Detecting Fingerprints of Waterborne Bacteria on a Sensor
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Bacteria
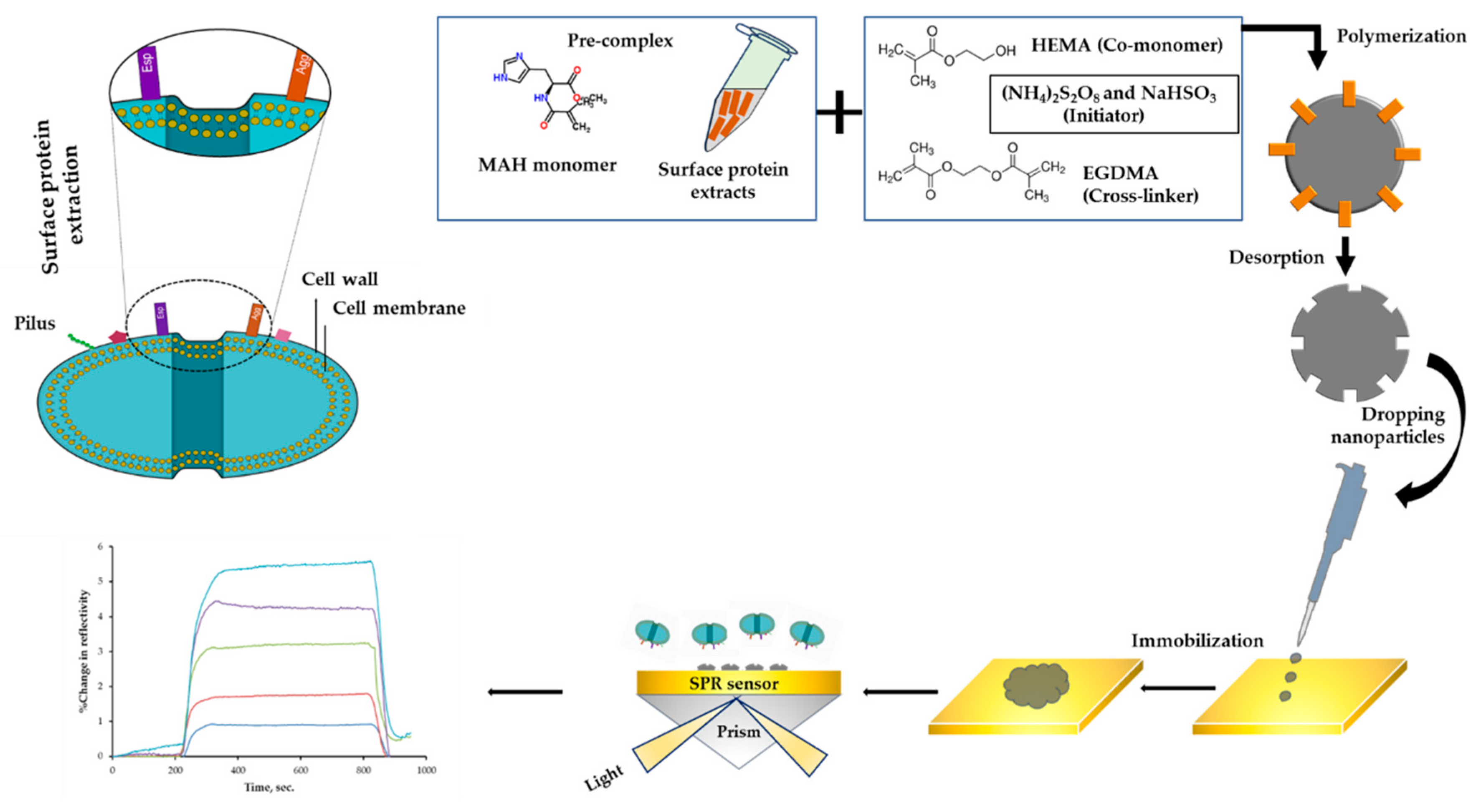
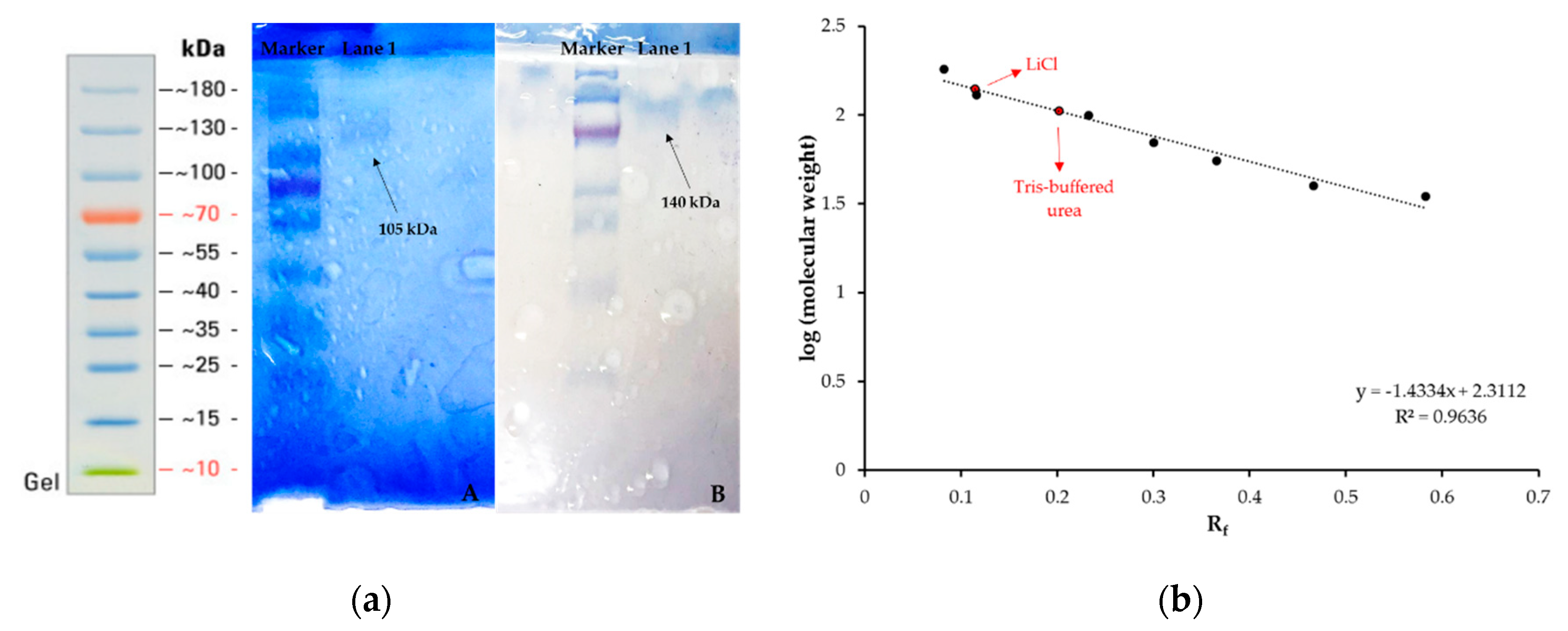
2.3. Preparation and Characterization of Surface Proteins
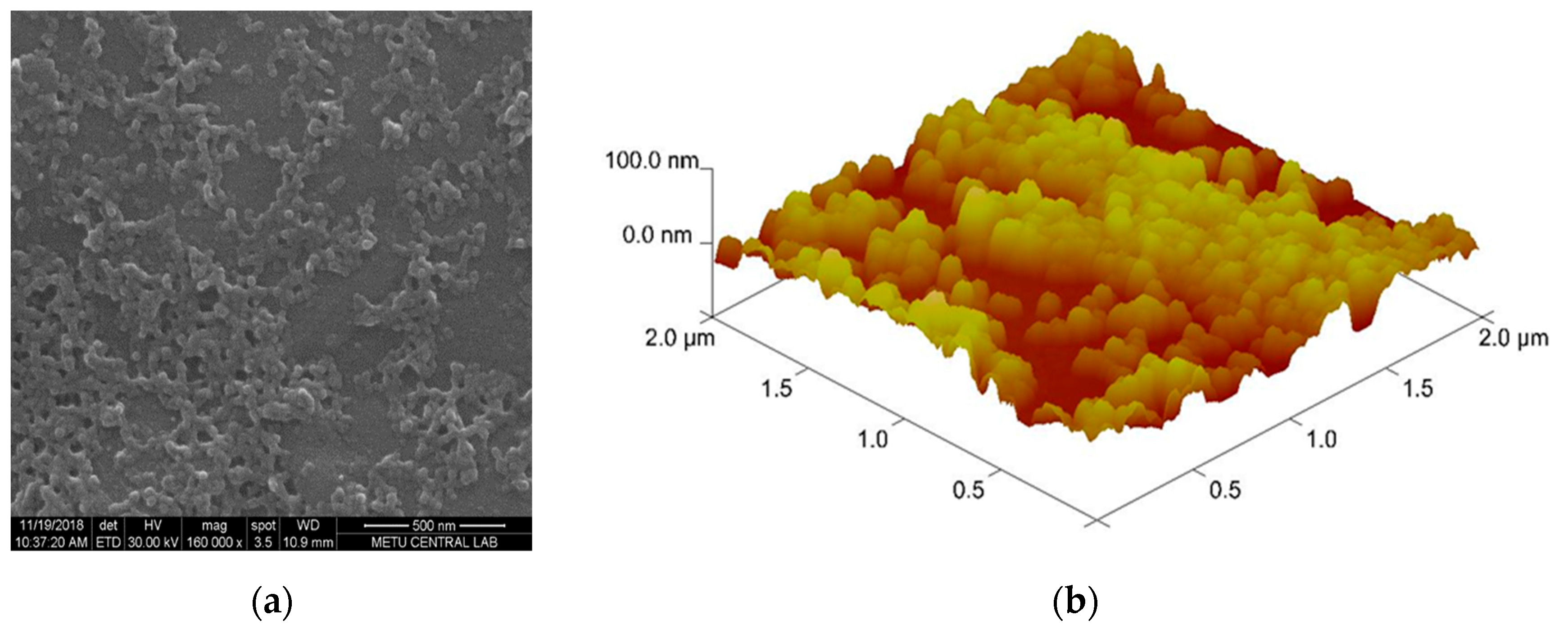
2.4. Preparation and Characterization of Nanoparticles
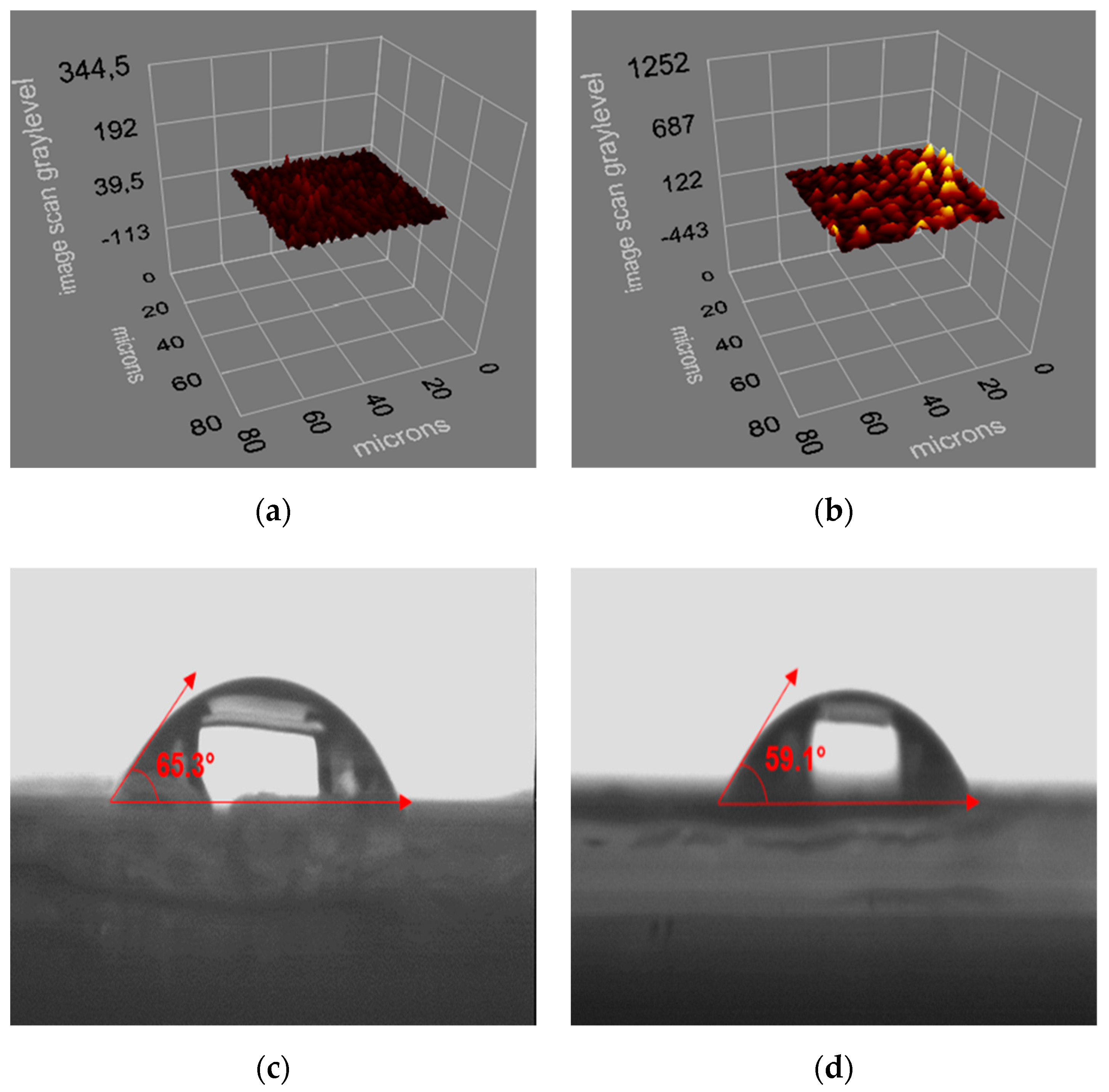
2.5. Preparation and Characterization of Sensor
2.6. Kinetic Analyses
2.7. Repeatability and Selectivity Analyses
2.8. Water Sample Analyses
3. Results and Discussion
3.1. Characterization Results
3.2. Kinetic Analyses Results
3.3. Adsorption Isotherm Models
3.4. Repeatability and Selectivity Analyses Results
3.5. Water Sample Analyses Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| k | Selectivity coefficient |
| k′ | Relative selectivity coefficient |
| ΔRmax | Maximum signal shift |
| ka (cfu/mL−1·s−1) | Forward kinetic rate constant |
| kd (1/s) | Reverse kinetic rate constant |
| KA (mL/cfu) | Forward equilibrium constant |
| KD (cfu/mL) | Reverse equilibrium constant |
| 1/n | Freundlich exponent |
| R2 | Correlation coefficient |
References
- Erdem, Ö.; Saylan, Y.; Cihangir, N.; Denizli, A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens. Biolectron. 2019, 126, 608–614. [Google Scholar] [CrossRef]
