy (SEM)
SEM measurements were recorded on a Supra 40VP Field Emission from Carl Zeiss Ltd. (Cambridge, UK) with an average chamber vacuum of 1.3 × 10−5 mbar and average gun vacuum of 1 × 10−9 mbar. To enhance the contrast of these images, a thin layer of Au/Pd (8 V, 30 s) was sputtered onto the electrodes with a SCP7640 from Polaron (Hertfordshire, UK).
2.7. HTM Measurements with MIP-Modified SPEs
The functionalised SPEs were cut into 1 × 1 cm squares, inserted into an additively manufactured/3D-printed flow cell [27] and sealed with an O-ring (RS Components, UK). Thermal measurements were carried out using the flow cell (Figure S2) of volume 110 µL in conjunction with the heat-transfer set-up which was designed in-house [28,29]. The flow cell is connected to a copper block which acts as a heat sink. The temperature of this block (T1) is controlled via a Proportional-Integral-Derivative (PID) controller. A type-K thermocouple (RS Components, UK) is used to measure the temperature of the sample solution (T2) 1.7 mm above the surface of the SPE. The thermal resistance (Rth) at the solid–liquid interface can be determined by dividing the measured temperature difference between the copper block and the solution (T1−T2) by the power provided to the heat source to maintain its temperature, . The aforementioned PID parameters can affect the power signal stability and therefore the sensitivity of the developed sensor. As such, the PID parameters were set to the optimized values determined for this heat source of P = 1, I = 14, D = 0.3 [21].
For all experiments, the flow cell was filled with PBS solution and the copper block was heated to a pre-set temperature of 37.00 ± 0.02 °C. The cell was left to stabilise for 15 min before a first injection of PBS was added to act as the blank measurement. Sequential amounts of amoxicillin (1–500 nM) were injected into the flow cell using an automated NE500 programmable syringe pump (Prosense, Oosterhout, the Netherlands) at a flow rate of 250 µL·min−1. Following each injection, the temperature was allowed to stabilise for 30 min, of which the last 10 min (600 data points) were averaged out and used to calculate the Rth. These experiments were carried out in triplicate to prove the reproducibility of the electrodes. The calculated Rth values were used to produce dose-response curves and calculate the limit of detection (LoD) using the three sigma method. Specificity tests were carried out by injecting the same concentrations of amoxicillin to an electrode modified with a NIP. Selectivity studies were carried out by injecting the same concentration of a competitive target, with a similar pharmacophore, nafcillin.
3. Results
3.1. Batch Rebinding Results
First, batch rebinding experiments with MIP1-3, that have similar compositions, but differ in the functional monomer, were performed and are presented in Table 2. The time of the experiments was fixed at 1 h since after this time no increase in the binding to the MIP particles was observed. In each experiment, 5 mL of amoxicillin solutions in PBS (Ci = 0.5–1.0 mM) were added to 10 mg of the MIP particles.

Table 2.
The amount of binding for MIP and Non-Imprinted Polymer (NIP) at Cf = 0.5 mM.
The results obtained from the batch rebinding experiments are presented in Table 2 and Figure 2. It was observed that MIP 2, made of acrylamide (AA) as the functional monomer, exhibited a significantly higher binding affinity for the amoxicillin target compared to MIP 1 (based on methacrylic acid) and MIP 3 (based on 2-vinyl pyridine). In addition to this, at a Cf value of 0.5 mM the calculated Imprint Factor (IF) for MIP 2 was significantly higher than either of the other MIPs at 6.47, exhibiting the MIPs affinity efficiency compared to the corresponding NIP. The highest affinity MIP using AA as a functional monomer was most likely due to the nature of the functional groups on the monomer when compared to the structure of the target. These functional groups would allow self-assembling of monomers around the target to form the non-covalent forces necessary to hold them together [30]. On the other hand, 2-vinylpyrdine lacks these functional groups required for binding [31].
