Abstract
The increasing incidence of organophosphate (OP) pesticide poisoning and the use of OP chemical warfare agents (CWA) in conflicts and terrorist acts need sustainable methods for sensing, decontamination, and detoxification of OP compounds. Enzymes can serve as specific, cost-effective biosensors for OPs. We will report on recent advancements in the use of carboxylesterases from the Hormone-Sensitive Lipase for the detection of OP compounds. In addition, enzymatic-based OP detoxification and decontamination offer long-term, environmentally friendly benefits compared to conventional methods such as chemical treatment, incineration, neutralization, and volatilization. Enzymatic detoxification has gained attention as an alternative to traditional OP-detoxification methods. This review provides an overview of the latest research on enzymatic sensing and detoxification of OPs, by exploiting enzymes, isolated from thermophilic/extremophilic Bacteria and Archaea that show exceptional thermal stability and stability in other harsh conditions. Finally, we will make examples of integration between sensing and decontamination systems, including protein engineering to enhance OP-degrading activities and detailed characterization of the best variants.
1. Introduction
Organophosphates (OPs) have gradually replaced organochlorides in agricultural production, becoming one of the primary chemicals for pest control [1,2,3,4]. The global market for organophosphate pesticides was anticipated to grow at an annual rate of 5.5% over the period from 2018 to 2023 [4] and it will be maintained until 2028. Additionally, the worldwide usage of organic phosphorus pesticides is estimated at around 10.2 million tons annually [2]. Although to date the use of synthetic OPs is decreasing due to toxicity and their likely involvement in diseases such as cancer, autism, and neurodegeneration [5], these compounds still represent a serious problem for populations all over the world and the environment, particularly for misuse in agricultural settings and for use in suicidal acts [6,7]. More than 100,000 people annually die of accidental or intentional poisoning by OP pesticides [8] mainly by paraoxon, parathion, coumaphos, diazinon, dimethoate, and chlorpyrifos. Worth noting is that the banned chemical warfare agents (CWA) (e.g., sarin, tabun, soman, and VX) are OPs that could potentially be exploited in terrorist acts like the recent dramatic events in Syria [9,10], other terrorist attacks in Iran and Japan [5], attempts at or assassinations of individuals mostly for political reasons [11,12]. Some OPs also bioaccumulate in the environment, causing damage to birds such as vultures [13] and bees. These compounds have been utilized as warfare agents, leading to the establishment of numerous international protocols and treaties aimed at their eradication, but many countries have yet to fulfill their commitments. Now, there is an increasing imperative to investigate suitable strategies for the sensing and decontamination of OPs [9].
In this review, our primary focus will be on the general aspects of biosensors, with particular emphasis on enzyme-based biosensors. We will explore the various characteristics that make these biosensors effective, such as their sensitivity and specificity, concentrating on fluorescence biosensors. In addition, we will then focus on thermostable enzymes, which have attracted considerable focus because of their exceptional stability and efficiency in challenging environments. These enzymes are particularly advantageous for both the detection of specific target compounds and the degradation of harmful substances, as they maintain their activity across a broad range of temperatures and environmental conditions. A detailed description of two classes of proteins thermostable Phosphotriesterase-like Lactonases (PLLs) [14] and carboxylesterases [15] is reported, considering their potentialities in sensing, decontamination, and detoxification of OP compounds.