- O’Mullan, G.D.; Juhl, A.R.; Reichert, R.; Schneider, E.; Martinez, N. Patterns of sediment-associated fecal indicator bacteria in an urban estuary: Benthic-pelagic coupling and implications for shoreline water quality. Sci. Total Environ. 2019, 656, 1168–1177. [Google Scholar] [CrossRef]
- Rochelle-Newall, E.; Nguyen, T.M.H.; Le, T.P.Q.; Sengtaheuanghoung, O.; Ribolzi, O. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Scott, T.M.; Jenkins, T.M.; Lukasik, J.; Rose, J.B. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 2015, 39, 283–287. [Google Scholar] [CrossRef]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016, 20, 127–133. [Google Scholar] [CrossRef]
- Waar, K.; Van Der Mei, H.C.; Harmsen, H.J.M.; Degener, J.E.; Busscher, H.J. Enterococcus faecalis surface proteins determine its adhesion mechanism to bile drain materials. Microbiology 2002, 148, 1863–1870. [Google Scholar] [CrossRef][Green Version]
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar] [CrossRef]
- Ali, Y.M.; Sim, R.B.; Schwaeble, W.; Sshabaan, M.A. Enterococcus faecalis escapes complement-mediated killing via recruitment of complement factor H. J. Infect. Dis. 2019. accepted manuscript. [Google Scholar] [CrossRef]