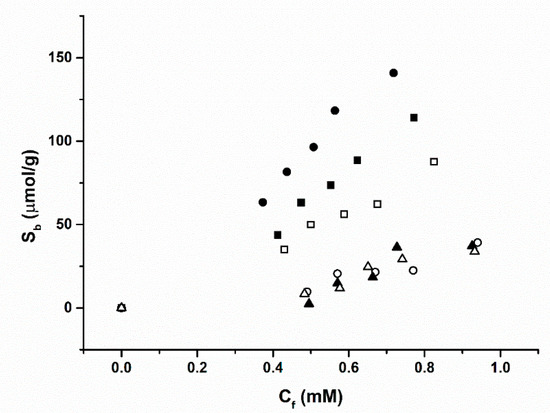
Figure 2.
Binding isotherms of the polymerised MIP/NIPs upon exposure to amoxicillin solutions in Phosphate Buffered Saline (PBS). MIP 1 (filled squares) and NIP 1 (hollow squares), MIP 2 (filled circles) and NIP 2 (hollow circles) and MIP 3 (filled triangles) and NIP 3 (hollow triangles).
Time dependent experiments were performed to determine the optimal time for rebinding. Readings were taken at 30 min, 2 h and 24 h after mixing MIP with target solution, resulting in absorbance values of 1.52, 1.51 and 1.47, respectively. This shows that there is no significant change over time and that 30 min is sufficient to gain an accurate representation of batch rebinding data.
Figure 3 displays the selectivity presented by MIP 2, which exhibits a higher binding affinity towards amoxicillin than towards nafcillin. This shows that the pores in the imprinted polymer have a higher affinity towards their intended target even if a very similar pharmacophore is used, thus confirming the polymers ability to recognise the target. The NIPs have no significant difference in binding affinity as expected, since they do not possess the stereoselective cavities present in the MIP.
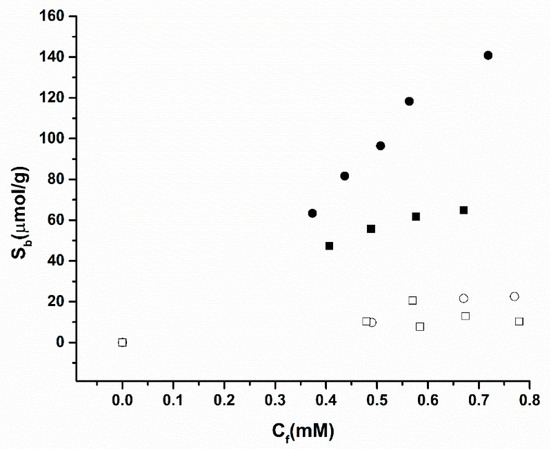
Figure 3.
Selectivity batch rebinding of MIP 2 (filled symbols) and NIP 2 (hollow symbols) with amoxicillin (squares) and nafcillin (circles) in PBS.
The relevant concentration range of amoxicillin in water samples is in the low nanomolar range [32]. Therefore, it was decided to evaluate the performance of the sensor to discriminate amoxicillin from its competitor molecule in the nanomolar concentration, for which it is required to use a more sensitive read-out strategy such as HTM.
3.2. Detection of Amoxicillin Using MIP Microparticles
Bulk polymerized MIPs (MIP 2) were functionalised onto SPEs (Figure S1A) as explained in Section 2.5, fitted inside the flow cell (Figure S2) and connected to the HTM equipment. SPEs were used in this work because they can be used as disposable sensors. This aligns well with the use of MIPs as multiple template removals is not advised for a reproducible platform. The flow cell was filled with PBS and allowed to stabilise for 45 min. Then, sequential injections of the target analyte amoxicillin (1–500 nM) in PBS were started, allowing the system to equilibrate for 30 min after each injection. The raw data output obtained for these additions is presented in Figure 4A. Specific values for the thermal resistance (Rth) values for each concentration of amoxicillin were calculated using the last 10 min of each stabilisation period. This resulted in a value and standard deviation being calculated over 600 data points. In PBS, the Rth of the MIP functionalised system stabilised at a value of 5.91 ± 0.05 °C/W. Upon the addition of 1 nM of amoxicillin the Rth increased from this baseline value to 6.06 ± 0.05 °C/W. The Rth value increased further until the addition of 500 nM of amoxicillin where it stabilised at a value of 6.42 ± 0.06 °C/W. An identical experiment was performed using an SPE modified with the NIP. In this case, Rth of the NIP functionalised system stabilised at a value of 5.51 ± 0.06 °C/W. After the addition of 1 nM of amoxicillin the Rth exhibited no statistically significant deviation from the baseline value to 5.55 ± 0.06 °C/W. The Rth value for the addition of 500 nM of amoxicillin showed a small increase due to some non-specific binding, where it stabilised at a value of 5.61 ± 0.06 °C/W.
