Spectroscopic Chemical Sensing and Imaging: From Plants to Animals and Humans
Abstract
1. Introduction
2. Fluorescence-Based Technologies
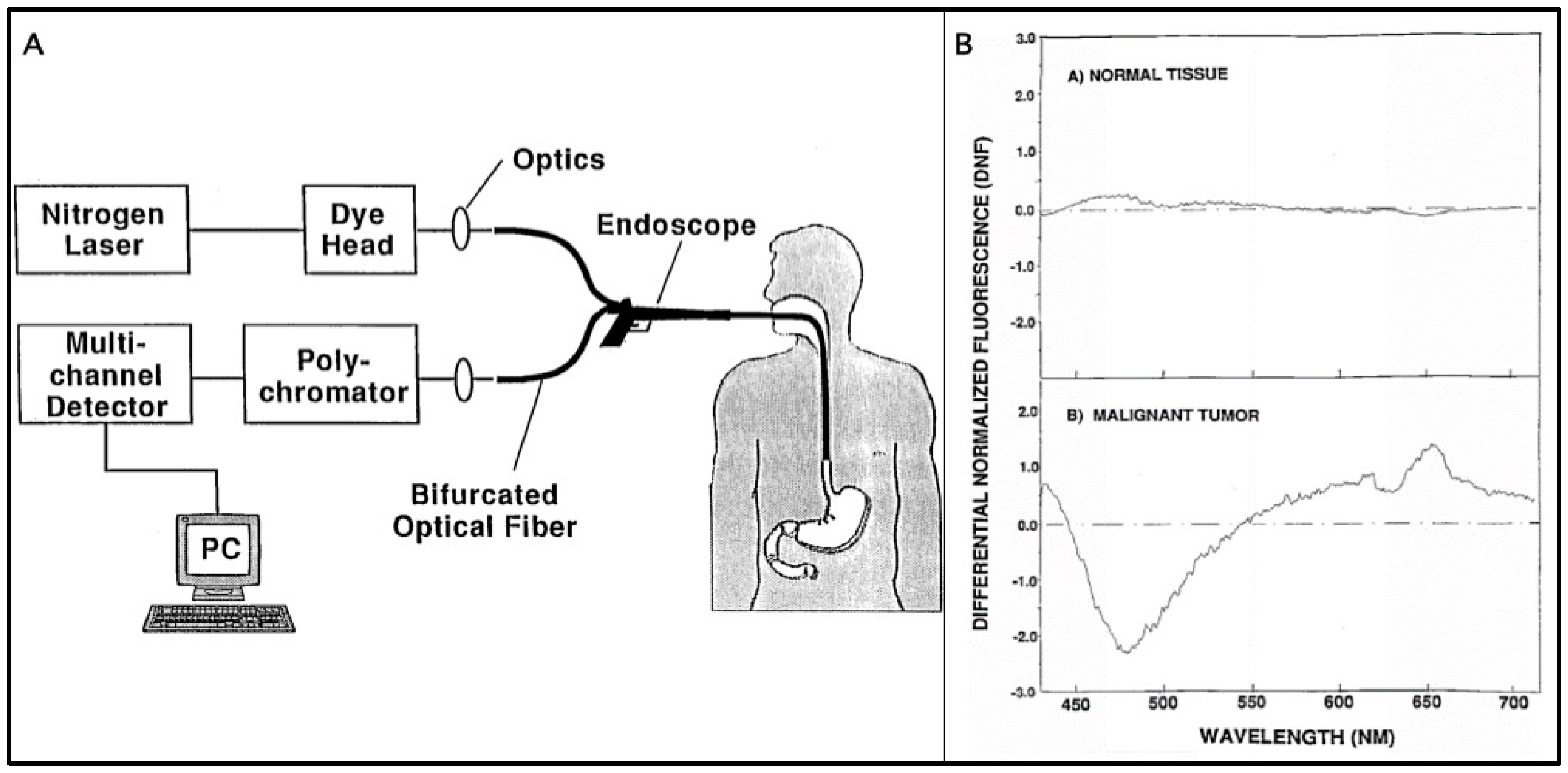
2.1. Differential Normalized Light Induced Fluorescence
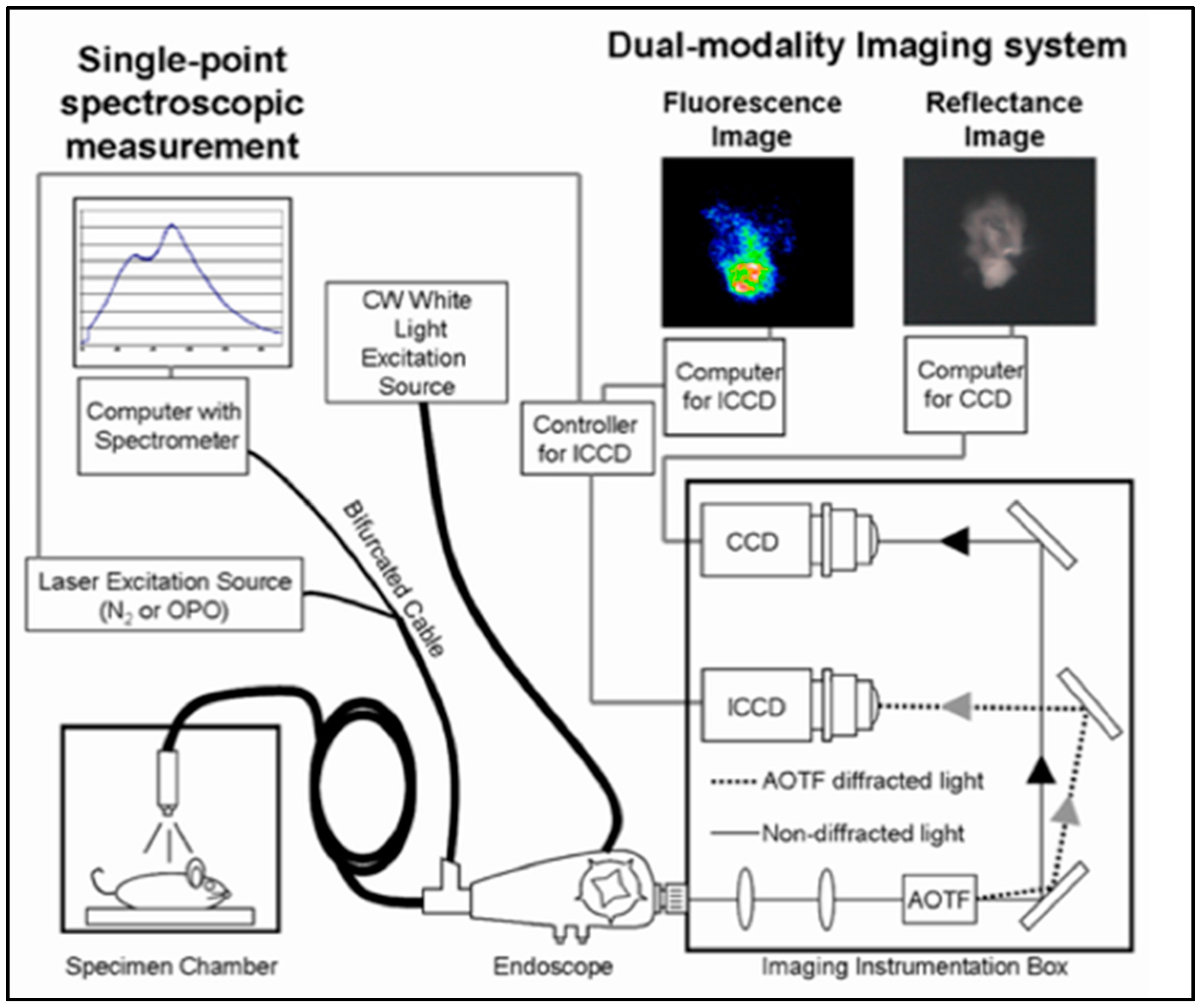
2.2. Hyperspectral Fluorescence Imaging
3. SERS-Based Technologies
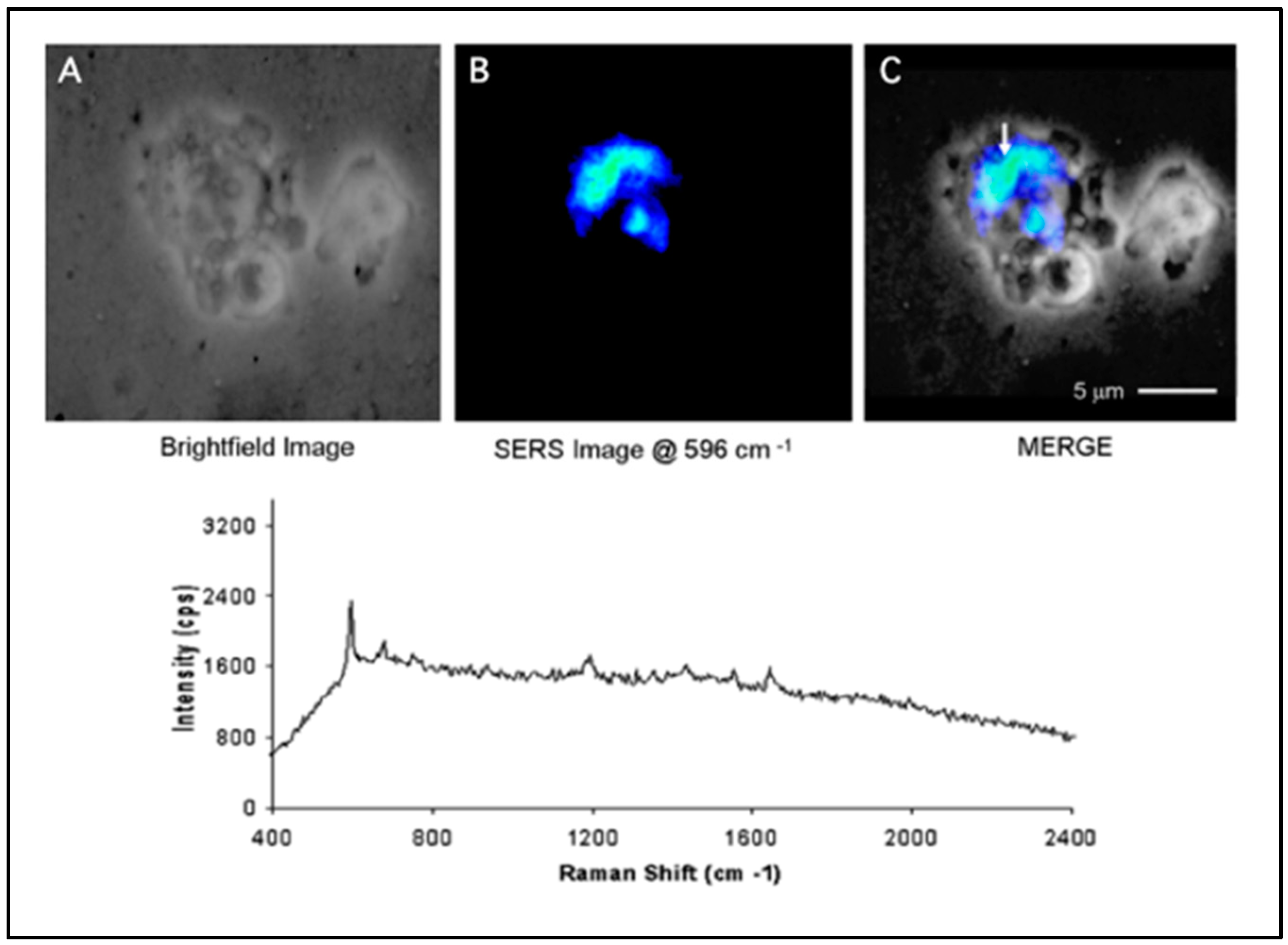
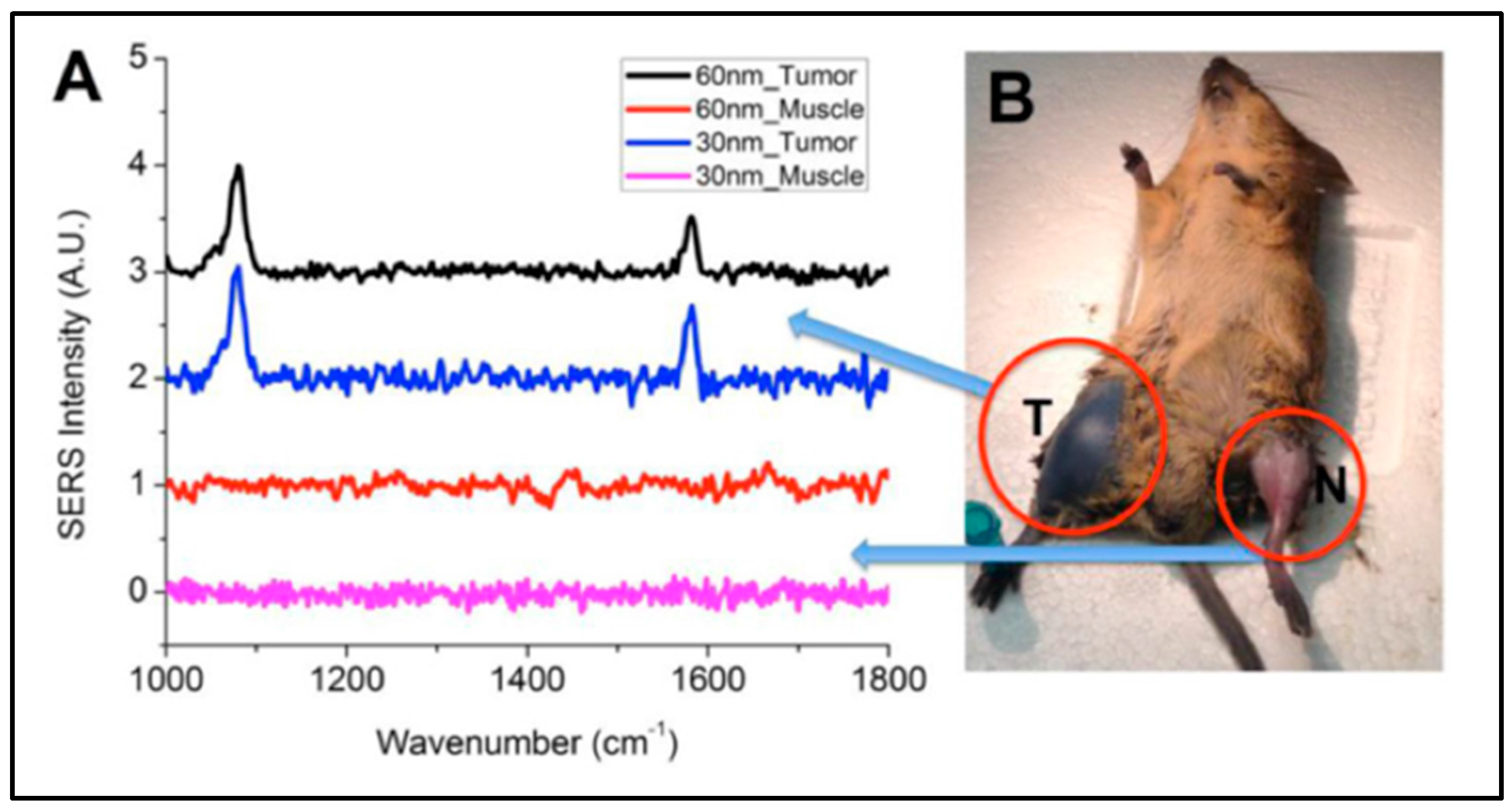
3.1. SERS Nanoprobes
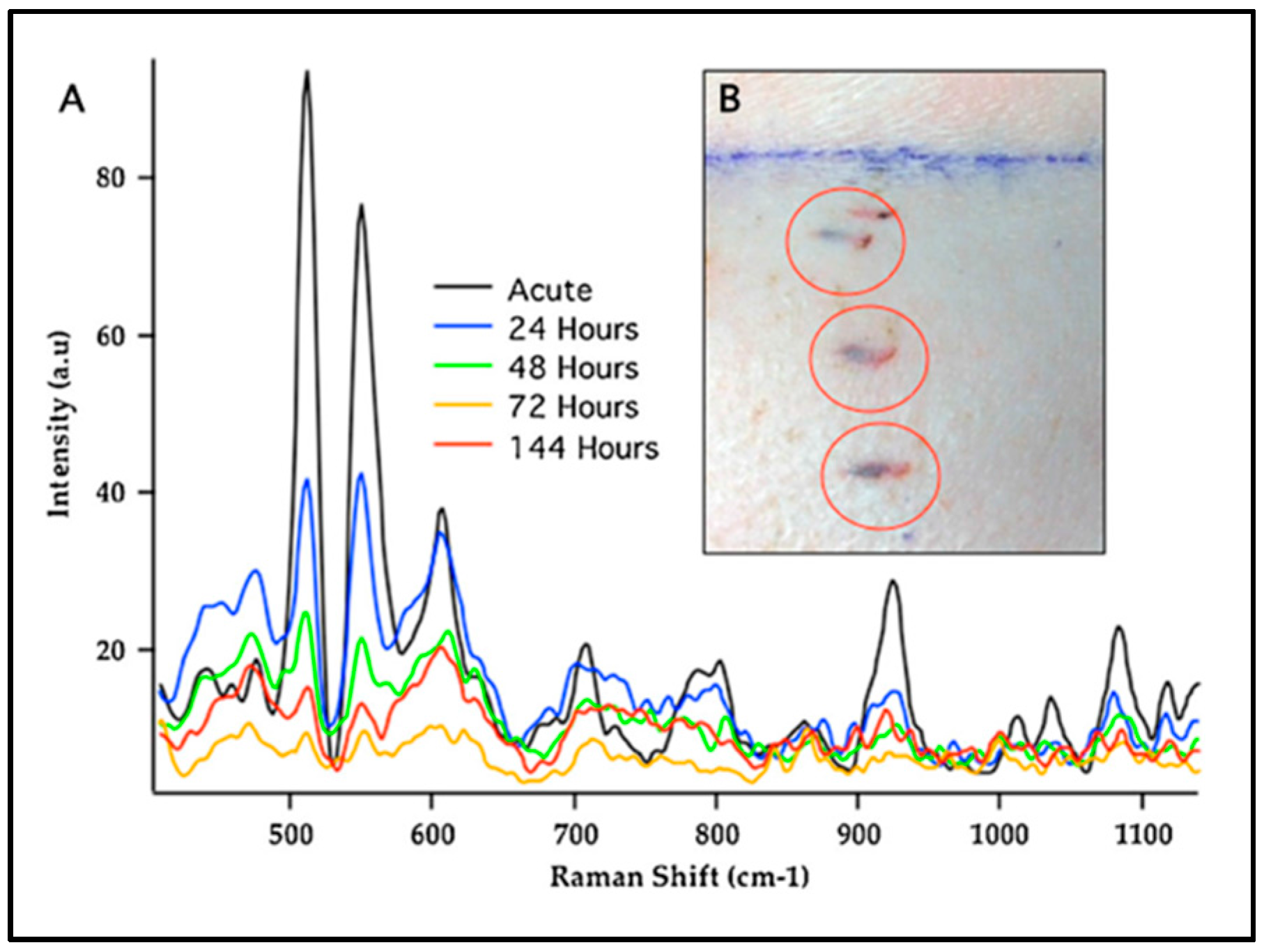
3.2. SERS Sensors
4. Conclusions and Perspective
Acknowledgments
Conflicts of Interest
References
- Vo-Dinh, T. Biomedical Photonics Handbook: Biomedical Diagnostics, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Lipson, R.L.; Baldes, E.J.; Olsen, A.M. The Use of a Derivative of Hematoporphyrin in Tumor Detection. JNCI: J. Natl. Cancer Inst. 1961, 26, 1–11. [Google Scholar] [PubMed]
- Andersson-Engels, S.; af Klinteberg, C.; Svanberg, K.; Svanberg, S. In vivo fluorescence imaging for tissue diagnostics. Phys. Med. Biol. 1997, 42, 815. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Panjehpour, M.; Overholt, B.F.; Buckley III, P. Laser-Induced Differential Fluorescence for Cancer Diagnosis without Biopsy. Appl. Spectrosc. 1997, 51, 58–63. [Google Scholar] [CrossRef]
- Liu, Q.; Grant, G.; Li, J.; Zhang, Y.; Hu, F.; Li, S.; Wilson, C.; Chen, K.; Bigner, D.; Vo-Dinh, T. Compact point-detection fluorescence spectroscopy system for quantifying intrinsic fluorescence redox ratio in brain cancer diagnostics. J. Biomed. Opt. 2011, 16, 037004. [Google Scholar] [CrossRef] [PubMed]