- Hashemi, E.; Forouzandeh, M. Designing a new biosensor “DNA ELISA” to detect Escherichia coli using genomic DNA and comparison of this method to PCR-ELISA. J. Enzyme Inhib. Med. Chem. 2018, 33, 722–725. [Google Scholar] [CrossRef]
- Zhu, L.; He, J.; Cao, X.; Huang, K.; Luo, Y.; Xu, W. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus cereus in food. Sci. Rep. 2016, 6, 16092. [Google Scholar] [CrossRef]
- Jordana-Lluch, E.; Giménez, M.; Quesada, M.D.; Ausina, V.; Martró, E. Improving the diagnosis of bloodstream infections: PCR coupled with mass spectrometry. BioMed Res. Int. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Ma, K.; Deng, Y.; Bai, Y.; Xu, D.; Chen, E.; Wu, H.; Li, B.; Gao, L. Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Control 2014, 42, 87–93. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Zheng, S.; Liu, Y.; He, Z.; Luo, F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sens. Actuators B Chem. 2016, 230, 191–198. [Google Scholar] [CrossRef]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Song, X.C.; Mrázek, J.; Lamačová, J.; Lynn, N.S., Jr.; Šedivákb, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Molecular fingerprints of hemoglobin on a nanofilm chip. Sensors 2018, 18, 3016. [Google Scholar] [CrossRef]