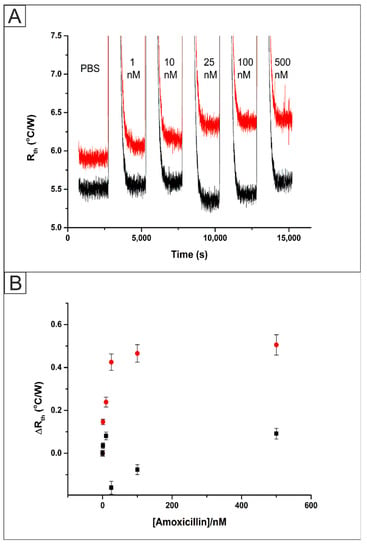
Figure 4.
(A) The raw Heat-Transfer Method (HTM) data plot of Rth versus time for the addition of amoxicillin (1–500 nM) in PBS to an Screen Printed Electrodes (SPE) functionalised with bulk UV polymerized MIP (red) and NIP (black). (B) Comparison between the absolute Rth values obtained from the HTM for a MIP functionalised SPE (red circles) and NIP functionalised SPE (black).
These Rth values were plotted against the concentration of amoxicillin injected into the system to produce a dose-response curve, Figure 4B. This was used to calculate a Limit of Detection (LOD) for the bulk UV polymer system of 1.90 ± 1.03 nM. These results were found to exhibit a lower LOD and wider detection range than that observed in previous work on caffeine detection [18]. This was expected due to the limited loading of MIP when incorporated directly into the SPE ink in comparison to drop casting. Additionally, template bleeding is a problem in the use of MIP microparticles. Up to 99% of the template is removed during the extraction step; however, the remaining template can cause issues when ‘leeching’ out of the polymer matrix during measurements giving erroneous quantification [33]. The removal of MIP over time is of considerable concern when using a drop casting method, therefore the direct formation of the MIP onto the electrode surface using UV polymerisation was investigated.
3.3. Detection of Amoxicillin Using MIP Thin Films
Screen-Printed Electrodes (SPEs) were functionalised with amoxicillin MIPs via UV-Vis polymerization (Figure S1B) as described in Section 2.4. These SPEs were inserted into a flow cell (Figure S2) and exposed to PBS solutions containing sequential amounts of the target analyte amoxicillin. The HTM raw data obtained for the addition of amoxicillin (1–500 nM) are presented in Figure 5A. In PBS, the Rth of the MIP functionalised system stabilised at a value of 7.58 ± 0.05 °C/W. The increase in the Rth value compared to previous work [18,21] was expected due to the presence of a thicker layer of polymer present on the electrode surface, which in turn will increase the resistance to heat flow through the interface. Upon the addition of 1 nM of amoxicillin an increase in the absolute Rth value was observed from its baseline of 7.58 ± 0.05 °C/W to 7.67 ± 0.06 °C/W. The Rth increased until the last addition of 500 nM amoxicillin where it reached a value of 8.16 ± 0.06 °C/W and an additional injection of PBS produced no significant change in the Rth. An identical experiment was performed using a NIP functionalised SPE where the Rth stabilised at a value of 7.59 ± 0.06 °C/W. Upon the addition of 1 nM of amoxicillin to the system there was no significant change to the Rth value with it remaining at 7.59 ± 0.06 °C/W. Once 500 nM of amoxicillin was added to the NIP modified SPE there was an increase in the Rth to 7.68 ± 0.07 °C/W. The increase in the Rth can be explained by some non-specific binding of the target molecule to the polymer on the surface of the SPE however, the increase is significantly less than observed for the MIP modified SPE. Both of these results were used to produce a dose-response curve (Figure 5B) which shows the improved response of the MIP based platform and was used to calculate the Limit of Detection (LOD) of 0.54 ± 0.10 nM.