- Ramanujam, N.; Mitchell, M.F.; Mahadevan, A.; Warren, S.; Thomsen, S.; Silva, E.; Richards-Kortum, R. In vivo diagnosis of cervical intraepithelial neoplasia using 337-nm-excited laser-induced fluorescence. Proc. Natl. Acad. Sci. USA 1994, 91, 10193. [Google Scholar] [PubMed]
- Schomacker, K.T.; Frisoli, J.K.; Compton, C.C.; Flotte, T.J.; Richter, J.M.; Deutsch, T.F.; Nishioka, N.S. Ultraviolet laser-induced fluorescence of colonic polyps. Gastroenterology 1992, 102, 1155–1160. [Google Scholar] [CrossRef]
- Deckelbaum, L.I.; Lam, J.K.; Cabin, H.S.; Clubb, K.S.; Long, M.B. Discrimination of normal and atherosclerotic aorta by laser-induced fluorescence. Lasers Surg. Med. 1987, 7, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.L.; Maher, C.O.; et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef] [PubMed]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Hiromoto, M.Y.K.; Begun, G.M.; Moody, R.L. Surface-enhanced Raman spectrometry for trace organic analysis. Anal. Chem. 1984, 56, 1667–1670. [Google Scholar] [CrossRef]
- Maier, S.A. Plasmonics: Fundamentals and Applications; Springer: New York, NY, USA, 2007. [Google Scholar]
- Kelly, K.L.; Coronado, E.; Zhao, L.L.; Schatz, G.C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Strobbia, P.; Languirand, E.; Cullum, B.M. Recent advances in plasmonic nanostructures for sensing: A review. Opt. Eng. 2015, 54, 100902. [Google Scholar] [CrossRef]
- Vo-Dinh, T. Surface-enhanced Raman spectroscopy using muctureetallic nanostrs. TrAC Trends Anal. Chem. 1998, 17, 557–582. [Google Scholar] [CrossRef]