- Farka, Z.; Juřík, T.; Pastucha, M.; Skládal, P. Enzymatic precipitation enhanced surface plasmon resonance immunosensor for the detection of Salmonella in powdered milk. Anal. Chem. 2016, 88, 11830–11836. [Google Scholar] [CrossRef]
- Saylan, Y.; Akgönüllü, S.; Yavuz, H.; Ünal, S.; Denizli, A. Molecularly imprinted polymer based sensors for medical applications. Sensors 2019, 19, 1279. [Google Scholar] [CrossRef]
- Shang, L.; Liu, C.; Chen, B.; Hayashi, K. Plant biomarker recognition by molecular imprinting based localized surface plasmon resonance sensor array: Performance improvement by enhanced hotspot of au nanostructure. ACS Sens. 2018, 3, 531–1538. [Google Scholar] [CrossRef]
- Yildiz, U.H.; Inci, F.; Wang, S.Q.; Toy, M.; Tekin, H.C.; Javaid, A.; Lau, D.T.Y.; Demirci, U. Recent advances in micro/nanotechnologies for global control of hepatitis B infection. Biotechnol. Adv. 2015, 33, 178–190. [Google Scholar] [CrossRef]
- Atale, S.S.; Dyawanapelly, S.; Jagtap, D.D.; Jain, R.; Dandekar, P. Understanding the nano-bio interactions using real-time surface plasmon resonance tool. Int. J. Biol. Macromol. 2019, 123, 97–107. [Google Scholar] [CrossRef]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular imprinting of macromolecules for sensor applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef]