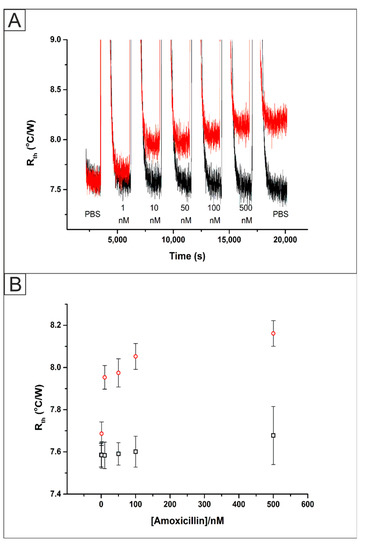
Figure 5.
(A) The raw HTM data plot of Rth versus time for the addition of amoxicillin (1–500 nM) in PBS to an SPE functionalised with a direct UV polymerized MIP (red) and NIP (black). (B) Comparison between the absolute Rth values obtained from the HTM for a MIP functionalised SPE (red circles) and NIP functionalised SPE (black).
The selectivity of the MIP was assessed through repeating the experiment with additions of a molecule with the same pharmacophore and similar size, nafcillin (1–500 nM). An SPE functionalised with a MIP via UV polymerization directly onto the surface was placed inside the flow cell (Figure S1) and attached to the HTM equipment. The data collected from the addition of the competitor (black) was plotted alongside the previous results for the addition of the target molecules (red) to MIP functionalised SPEs.
The raw data plot that was obtained is presented in Figure 6A (black) where there is a clear difference between the stabilised Rth values. This can be explained as the Type K thermocouples used in the experiment have a temperature error of up to 2.5 °C. The Rth value of the system in PBS stabilised at a value of 6.47 ± 0.05 °C/W. Upon the addition of 1 nM of nafcillin, there was no observed statistically significant change in the stabilised Rth value at 6.46 ± 0.05 °C/W. Once 500 nM of nafcillin had been injected into the flow cell an increase in the Rth was observed from the baseline level of 6.47 ± 0.05 °C/W up to 6.60 ± 0.05 °C/W. These values, along with those obtained for the addition of the target molecule were used to produce dose-response curves, Figure 6B, showing the increase in the Rth from the baseline stabilised levels for both the target (red circles) and competitor (black squares). This indicated that there was some non-specific binding of the competitor antibiotic to the surface of the polymer. However, the increase was significantly lower than that observed for the injections of the target antibiotic, showing the high selectivity of the sensing platform.
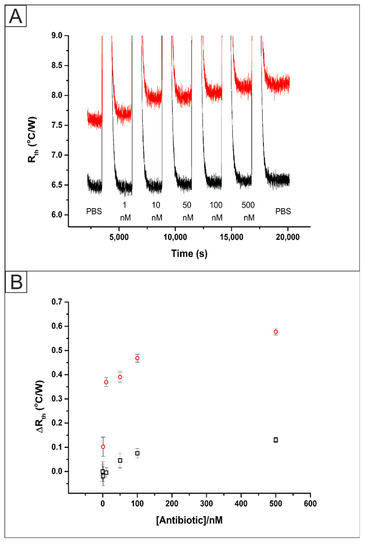
Figure 6.
(A) The raw HTM data plot of Rth versus time for the addition of amoxicillin (1–500 nM, red) and nafcillin (1–500 nM, black) in PBS to an SPE functionalised with a direct UV polymerized MIP. (B) Comparison between the absolute Rth values obtained from the HTM for a MIP functionalised SPE with additions of amoxicillin (red) and nafcillin (black).