- Dhawan, A.; Norton, S.J.; Gerhold, M.D.; Vo-Dinh, T. Comparison of FDTD numerical computations and analytical multipole expansion method for plasmonics-active nanosphere dimers. Opt. Express 2009, 17, 9688–9703. [Google Scholar] [CrossRef] [PubMed]
- Khoury, C.G.; Norton, S.J.; Vo-Dinh, T. Plasmonics of 3-D Nanoshell Dimers Using Multipole Expansion and Finite Element Method. ACS Nano 2009, 3, 2776–2788. [Google Scholar] [CrossRef] [PubMed]
- Norton, S.J.; Vo-Dinh, T. Plasmon Resonances of Nanoshells of Spheroidal Shape. IEEE Trans. Nanotechnol. 2007, 6, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Norton, S.J.; Vo-Dinh, T. Optical response of linear chains of metal nanospheres and nanospheroids. J. Opt. Soc. Am. A 2008, 25, 2767–2775. [Google Scholar] [CrossRef]
- Norton, S.J.; Vo-Dinh, T. Spectral bounds on plasmon resonances for Ag and Au prolate and oblate nanospheroids. J. Nanophotonics 2008, 2, 029501. [Google Scholar] [CrossRef] [PubMed]
- Norton, S.J.; Vo-Dinh, T. Plasmonics enhancement of a luminescent or Raman-active layer in a multilayered metallic nanoshell. Appl. Opt. 2009, 48, 5040–5049. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Dhawan, A.; Norton, S.J.; Khoury, C.G.; Wang, H.-N.; Misra, V.; Gerhold, M.D. Plasmonic Nanoparticles and Nanowires: Design, Fabrication and Application in Sensing. J. Phys. Chem. C 2010, 114, 7480–7488. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.E.; Holthoff, E.L.; Pellegrino, P.M. Surface-Enhanced Raman Scattering Detection of Ammonium Nitrate Samples Fabricated Using Drop-on-Demand Inkjet Technology. Appl. Spectrosc. 2014, 68, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Bello, J.M.; Narayanan, V.A.; Stokes, D.L.; Vo-Dinh, T. Fiber-optic remote sensor for in situ surface-enhanced Raman scattering analysis. Anal. Chem. 1990, 62, 2437–2441. [Google Scholar] [CrossRef]