2. OP Structures and Mechanism of Toxicity
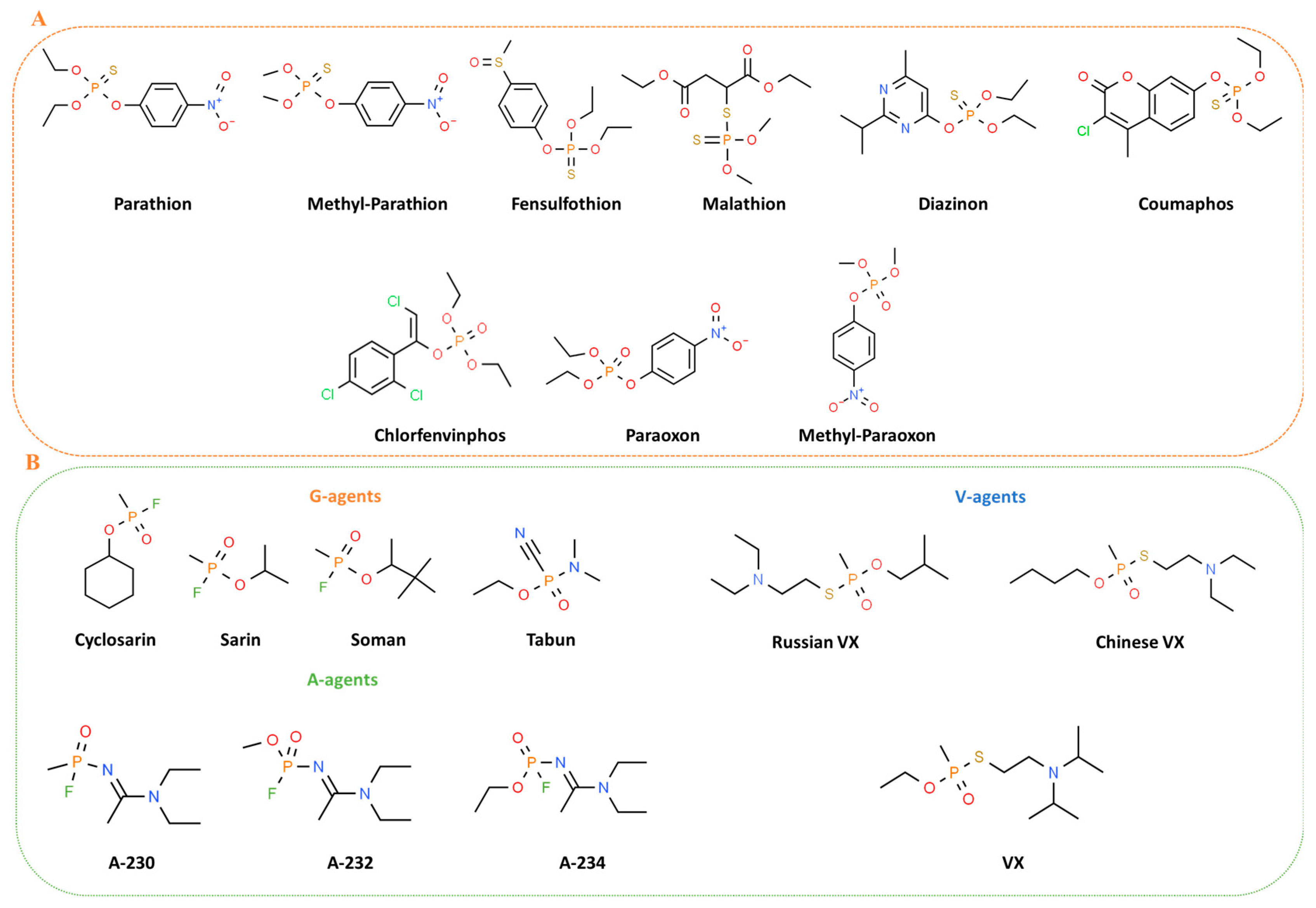
OPs stand for a class of organic chemicals that arise from phosphoric acids and their derivatives, characterized by at least one carbon-phosphorus bond. Many OPs are phosphotriesters that hold various combinations of oxygen, carbon, sulfur, and nitrogen. In oxo OPs, the central phosphorus atom is linked to oxygen by a double bond, while in thio OPs, it is linked to a sulfur atom. Additionally, the phosphorus atom can be bonded to alkoxy or amino groups via ester linkages, and to other chemical groups such as halogen, aliphatic, or aromatic compounds [16]. The toxicity of these compounds is significantly affected by the substituents attached to the phosphorus atom in phosphoric acid esters [17]. The class of OP pesticides falls into various categories, including thiols, amides, or esters derived from phosphonic, phosphinic, phosphoric, or thiophosphoric acids, often combined with organic side chains such as phenoxy, cyanide, or thiocyanate groups [18] (Figure 1A). Certain OPs like phosphonothioates (S-substituted) and phosphonofluoridates, which also are defined nerve agents (NA), are often recognized and used as chemical warfare agents [19]. These agents are grouped into four categories: the G-, the V-, the GV-, and the Novichok agents. The G-series agents, developed by Germans, encompass tabun (GA), sarin (GB), soman (GD), and cyclosarin (GF). V-series agents, denoted as venomous, include VE, VG, VM, VX [20], Chinese VX, and Russian VX [21]. GV-series combines properties of both G and V series, exemplified by GV, 2-dimethylaminoethyl-(dimethylamido)-fluorophosphate [22]. In general, G-series agents are less toxic than those in the V-series [23]. However, the secrecy surrounding research on these compounds has sparked significant debate, resulting in numerous speculative versions of their structures. The novel A-series compounds [24] are less known and called “Novichok”, an equivalent word for “newcomer” in Russian [25]. The Novichok series includes substances like substance-33, A230, A232, A234, Novichok-5, and Novichok-7 [26,27] (Figure 1B). The central part of the information we know about these agents is from Dr. Vil Mirzayanov, a Russian scientist [28], who detailed the development of the first three compounds at the GosNIIOKhT facility in Russia [29]. These compounds were initially unitary agents, with Novichok-5 as the first synthesized binary agent in 1989, based on Unitary A232 [29]. Novichok agents are typically liquid but can convert to a dusty formulation through the adsorption of liquid droplets on carriers like silica gel, pumice, fuller’s earth, or talc [30]. Hydrolysis of A230, A232, and A234 is slower compared to G- and V-series agents. OPs such as fluoride-releasing volatile soman and sarin, cyanide-releasing tabun, and thiocholine-releasing VX feature a stereogenic phosphorus atom; they have two enantiomers, P(−) and P(+), except Soman, which has two chiral atoms—one carbon center and one phosphorus—with four enantiomeric forms: C(+)P(+), C(+)P(−), C(−)P(+), and C(−)P(−) [31]. Stereoisomers play a crucial role in determining the range of toxicity of the compounds, with P(−) enantiomers generally exhibiting higher toxicity [31].