- Garipcan, B.; Denizli, A. A novel affinity support material for the separation of immunoglobulin G from human plasma. Macromol. Biosci. 2002, 2, 135–144. [Google Scholar] [CrossRef]
- Tiong, H.K.; Hartson, S.; Muriana, P.M. Comparison of five methods for direct extraction of surface proteins from Listeria monocytogenes for proteomic analysis by orbitrap mass spectrometry. J. Microbiol. Methods 2015, 110, 54–60. [Google Scholar] [CrossRef]
- Templier, V.; Roux, A.; Roupioz, Y.; Livache, T. Ligands for label-free detection of whole bacteria on biosensors: A review. Trends Anal. Chem. 2016, 79, 71–79. [Google Scholar] [CrossRef]
- Saylan, Y.; Tamahkar, E.; Denizli, A. Recognition of lysozyme using surface imprinted bacterial cellulose nanofibers. J. Biomater. Sci. Polym. 2017, 28, 1950–1965. [Google Scholar] [CrossRef]
- Jia, M.; Zhang, Z.; Li, J.; Ma, X.; Chen, L.; Yang, X. Molecular imprinting technology for microorganism analysis. Trends Anal. Chem. 2018, 106, 190–201. [Google Scholar] [CrossRef]
- Masdor, N.; Altintas, Z.; Tothill, I. Surface plasmon resonance immunosensor for the detection of Campylobacter jejuni. Chemosensors 2017, 5, 16. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, Y.P.; Wang, W.; Shen, Y.; Guo, J.S. Surface plasmon resonance for water pollutant detection and water process analysis. Trends Anal. Chem. 2016, 85, 153–165. [Google Scholar] [CrossRef]