- Lussier, F.; Brulé, T.; Vishwakarma, M.; Das, T.; Spatz, J.P.; Masson, J.-F. Dynamic-SERS Optophysiology: A Nanosensor for Monitoring Cell Secretion Events. Nano Lett. 2016, 16, 3866–3871. [Google Scholar] [CrossRef] [PubMed]
- Karabeber, H.; Huang, R.; Iacono, P.; Samii, J.M.; Pitter, K.; Holland, E.C.; Kircher, M.F. Guiding Brain Tumor Resection Using Surface-Enhanced Raman Scattering Nanoparticles and a Hand-Held Raman Scanner. ACS Nano 2014, 8, 9755–9766. [Google Scholar] [CrossRef] [PubMed]
- Noonan, J.; Grassia, G.; Macritchie, N.; Asiala, S.; Gracie, K.; Faulds, K.; McInnes, I.B.; Garside, P.; Graham, D.; Maffia, P. Multiplex detection of endothelial cell activation biomarkers using surface enhanced raman spectroscopy (sers). Atherosclerosis 2017, 263, e1–e18. [Google Scholar] [CrossRef]
- Prosst, R.; Gahlen, J. Fluorescence diagnosis of colorectal neoplasms: A review of clinical applications. Int. J. Colorectal Dis. 2002, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- DSouza, A.V.; Lin, H.; Henderson, E.R.; Samkoe, K.S.; Pogue, B.W. Review of fluorescence guided surgery systems: Identification of key performance capabilities beyond indocyanine green imaging. J. Biomed. Opt. 2016, 21, 080901. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.H.; Kwok, S.J.J. Light in diagnosis, therapy and surgery. Nat. Biomed. Eng. 2017, 1, 0008. [Google Scholar] [CrossRef] [PubMed]