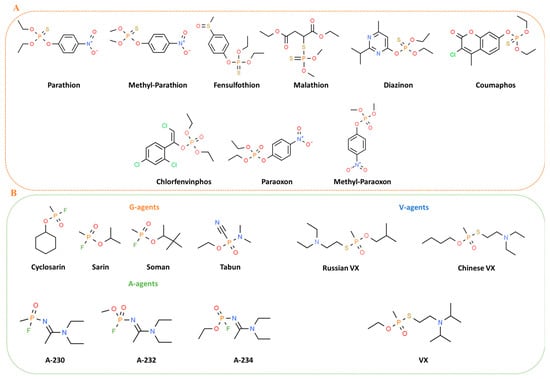
Figure 1.
Chemical structures of main pesticides (A) and chemical warfare (B) OPs.
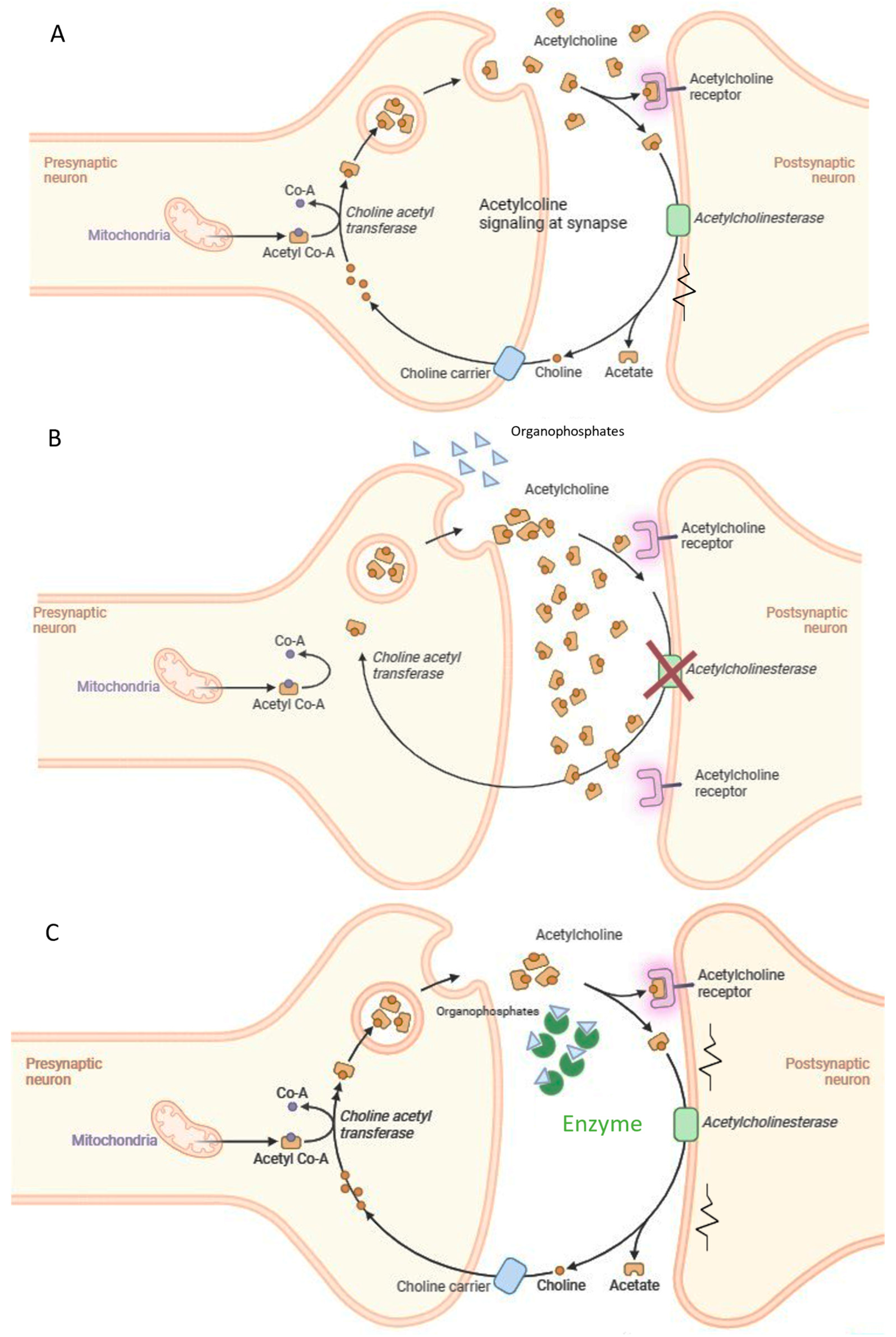
Toxicity of nerve agents, including OP pesticides is the consequence of targeting acetylcholinesterase (AChE), one of the key enzymes in cholinergic transmission [32]. The irreversible inhibition of acetylcholinesterase (AChE) leads to the buildup of acetylcholine at synapses between neurons or neuromuscular junctions, causing a range of clinical issues and potentially resulting in death [33] (Figure 2). Numerous toxicological studies have also demonstrated the genotoxic and carcinogenic potential of OP pesticides, showing that they can induce gene mutations, chromosomal alterations, and DNA damage in mammals [34], as well as negatively affect semen quality in humans [35]. A 2015 study published in Lancet Oncology [36] identified tetrachlorvinphos, parathion, malathion, diazinon, and glyphosate as probable carcinogens in mammals, with some already banned within the EU. These concerns surrounding organophosphorus poisoning have highlighted the necessity for environmental monitoring (biosensors) and decontamination or detoxification strategies for OP-contaminated soil, surfaces, air, water, and waste (including obsolete pesticide stocks and nerve agents). Additionally, there is an immediate need for effective solutions to rapidly detoxify humans exposed to OP nerve gasses.