- Aryantini, N.P.D.; Kondoh, D.; Nishiyama, K.; Yamamoto, Y.; Mukai, T.; Sujaya, I.N.; Urashima, T.; Fukuda, K. Anchorless cell surface proteins function as laminin-binding adhesins in Lactobacillus rhamnosus FSMM22. FEMS Microbiol. Lett. 2017, 364, fnx056. [Google Scholar] [CrossRef]
- Moreno-Gordaliza, E.; Stigter, E.C.; Lindenburg, P.W.; Hankemeier, T. Bacterial surface layer proteins as a novel capillary coating material for capillary electrophoretic separations. Anal. Chim. Acta 2016, 923, 89–100. [Google Scholar] [CrossRef]
- McBirney, S.E.; Trinh, K.; Wong-Beringer, A.; Armani, A.M. Wavelength-normalized spectroscopic analysis of Staphylococcus aureus and Pseudomonas aeruginosa growth rates. Biomed. Opt. Express 2016, 7, 4034–4042. [Google Scholar] [CrossRef]
- Hancock, L.E.; Murray, B.E.; Sillanpää, J. Enterococcal Cell Wall Components and Structures. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Master’s Thesis, Massachusetts Eye and Ear Infirmary, Boston, MA, USA, 2014; pp. 1–44. [Google Scholar]
- Khan, M.A.R.; Moreira, F.T.; Riu, J.; Sales, M.G.F. Plastic antibody for the electrochemical detection of bacterial surface proteins. Sens. Actuators B Chem. 2016, 233, 697–704. [Google Scholar] [CrossRef]