- Henry, A.-I.; Sharma, B.; Cardinal, M.F.; Kurouski, D.; Van Duyne, R.P. Surface-Enhanced Raman Spectroscopy Biosensing: In Vivo Diagnostics and Multimodal Imaging. Anal. Chem. 2016, 88, 6638–6647. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-enhanced Raman spectroscopy for in vivo biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar] [CrossRef]
- Wagnieres, G.A.; Star, W.M.; Wilson, B.C. In Vivo Fluorescence Spectroscopy and Imaging for Oncological Applications. Photochem. Photobiol. 1998, 68, 603–632. [Google Scholar] [CrossRef] [PubMed]
- Panjehpour, M.; Overholt, B.F.; Vo-Dinh, T.; Coppola, D. The effect of reactive atypia/inflammation on the laser-induced fluorescence diagnosis of non-dysplastic Barrett's esophagus. Lasers Surg. Med. 2012, 44, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T. A hyperspectral imaging system for in vivo optical diagnostics. IEEE Eng. Med. Biol. Mag. 2004, 23, 40–49. [Google Scholar] [PubMed]
- Kong, S.G.; Martin, M.; Vo-Dinh, T. Hyperspectral Fluorescence Imaging for Mouse Skin Tumor Detection. Etri J. 2006, 28, 770–776. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced Raman spectroscopy: A brief retrospective. J. Raman Spectrosc. 2005, 36, 485–496. [Google Scholar] [CrossRef]
- Wabuyele, M.B.; Yan, F.; Griffin, G.D.; Vo-Dinh, T. Hyperspectral surface-enhanced Raman imaging of labeled silver nanoparticles in single cells. Rev. Sci. Instrum. 2005, 76, 063710. [Google Scholar] [CrossRef]
- Jackson, J.B.; Halas, N.J. Surface-enhanced Raman scattering on tunable plasmonic nanoparticle substrates. Proc. Natl. Acad. Sci. USA 2004, 101, 17930. [Google Scholar] [CrossRef] [PubMed]