4. Conclusions
MIPs for the detection of amoxicillin were synthesized using bulk UV polymerisation with the functional monomers methacrylic acid, acrylamide (AA) and 2-vinylpyridine. Batch rebinding experiments evaluated via spectroscopic methods determined that the MIPs using AA (MIP 2) as the functional monomer exhibited the highest recognition capability (IF = 6.47) for amoxicillin. Subsequently, MIP 2 was drop cast onto a SPE, fitted to the HTM and subjected to injections of increasing concentrations of amoxicillin (1–500 nM). An increase in the measured Rth was observed from the baseline of 5.91 ± 0.05 °C/W to 6.42 ± 0.06 °C/W after the addition of 500 nM amoxicillin for the MIP functionalised SPE; whereas, the NIP functionalised exhibited an Rth of 5.52 ± 0.05 °C/W and 5.61± 0.06 °C/W for the baseline and 500 nM amoxicillin respectively. These results were used to construct a dose-response curve and calculate a LOD of 1.89 ± 1.03 nM. This was then compared to a system where the MIP was formed directly onto the SPE surface through UV polymerisation. This methodology dramatically reduced the preparation time and increased the reliability of the sensor through the attachment of the polymer to the electrode surface. An increase in the measured Rth was observed for the addition of amoxicillin (1–500 nM) to the MIP functionalised SPE from the baseline value of 7.58 ± 0.06 °C/W to 8.16 ± 0.06 °C/W at 500 nM amoxicillin. The NIP produced a significantly smaller increase in the Rth from 7.59 ± 0.06 °C/W to 7.68 ± 0.07 °C/W after 500 nM amoxicillin. The dose-response curve for the MIP allowed for the determination of the LOD of 0.54 ± 0.1 nM which is a big improvement over the drop cast method. The direct UV polymerized SPE was also tested for interference from a competitive pharmacophore, nafcillin. This exhibited a rise in the measured Rth from of 6.47 ± 0.05 °C/W in PBS to of 6.60 ± 0.05 °C/W after 500 nM nafcillin, showing the excellent specificity the MIP sensing platform has for amoxicillin.
The developed sensor platform has shown the ability to detect amoxicillin at relevant concentration ranges for environmentally contaminated aqueous samples. Molecular imprinting is versatile and by adapting the MIP, other antibiotics and pollutants can be targeted. The polymerisation methodology was significantly shortened in comparison to the bulk synthesis method and in conjunction with the thermal set-up, offers a portable and low-cost sensing platform suitable for on-site measurements.
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-9040/8/1/5/s1, Figure S1: (A) SEM image (2 k magnification) of MIP 2 microparticles immobilized onto an SPE using polypyrrole. (B) SEM image (1 k magnification) of thin film of polymer made directly on an SPE. Figure S2: Schematic image of the flow cell used in the experiments. The SPE is placed in the central chamber, which is sealed with an O-ring. The thermocouple (T2) is measured throughout the experiment (1 second = 1 data point). Solutions of analyte are injected into the inflow and the system is allowed to stabilise for 30 min before the next injection.
Author Contributions
Conceptualization M.P. and R.D.C.; Data curation O.J., T.C.C.S., B.A.d.F., A.H., F.M. and R.D.C.; Formal analysis O.J., M.P. and R.D.C.; Methodology A.H., S.J.R.-N., M.P., C.E.B. and R.D.C.; Resources S.J.R.-N., C.E.B., J.G., K.N. and M.P., Supervision C.E.B., J.G., K.N., M.P. and R.D.C.; Writing – Original Draft O.J., A.H., M.P. and R.D.C.; Writing – Review and Editing C.E.B., J.G., K.N., M.P. and R.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by EPSRC grant number EP/R029296/1 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant numbers 306147/2016-5 and 424027/2018-6.