- Lahcen, A.A.; Arduini, F.; Lista, F.; Amine, A. Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection. Sens. Actuators B Chem. 2018, 276, 114–120. [Google Scholar] [CrossRef]
- Khan, M.A.R.; Cardoso, A.R.A.; Sales, M.G.F.; Merino, S.; Tomás, J.M.; Rius, F.X.; Riu, J. Artificial receptors for the electrochemical detection of bacterial flagellar filaments from Proteus mirabilis. Sens. Actuators B Chem. 2017, 244, 732–741. [Google Scholar] [CrossRef]

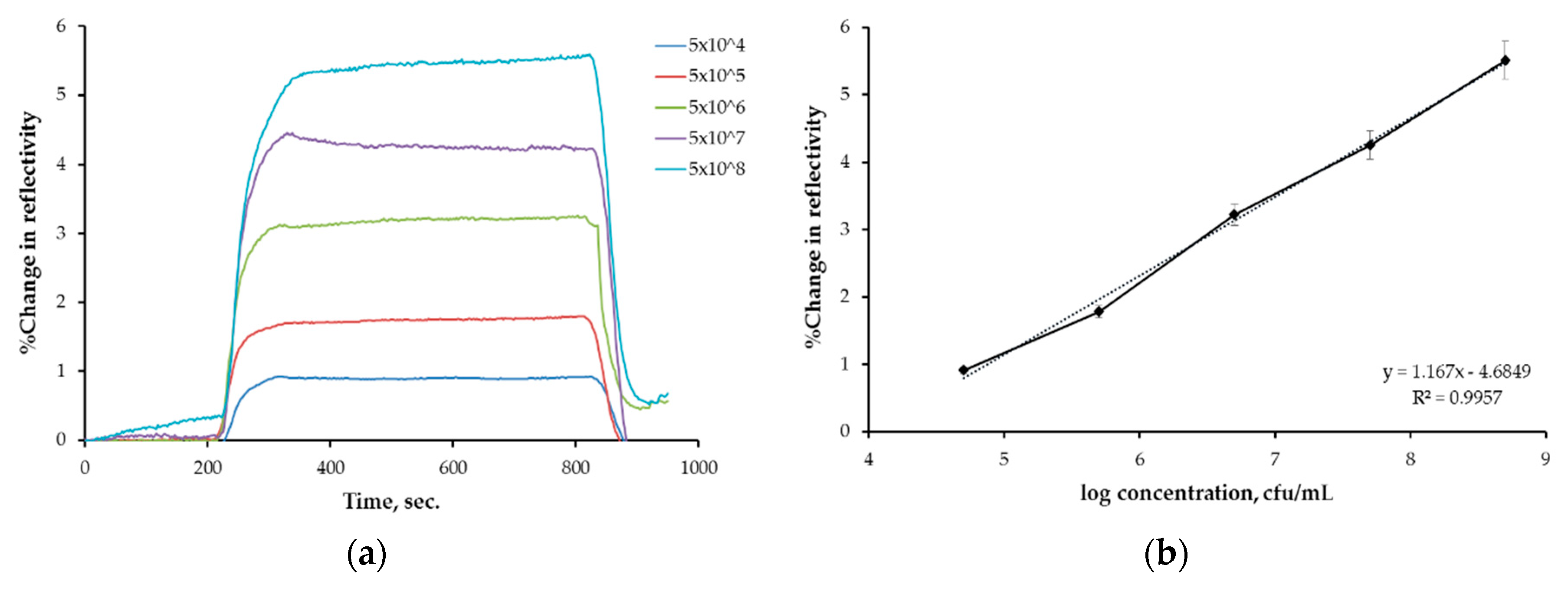
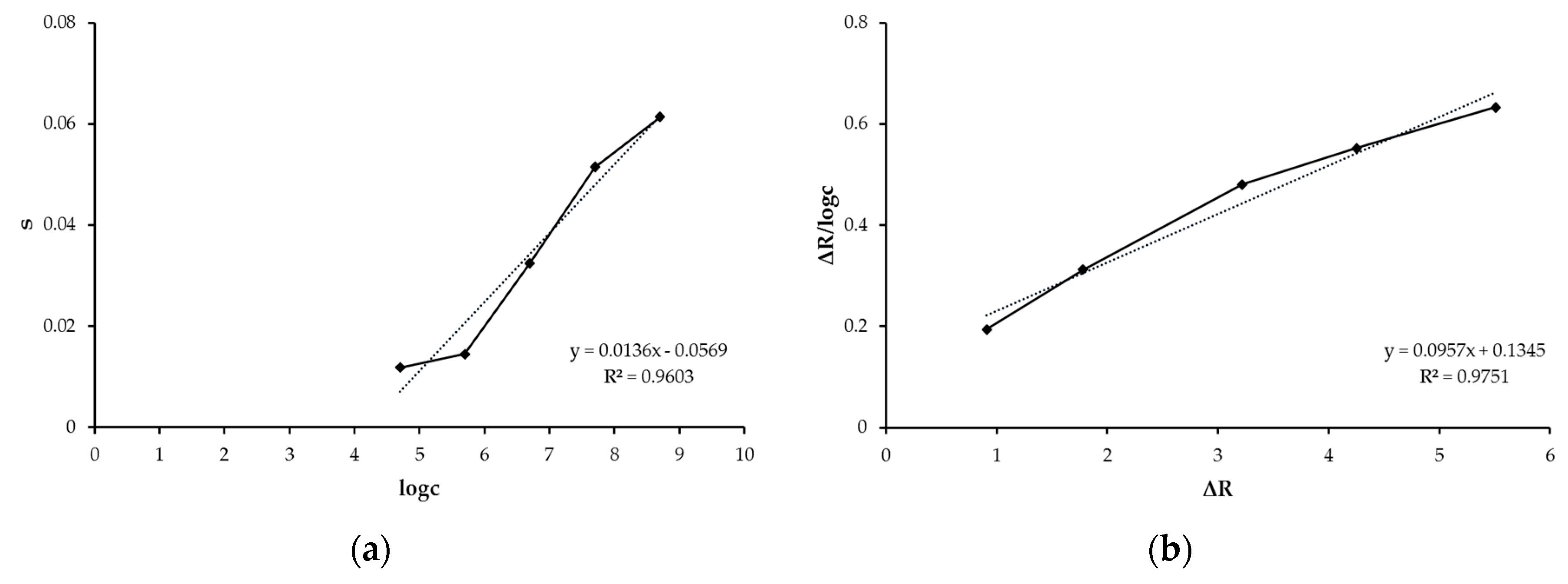


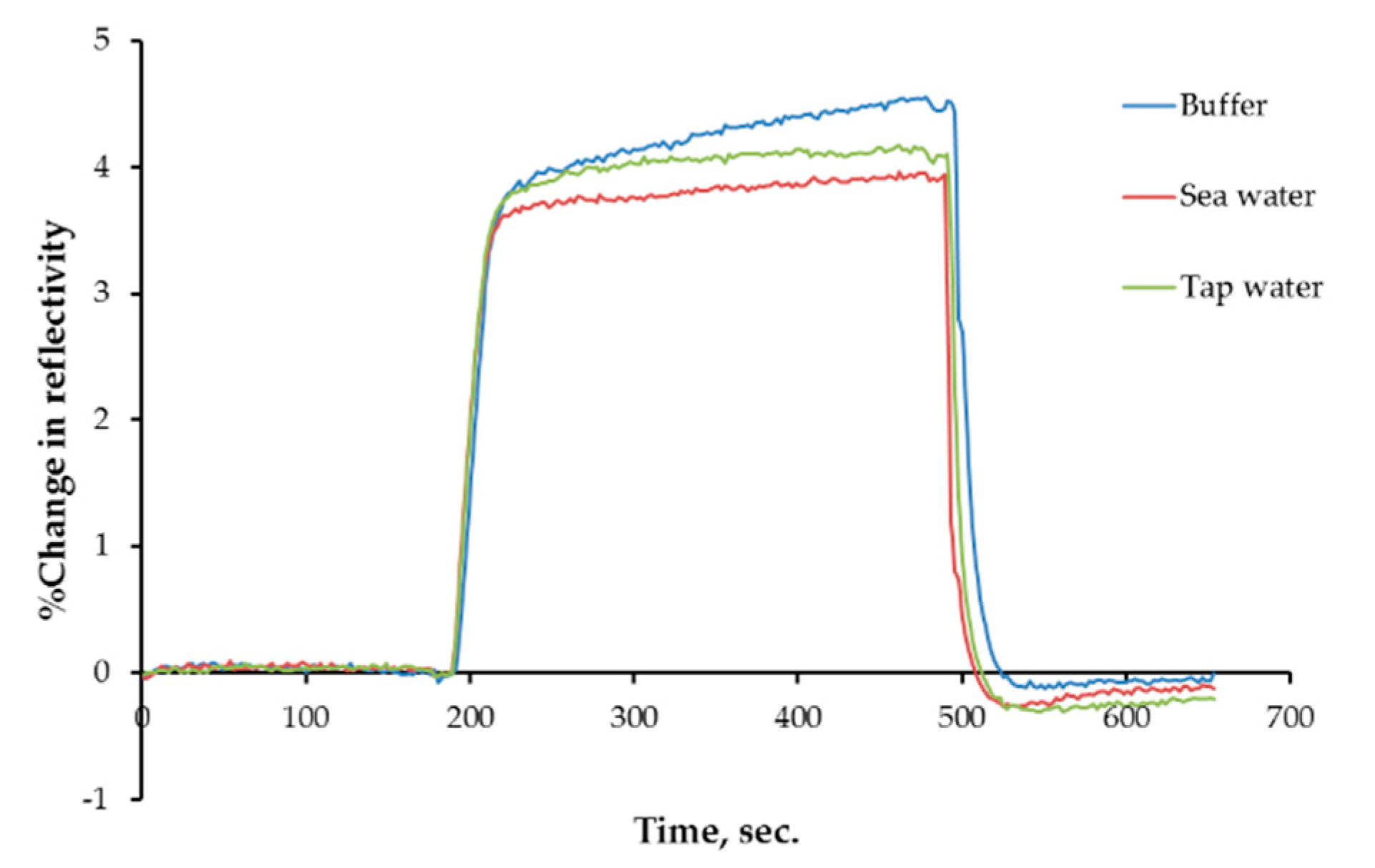
| Association | Scatchard | Langmuir | Freundlich |
|---|---|---|---|
| ka = 0.014 | Ka = 0.096 | Ka = 0.106 | 1/n = 0.95 |
| kd = 0.057 | Kd = 10.45 | Kd = 9.466 | ΔRmax = 97.92 |
| Ka = 0.239 | ΔRmax = 1.41 | ΔRmax = 1.01 | R2 = 0.983 |
| Kd = 4.184 R2 = 0.960 | R2 = 0.975 | R2 = 0.936 |
| Bacteria | ΔRImprinted | kImprinted | ΔRNon-Imprinted | kNon-Imprinted | k′ |
|---|---|---|---|---|---|
| E. faecalis | 5.51 | − | 0.81 | − | − |
| S. aureus | 2.04 | 2.71 | 0.75 | 1.08 | 2.51 |
| B. subtilis | 1.82 | 3.03 | 0.74 | 1.09 | 2.77 |
| E. coli | 1.56 | 3.53 | 0.72 | 1.13 | 3.14 |
| Bacteria | Method | Linearity | Limit of Detection | Reference |