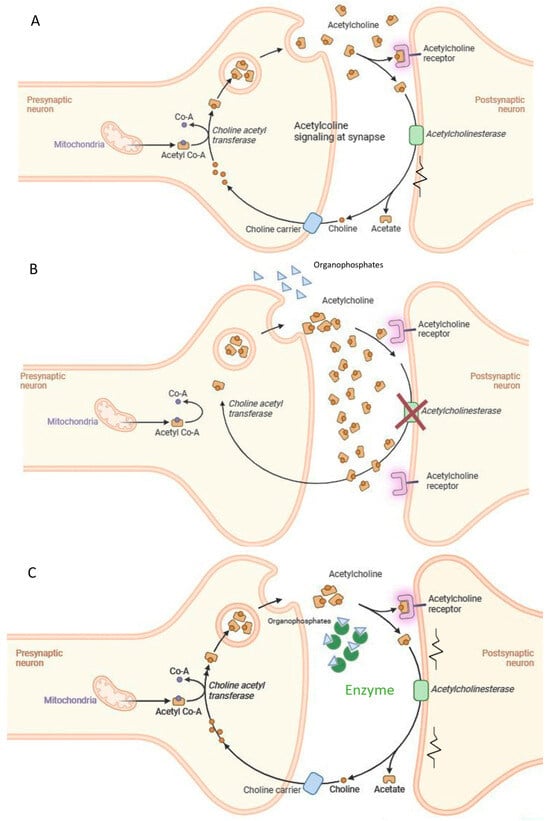
Figure 2.
Acute toxicity mechanism of organophosphate compounds and suggested intervention. (A) Acethylcholine signaling at the synapse; (B) OPs inhibit Acetylcholinesterase activity; (C) the enzymes degrade the OPs.
3. Sensing of OP Compounds: An Enzymatic Approach
Detecting organophosphates in various environmental samples typically requires advanced analytical techniques like gas and liquid chromatography combined with mass spectrometry (GC- and LC-MS) [37,38,39]. Nevertheless, to complement existing technologies and enhance the efficiency of OP monitoring, several alternative approaches have been developed in recent decades. There is increasing interest in developing sensors and biosensors for detecting and measuring OPs in various environments such as water, soil, and food, as well as in human samples like blood, urine, and tissue [40]. Enzymatic biosensors appear as a promising method due to their potential for being faster, more cost-effective, and user-friendly [41,42]. Additionally, enzymes have garnered significant attention for OP decontamination and detoxification, especially from an environmental perspective, as they offer a non-corrosive, safe, eco-friendly option that is also applicable to human treatment [43,44,45]. Injectable enzymes, known as bioscavengers, which can neutralize OPs in the bloodstream, are being explored as an alternative to traditional pre- and post-exposure therapies [46]. Despite challenges related to enzyme efficiency, stability, and production costs, recent advancements in the discovery and engineering of highly thermostable carboxylesterases and OP-degrading enzymes [47,48] are likely to make these enzymes competitive and cost-effective solutions for OP detection and detoxification in the near future. Moreover, enzymes can be easily immobilized on various substrates, such as foams, offering additional benefits like reusability [49,50]. In this review two classes of enzymes will be explored: the Hormone-Sensitive Lipase (HSL) Carboxylesterases and the Phosphotriesterase-like Lactonase (PLL) enzymes for detection and the detoxification of the OPs, respectively. In particular, biosensors that utilize enzymes, such as carboxylesterases or PLLs, for the detection and detoxification of OPs offer significant advantages over traditional methods.