Acknowledgments
All authors would like to thank Heitor Valarini. MP would like to acknowledge the EPSRC for funding under grant number EP/R029296/1. JG thanks Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support (grant numbers: 306147/2016-5 and 424027/2018-6). Data available from https://data.ncl.ac.uk.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance and Primary Health Care: Brief; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, A.; Murby, E.; Kolpin, D.W.; Costanzo, S. The occurrence of antibiotics in an urban watershed: From wastewater to drinking water. Sci. Total Environ. 2009, 407, 2711–2723. [Google Scholar] [CrossRef]
- Hall, W. Superbugs: An Arms Race against Bacteria. Harvard University Press: Cambridge, MA, USA, 2018.
- Mutiyar, P.K.; Mittal, A.K. Risk assessment of antibiotic residues in different water matrices in india: Key issues and challenges. Environ. Sci. Pollut. Res. 2014, 21, 7723–7736. [Google Scholar] [CrossRef] [PubMed]
- Fick, J.; Söderström, H.; Lindberg, R.H.; Phan, C.; Tysklind, M.; Larsson, D.J. Contamination of surface, ground, and drinking water from pharmaceutical production. Environ. Toxicol. Chem. 2009, 28, 2522–2527. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 2004, 198, 229–238. [Google Scholar] [CrossRef] [PubMed]
- European Union. Directive 2008/105/EC of the european parliament and of the council of 16 december 2008 on environmental quality standards in the field of water policy. Off. J. Eur. Union 2008, 348, 84–97. [Google Scholar]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Rezaei, B.; Damiri, S. Electrochemistry and adsorptive stripping voltammetric determination of amoxicillin on a multiwalled carbon nanotubes modified glassy carbon electrode. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2009, 21, 1577–1586. [Google Scholar] [CrossRef]
- Prado, T.M.; Cincotto, F.H.; Moraes, F.C.; Machado, S.A. Electrochemical sensor-based ruthenium nanoparticles on reduced graphene oxide for the simultaneous determination of ethinylestradiol and amoxicillin. Electroanalysis 2017, 29, 1278–1285. [Google Scholar] [CrossRef]
- Muhammad, A.; Yusof, N.; Hajian, R.; Abdullah, J. Construction of an electrochemical sensor based on carbon nanotubes/gold nanoparticles for trace determination of amoxicillin in bovine milk. Sensors 2016, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.E.; Carpico, G.; Coni, E. Electrochemical sensor for the detection and presumptive identification of quinolone and tetracycline residues in milk. Anal. Chim. Acta 2004, 520, 13–18. [Google Scholar] [CrossRef]
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019, 37, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhao, F. Molecularly imprinted polymer grown on multiwalled carbon nanotube surface for the sensitive electrochemical determination of amoxicillin. Electrochim. Acta 2015, 174, 33–40. [Google Scholar] [CrossRef]
- Zeinali, S.; Khoshsafar, H.; Rezaei, M.; Bagheri, H. Fabrication of a selective and sensitive electrochemical sensor modified with magnetic molecularly imprinted polymer for amoxicillin. Anal. Bioanal. Chem. Res. 2018, 5, 195–204. [Google Scholar]
- Betlem, K.; Mahmood, I.; Seixas, R.; Sadiki, I.; Raimbault, R.; Foster, C.; Crapnell, R.; Tedesco, S.; Banks, C.; Gruber, J.; et al. Evaluating the temperature dependence of heat-transfer based detection: A case study with caffeine and molecularly imprinted polymers as synthetic receptors. Chem. Eng. J. 2019, 359, 505–517. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.-T.; Li, D.-W.; Long, Y.-T. Recent developments and applications of screen-printed electrodes in environmental assays—A review. Anal. Chim. Acta 2012, 734, 31–44. [Google Scholar] [CrossRef]