- Orendorff, C.J.; Gearheart, L.; Jana, N.R.; Murphy, C.J. Aspect ratio dependence on surface enhanced Raman scattering using silver and gold nanorod substrates. Phys. Chem. Chem. Phys. 2006, 8, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Khoury, C.G.; Vo-Dinh, T. Gold Nanostars For Surface-Enhanced Raman Scattering: Synthesis, Characterization and Optimization. J. Phys. Chem. C 2008, 112, 18849–18859. [Google Scholar] [CrossRef]
- Li, H.; Cullum, B.M. Dual Layer and Multilayer Enhancements from Silver Film over Nanostructured Surface-Enhanced Raman Substrates. Appl. Spectrosc. 2005, 59, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Fales, A.M.; Khoury, C.G.; Liu, J.; Vo-Dinh, T. Spectral characterization and intracellular detection of Surface-Enhanced Raman Scattering (SERS)-encoded plasmonic gold nanostars. J. Raman Spectrosc. 2013, 44, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Fales, A.M.; Yuan, H.; Vo-Dinh, T. Development of Hybrid Silver-Coated Gold Nanostars for Nonaggregated Surface-Enhanced Raman Scattering. J. Phys. Chem. C 2014, 118, 3708–3715. [Google Scholar] [CrossRef] [PubMed]
- Fales, A.M.; Crawford, B.M.; Vo-Dinh, T. Folate Receptor-Targeted Theranostic Nanoconstruct for Surface-Enhanced Raman Scattering Imaging and Photodynamic Therapy. ACS Omega 2016, 1, 730–735. [Google Scholar]
- Cho, E.H.; Boico, A.; Wisniewski, N.A.; Gant, R.; Helton, K.L.; Brown, N.L.; Register, J.K.; Vo-Dinh, T.; Schroeder, T.; Klitzman, B. Micovascular integration into porous polyHEMA scaffold. In Proceedings of the SPIE, Bioinspired, Biointegrated, Bioengineered Photonic Devices II, San Francisco, CA, USA, 3 March 2014; Volume 8958, p. 4. [Google Scholar]