3.1. Sensing of OPs
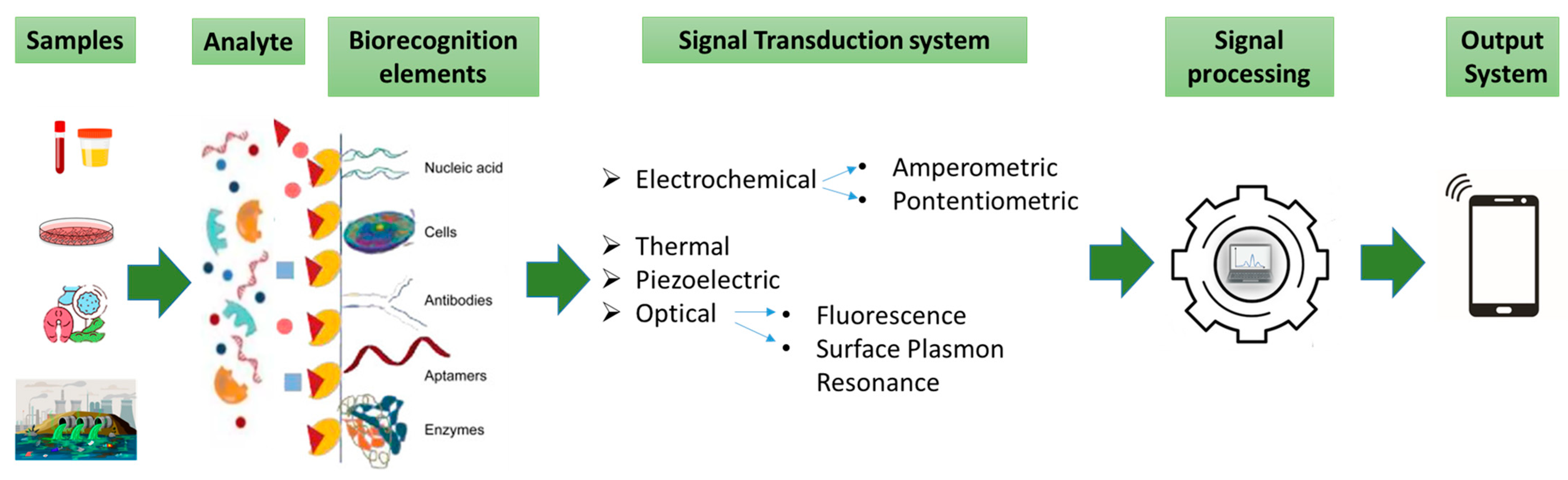
A typical biosensor is a device that integrates biological components with transducers to transform biological responses into electrical signals for detecting and measuring analytes. They are versatile tools employed in diverse areas like healthcare, disease diagnosis, environmental monitoring, and food quality control. Biosensors include an analyte, bioreceptor (in this case, enzymes), transducer, electronics, and display (Figure 3) [51]. The classification of biosensors depends on the type of bioreceptor and transducer [51], and includes immunosensors (using antibodies) [52], aptamer-based sensors (using DNA/RNA) [53], microbial sensors (using living cells) [54], and enzymatic sensors (using enzymes) [55]. Microbial or whole-cell biosensors utilize enzymes engineered in living cells to produce detectable signals in response to specific targets. Their adaptability to emerging threats has proven critical, exemplified by their use in colorimetric biosensors for visual detection of SARS-CoV-2 RNA during the COVID-19 pandemic. These systems highlight the potential of cell-based technologies in rapid and versatile diagnostics [54]. The enzyme-based biosensors being one of the most prevalent types work by detecting a target analyte through catalytic reactions [55]. These biosensors not only provide high sensitivity and specificity in detecting OPs, but the ability to immobilize enzymes on different materials enhances their reusability, making them even more cost effective. Thus, advancements in enzyme technology are directly linked to the progress of more efficient and sustainable biosensors for OP detection and detoxification. Each type has distinct advantages and applications. There are two types of biosensors, based on their recognition mechanisms: catalytic and affinity (non-catalytic) types [56]. Catalytic biosensors use biochemical reactions, often involving enzymes, to detect target analytes [57,58,59,60]. While enzyme-based biosensors face challenges like enzyme sensitivity and stability, they are highly effective in applications such as glucose and urea detection [60]. Recent innovations, such as enzyme-based sensors using nanoparticles [61], have improved sensor stability and response times. Incorporating nanomaterials into enzyme-based sensors has expanded their use as recognition components in biosensors [62]. Biosensors offer significant advantages over traditional detection methods for substances like organophosphates (OPs), including simplicity, cost-effectiveness, and rapid, on-site monitoring [63]. Fluorescence-based enzymatic biosensors are particularly sensitive, with strong signal-to-noise ratios and quick responses, even in complex environments [64,65,66] (for further details, see the review in [67]). These biosensors use fluorescent markers to detect interactions between target molecules and sensing probes. Advances in sensor materials and techniques, including signal amplification, have enhanced their sensitivity and functionality [68]. These biosensors find applications in environmental pollution monitoring, clinical diagnostics, veterinary/agricultural contexts, and industrial processing and monitoring. A 2023 study by Xiaoyang Wang et al. introduced a new OP detection system that used the mobile app “RGB Color Picker” to analyze fluorescent color shifts corresponding to different OP concentrations, enabling visual detection. By establishing an RGB standard curve, they programmed a WeChat mini app to quickly assess OP levels. The developed sensor offers advantages such as short detection time, high efficiency, and reliable results. Increasing OP concentration leads to decreased cyan fluorescence and increased orange fluorescence, enabling visual OP detection. The mechanism for detecting OPs using Tetraphenylethylene-ferrocene (TPE-Fc) molecules and glutathione-stabilized gold nanoclusters (GSH-AuNCs) operates as follows: acetylcholine is catalyzed by AChE and choline oxidase (ChOx) to produce hydrogen peroxide (H2O2), which oxidizes TPE-Fc to TPE-Fc+, boosting fluorescence at 472 nm and enhancing cyan fluorescence. The hydroxyl radicals (•OH) generated during this process damage GSH-AuNCs, reducing the orange fluorescence at 615 nm. The sensor exhibits a linear dynamic range of 10–2000 ng/mL, with a detection limit of 2.05 ng/mL. Smartphone-based color identification and a WeChat mini program were utilized for rapid OP analysis with promising results [69]. Despite the ongoing research in biosensor technology for OP detection, challenges remain, including limited stability and therefore reproducibility, complex manufacturing processes, and labor-intensive sample preparation with resulting high costs. Furthermore, environmental samples, which often contain multiple pesticides, make it difficult to identify individual compounds due to interference and inhibition. The focus on thermostable enzymes can afford at the same time the required stability and selectivity.