|---|---|---|---|---|
| Staphylococcus aureus (protein A) | Electrochemical impedance spectroscopy | 23.8 pM–4.76 µM | 16.83 nM | [33] |
| Bacillus cereus (spores) | Electrochemical sensor | 102–105 cfu/mL | 102 cfu/mL | [34] |
| Proteus mirabilis (flagellar filaments) | Electrical impedance spectroscopy | 0.01 ± 60 µg/mL | 0.7 ng/mL | [35] |
| Enterococcus faecalis (surface protein) | Surface plasmon resonance | 5 × 104–5 × 108 cfu/mL | 3.4 × 104 cfu/mL | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saylan, Y.; Erdem, Ö.; Cihangir, N.; Denizli, A. Detecting Fingerprints of Waterborne Bacteria on a Sensor. Chemosensors 2019, 7, 33. https://doi.org/10.3390/chemosensors7030033
Saylan Y, Erdem Ö, Cihangir N, Denizli A. Detecting Fingerprints of Waterborne Bacteria on a Sensor. Chemosensors. 2019; 7(3):33. https://doi.org/10.3390/chemosensors7030033
Chicago/Turabian StyleSaylan, Yeşeren, Özgecan Erdem, Nilüfer Cihangir, and Adil Denizli. 2019. "Detecting Fingerprints of Waterborne Bacteria on a Sensor" Chemosensors 7, no. 3: 33. https://doi.org/10.3390/chemosensors7030033
APA StyleSaylan, Y., Erdem, Ö., Cihangir, N., & Denizli, A. (2019). Detecting Fingerprints of Waterborne Bacteria on a Sensor. Chemosensors, 7(3), 33. https://doi.org/10.3390/chemosensors7030033