- Register, J.K.; Fales, A.M.; Wang, H.-N.; Norton, S.J.; Cho, E.H.; Boico, A.; Pradhan, S.; Kim, J.; Schroeder, T.; Wisniewski, N.A.; Klitzman, B.; Vo-Dinh, T. In vivo detection of SERS-encoded plasmonic nanostars in human skin grafts and live animal models. Anal. Bioanal.Chem. 2015, 407, 8215–8224. [Google Scholar] [CrossRef] [PubMed]
- Greish, K. Enhanced Permeability and Retention (EPR) Effect for Anticancer Nanomedicine Drug Targeting. In Cancer Nanotechnology: Methods and Protocols; Grobmyer, R.S., Moudgil, M.B., Eds.; Humana Press: Totowa, NJ, USA, 2010; p. 25. [Google Scholar]
- Liu, Y.; Ashton, J.R.; Moding, E.J.; Yuan, H.; Register, J.K.; Fales, A.M.; Choi, J.; Whitley, M.J.; Zhao, X.; Qi, Y.; et al. A Plasmonic Gold Nanostar Theranostic Probe for In Vivo Tumor Imaging and Photothermal Therapy. Theranostics 2015, 5, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Houck, K.; Stokes, D.L. Surface-Enhanced Raman Gene Probes. Anal. Chem. 1994, 66, 3379–3383. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-N.; Vo-Dinh, T. Multiplex detection of breast cancer biomarkers using plasmonic molecular sentinel nanoprobes. Nanotechnology 2009, 20, 065101. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Plasmonics-based SERS nanobiosensor for homogeneous nucleic acid detection. Nanomedicine 2015, 11, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-N.; Crawford, B.M.; Fales, A.M.; Bowie, M.L.; Seewaldt, V.L.; Vo-Dinh, T. Multiplexed Detection of MicroRNA Biomarkers Using SERS-Based Inverse Molecular Sentinel (iMS) Nanoprobes. J. Phys. Chem. C 2016, 120, 21047–21055. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Label-Free DNA Biosensor Based on SERS Molecular Sentinel on Nanowave Chip. Anal. Chem. 2013, 85, 6378–6383. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Nicholson, B.P.; Woods, C.W.; Vo-Dinh, T. DNA bioassay-on-chip using SERS detection for dengue diagnosis. Analyst 2014, 139, 5655–5659. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Plasmonic SERS biosensing nanochips for DNA detection. Anal. Bioanal.Chem. 2016, 408, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-N.; Register, J.K.; Fales, A.M.; Gandra, N.; Cho, E.H.; Boico, A.; Palmer, G.M.; Klitzman, B.; Vo-Dinh, T. Surface-enhanced Raman scattering nanosensors for in vivo detection of nucleic acid targets in a large animal model. Nano Res. 2018, in press. [Google Scholar] [CrossRef]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strobbia, P.; Odion, R.A.; Vo-Dinh, T. Spectroscopic Chemical Sensing and Imaging: From Plants to Animals and Humans. Chemosensors 2018, 6, 11. https://doi.org/10.3390/chemosensors6010011
Strobbia P, Odion RA, Vo-Dinh T. Spectroscopic Chemical Sensing and Imaging: From Plants to Animals and Humans. Chemosensors. 2018; 6(1):11. https://doi.org/10.3390/chemosensors6010011
Chicago/Turabian StyleStrobbia, Pietro, Ren A. Odion, and Tuan Vo-Dinh. 2018. "Spectroscopic Chemical Sensing and Imaging: From Plants to Animals and Humans" Chemosensors 6, no. 1: 11. https://doi.org/10.3390/chemosensors6010011
APA StyleStrobbia, P., Odion, R. A., & Vo-Dinh, T. (2018). Spectroscopic Chemical Sensing and Imaging: From Plants to Animals and Humans. Chemosensors, 6(1), 11. https://doi.org/10.3390/chemosensors6010011