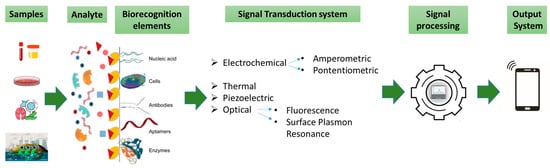
Figure 3.
Elements and selected components of a typical biosensor.
3.2. Thermostable Carboxylesterases of the Hormone-Sensitive Lipase (HSL) Family: Structure and Function
Carboxylesterases (EC 3.1.1.1) is a class of enzymes that catalyzes the hydrolysis of the carboxyl ester bond, in substrates with short to medium-acyl chains. Esterases, lipases, and cholinesterases form a diverse group of proteins with representatives across Eukarya, Bacteria, and Archaea [70,71,72,73,74], connected by the common α/β hydrolase fold [73]. Initially, they were categorized into three groups (C, L, H) based on sequence identity and conserved motifs. The “C” group consisted of cholinesterases, fungal lipases, various esterases, and certain non-enzymatic proteins. The “L” group included lipoprotein lipases, phospholipases, and related non-enzymatic proteins, while the “H” group, named after the mammalian Hormone-Sensitive Lipase (HSL), comprised proteins that shared sequence similarity with HSL [71,72,75]. Subsequently, new classifications appeared, proposing an “X” group which incorporated a wide range of proteins such as esterases, lipases, thioesterases, carboxylactonases, cutinases, endopeptidases, acyltransferases, and additional non-enzymatic proteins. These proteins share the alpha/beta hydrolase structural motif but do not fit into the earlier categories [15]. The “C” group included cholinesterases (CE) that are enzymes responsible for hydrolyzing esters of choline. Within this group, two distinct enzymes were found: Acetylcholinesterase (AChE), found in the nervous system where it terminates neurotransmission, and Butyrylcholinesterase (BChE), which is present in serum, although its specific function remains undisclosed [76,77]. Both enzymes exhibit sensitivity to a wide range of molecules, many of which are of artificial origin, such as organophosphate and carbamate pesticides or nerve agents [78]. Extensive research on natural inhibitors, leading to the discovery of substances like physostigmine or galantamine, has a long history, with recent introductions of their synthetic counterparts as well [79,80].
BuChE catalyzes the hydrolysis of esters of choline, such as butyrylcholine, succinylcholine, and acetylcholine [81]. It shares structural and functional similarities with AChE, which specializes in acetylcholine hydrolysis [82], and relies on a catalytic serine residue for activity [83]. BChE shares a similar α/β-fold family of proteins with a central β-sheet surrounded by α-helices [84]. This structural motif is also found in various other proteins, including hormone precursors like thyroglobulin and cell adhesion molecules such as neuroligin [85]. Although less efficient than AChE at hydrolyzing acetylcholine [86], BChE demonstrates broader substrate specificity, acting on compounds such as cocaine, acetylsalicylic acid, and heroin [87]. Additionally, it plays a crucial role in scavenging anticholinesterase agents, including natural inhibitors like physostigmine and synthetic organophosphate [88]. Some years ago, some of us started to study a thermostable enzyme with carboxylesterase activity, namely EST2, from the thermoacidophilic eubacterium Alicyclobacillus acidocaldarius; it belongs to the Hormone-Sensitive Lipase (HSL) family [89,90] and recently has been studied as a potential biosensor for the detection of OP pesticides [90]. EST2 is a monomeric carboxylesterase of about 34 kDa, which shows remarkable temperature stability, optimal temperature at 70 °C and maximal activity toward p-nitrophenyl- (pNP-) esters with acyl chains from six to eight carbon atoms [89]. It exhibited stability in the presence of water-miscible organic solvents (up to 20% v/v) and activated up to 3–4 times by low concentrations (4–5%) of them [91]. Under certain conditions it was enantioselective [89] and regioselective [92]. This structure is characterized by the typical α/β hydrolase fold, consisting of a central eight-stranded mixed β-sheet flanked by five helices. Two distinct helical regions form a “cap” at the C-terminal end of the central β-sheet. One portion of this cap includes the first 45 N-terminal residues (comprising helices α1 and α2), while the other part is made up of approximately 40 residues, organized into two short helices located between strands β6 and β7. Our group solved the crystal structure of EST2, revealing a covalently bound 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) adduct [93]. This enzyme has a high binding affinity for the organophosphate paraoxon (POX), leading to irreversible inhibition of its activity [94]. Previous research has also examined various EST2 mutants for their biochemical properties [95] and potential applications in biosensors [96]. The mechanism of action of the enzyme-based biosensors depends on the enzyme; in the Cholinesterase-based Biosensors (Acetylcholinesterase and Butyrylcholinesterase) the OP inhibits cholinesterase activity, causing a reduction in enzyme activity. This inhibition can be measured through changes in electrical signals, optical signals, or other responses [97]. A selection of enzyme-based biosensors is presented in Table 1.