- Almeida, E.S.; Silva, L.A.; Sousa, R.M.; Richter, E.M.; Foster, C.W.; Banks, C.E.; Munoz, R.A. Organic-resistant screen-printed graphitic electrodes: Application to on-site monitoring of liquid fuels. Anal. Chim. Acta 2016, 934, 1–8. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Canfarotta, F.; Czulak, J.; Johnson, R.; Betlem, K.; Mecozzi, F.; Down, M.P.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; et al. Thermal detection of cardiac biomarkers h-fabp and st2 using a molecularly imprinted nanoparticle-based multiplex sensor platform. ACS Sens. 2019, 4, 2838–2845. [Google Scholar] [CrossRef]
- Hudson, A.D.; Solà, R.; Ueta, J.T.; Battell, W.; Jamieson, O.; Dunbar, T.; Maciá, B.; Peeters, M. Synthesis of optimized molecularly imprinted polymers for the isolation and detection of antidepressants via HPLC. Biomimetics 2019, 4, 18. [Google Scholar] [CrossRef]
- Peeters, M.M.; van Grinsven, B.; Foster, C.W.; Cleij, T.J.; Banks, C.E. Introducing thermal wave transport analysis (twta): A thermal technique for dopamine detection by screen-printed electrodes functionalized with molecularly imprinted polymer (mip) particles. Molecules 2016, 21, 552. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.W.; Metters, J.P.; Kampouris, D.K.; Banks, C.E. Ultraflexible screen-printed graphitic electroanalytical sensing platforms. Electroanalysis 2014, 26, 262–274. [Google Scholar] [CrossRef]
- Cumba, L.R.; Foster, C.W.; Brownson, D.A.; Smith, J.P.; Iniesta, J.; Thakur, B.; Do Carmo, D.R.; Banks, C.E. Can the mechanical activation (polishing) of screen-printed electrodes enhance their electroanalytical response? Analyst 2016, 141, 2791–2799. [Google Scholar] [CrossRef]
- Blanco, E.; Foster, C.W.; Cumba, L.R.; Do Carmo, D.R.; Banks, C.E. Can solvent induced surface modifications applied to screen-printed platforms enhance their electroanalytical performance? Analyst 2016, 141, 2783–2790. [Google Scholar] [CrossRef] [PubMed]
- Casadio, S.; Lowdon, J.W.; Betlem, K.; Ueta, J.T.; Foster, C.W.; Cleij, T.J.; van Grinsven, B.; Sutcliffe, O.B.; Banks, C.E.; Peeters, M. Development of a novel flexible polymer-based biosensor platform for the thermal detection of noradrenaline in aqueous solutions. Chem. Eng. J. 2017, 315, 459–468. [Google Scholar] [CrossRef]
- van Grinsven, B.; Vanden Bon, N.; Strauven, H.; Grieten, L.; Murib, M.; Jiménez Monroy, K.L.; Janssens, S.D.; Haenen, K.; Schöning, M.J.; Vermeeren, V.; et al. Heat-transfer resistance at solid–liquid interfaces: A tool for the detection of single-nucleotide polymorphisms in DNA. ACS Nano 2012, 6, 2712–2721. [Google Scholar] [CrossRef]
- Canfarotta, F.; Czulak, J.; Betlem, K.; Sachdeva, A.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; Peeters, M. A novel thermal detection method based on molecularly imprinted nanoparticles as recognition elements. Nanoscale 2018, 10, 2081–2089. [Google Scholar] [CrossRef]
- Vasapollo, G.; Sole, R.D.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef]
- Piletska, E.V.; Guerreiro, A.R.; Romero-Guerra, M.; Chianella, I.; Turner, A.P.; Piletsky, S.A. Design of molecular imprinted polymers compatible with aqueous environment. Anal. Chim. Acta 2008, 607, 54–60. [Google Scholar] [CrossRef]
- Martinez Bueno, M.J.; Hernando, M.D.; Herrera, S.; Gómez, M.J.; Fernández-Alba, A.R.; Bustamante, I.; Garcia-Calvo, E. Pilot survey of chemical contaminants from industrial and human activities in river waters of spain. Int. J. Environ. Anal. Chem. 2010, 90, 321–343. [Google Scholar] [CrossRef]
- Bui, B.T.S.; Haupt, K. Molecularly imprinted polymers: Synthetic receptors in bioanalysis. Anal. Bioanal. Chem. 2010, 398, 2481–2492. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).