Table 1.
Performance of some examples of enzyme-based biosensors.
3.3. EST2 Used as Biosensor
The carboxylesterase EST2, from Alicyclobacillus acidocaldarius, has been used as a proof-of-concept of diverse types of biosensors. Owing to its exceptional properties, it has been immobilized on different matrices such as polyvinylidene difluoride (PVDF) hydrophobic fluoropolymer membrane [102], nitrocellulose strips, and the residual activity tested in situ after interaction with the irreversible inhibitor paraoxon [103]. Research on the stability of EST2 enzymatic activity, following its immobilization onto nitrocellulose membranes, was conducted by storing the immobilized enzyme for 60 days at room temperature or at 4 °C, without specific precautions for environmental factors aside from normal atmospheric dust exposure. Activity assays performed at different intervals demonstrated that EST2 retained full activity throughout the two-month monitoring period [104]. These results support the feasibility of creating simple and cost-effective biosensors for paraoxon detection using immobilized EST2 in colorimetric assays. Inhibition has also been tested spectrophotometrically, as residual enzyme activity or as p-nitrophenol released from the inhibitor paraoxon [94]. Remarkably, EST2 activity remained completely unaffected by certain organophosphates, such as parathion, methyl parathion, dursban, coumaphos, fensulfothion, and diazinon, maintaining 100% activity even after prolonged exposure. Moreover, increasing concentrations of these OP inhibitors had no impact on EST2 activity, suggesting that these compounds do not act as irreversible or reversible inhibitors. Further investigations ruled out the possibility of EST2 breaking down these OP compounds via hydrolysis, confirming that the tested inhibitors are not substrates for the enzyme [104,105]. Some of us also exploited electrochemistry to unravel EST2 inhibition levels. In this case, EST2 has been immobilized on an electrode, and activity tested with an electroactive substrate. By using cyclic voltammetry and β-naftilacetate, we could measure the activity of EST2 before and after the treatment with an OP [96]. The biosensor employs a cost-effective multichannel electrochemical platform (MEP) designed with screen-printing technology for large-scale production. The MEP consists of eight independent individual electrochemical cells, each featuring a carbon working electrode (WE), a silver pseudo-reference electrode (RE), and a graphite counter-electrode (CE). This setup enables the biosensor to function as a disposable tool for short-term time-specific monitoring. The experiments utilized several enzymes including the wild-type EST2, various EST2 mutants, and a double mutant K42R/K61R [96]. To develop the MEP into a functional biosensor, the K42R/K61R mutant of EST2 was immobilized on the surface of the working electrode (WE) via a cross-linking process using carboxymethylcellulose (CMC) and bovine serum albumin (BSA). The immobilized enzyme catalyzes the conversion of 2-Naphtylacetate (NaAc) into its electroactive product, 2-naphthol (2-Na), which is detected using Differential Pulse Voltammetry (DPV). The electrochemical oxidation of 2-Na generates a peak current that serves as the analytical signal. In the presence of elevated OPs, enzyme inhibition occurs, reducing the production of 2-Na and subsequently lowering the anodic peak current. This decrease in current correlates with the concentration of the OPs. Enzyme inhibition tests at 1 μM OP concentration revealed distinct responses among the enzymes tested, including K42R/K61R mutant, WT, and five other mutants (E50A, R31A, K61A, D145A, S36C). Notably, only K42R/K61R mutant displayed complete inhibition by 1 μM Paraoxon under the specified conditions [96]. This mutant also exhibited increased sensitivity to various OPs, showing a higher inhibition percentage compared to the other mutants. This inhibition pattern highlights the potential of K42R/K61R in biosensor applications as it remains active until interacting with OPs, which covalently and stably bind to the crucial S155 residue in the active site, thereby disrupting the electrochemical signal [96]. Some of us have engineered EST2 by mutation of a cysteine at position 35 in the active site, where the fluorophore 5-({2-[(iodoacetyl)amino] ethyl} amino) naphthalene-1-sulfonic acid (IAEDANS) can be conjugated. After conjugation, the modified enzyme was effectively purified using size exclusion chromatography to remove excess unbound probes. The fluorescence of the labeled enzyme was then assessed through fluorescence spectroscopy [106,107]. This enzyme demonstrates a high affinity for paraoxon [94], leading to the irreversible inhibition of its enzymatic activity. This characteristic, along with the heat tolerance of EST2 and the enzyme’s overall stability, positions it as an excellent candidate for further biosensor development. Various mutants of EST2 have been investigated for their properties [95] and their biosensor capabilities [96,108]. Recently, some of us studied a double mutant of EST2, named “2m-EST2”; this variant incorporates a cysteine at position 35 for conjugation with the fluorophore IAEDANS and features an additional mutation aimed at enhancing selectivity towards OPs [109]. The enzyme was characterized, and optimal conditions were established. The impact of different organophosphate compounds on the enzymatic activity of 2m-EST2 was evaluated through activity assays involving the organophosphate pesticides paraoxon, parathion, tolclofos, chlorpyrifos, and glyphosate. Paraoxon completely inhibited the enzyme, whereas the other four pesticides showed no inhibition [109]. The enzyme, labeled with the fluorescent probe IAEDANS, was used to screen 16 different pesticides for their fluorescence quenching effects using a microplate reader. Paraoxon and methyl-paraoxon exhibited fluorescence quenching of 18% and 14%, respectively, when tested in equimolar amounts with the enzyme at nanomolar concentrations [110]. The estimated limit of detection (LOD) for paraoxon was relatively low, around 15 pmol of the pesticide [110]. This kind of fluorescence-bioreceptor was able to detect the presence of the pesticide paraoxon in real food (grapes, cherries, and fruit juice) at concentrations below 100 pmoles [110]. Given its high sensitivity, it has significant potential for applications in food traceability and environmental monitoring, particularly for controlling toxic chemical levels, especially organophosphate pesticides. The approach of inverse virtual screening technique has been employed to discover novel bioreceptors for the development of pesticide biosensors. Various pesticides were docked onto a limited database of proteins with known 3D structures. Compounds of interest were selected from diverse classes, including organophosphates, organo-chlorurates, carbamates, anilides, neonicotinoids, aromatic, and heterocyclic azoto-organics. Through the automated docking of hundreds of molecules on hundreds of structures, many potential binders were identified. The focus narrowed down to an E. coli Helicase RecQ that demonstrated the ability to bind the organophosphate Coumaphos at multiple sites [111]. In the article by Tortora F. et al. (2021) [111], the binding of the HRDC domain to the fluorescence probe IEDANS was described. Specific probes that target cysteine residues, such as IAEDANS, were selected for their advantageous properties. Notably, the excitation and emission wavelengths of this probe, along with the decay time of the emission signal, remained relatively stable despite variations in pH and other environmental factors like temperature [106]. After refining the conditions for specific fluorescence intensity and quenching reactivity at room temperature, they found that labeling in the presence of imidazole (6 mM) resulted in an increase in specific fluorescence intensity. The labeled protein remained stable for at least 30 days at 4 °C, and the specific fluorescence activity was approximately 380 AUF (Arbitrary Fluorescence Units) per μg of protein. The presence of Coumaphos at 300 nM resulted in a 30% fluorescence quenching [111]. Finding new bioreceptors is, therefore, a powerful tool to be exploited for the development of specific biosensors.
4. OP Decontamination and Detoxification
The use of various antidotes to address NA poisoning and effectively restore aged AChE remains a significant challenge. Bioscavengers introduced through intravenous or intramuscular means interact with OPs, starting a neutralization reaction in the bloodstream [112]. These bioscavengers can also be used topically as active skin protectants [113] and demonstrated a neutralization effect preventing OP poisoning in macaques when delivered via aerosols or intravenously [114,115]. These bioscavengers can be categorized into three main types: (1) stoichiometric enzymes, such as acetylcholinesterase and butyrylcholinesterase, which bind to and inactivate OPs; (2) catalytic bioscavengers which hydrolyze the OPs (Phosphotriesterase, PLL, methyl parathion hydrolase, organophosphorus acid anhydrolase, diisopropyl fluorophosphatase, some carboxylesterases, and paraoxonase) and degrade them to less toxic components; and (3) pseudo-catalytic bioscavengers (Acetylcholinesterase + HI-6) which are a mixture of stoichiometric bioscavengers with a chemical reactivator such as oxime [116]. Currently, various conventional technologies are available for the detoxification of OP nerve agent stockpiles and the decontamination of surfaces. For example, chemical treatments often involve sodium hydroxide [117,118,119], sodium hypochlorite, or similar formulations [120] for the removal of OPs. These methods generally involve harsh chemical conditions and are not compatible with the care of personnel or sensitive materials [121]. Physical methods such as incineration have been used for the disposal of stocks of OP nerve agents [122] but are expensive in most cases and not effective in removing contaminants at low concentrations [123]. While emergency pharmacological treatments for OP poisoning remain less than ideal, the concurrent development of nanotherapies and bioscavengers offers new opportunities [124]. Recent advancements in nanodetoxification strategies aim to develop antidotal nanoparticles specifically designed for detoxification [125,126,127,128]. These nanoparticles are engineered to encapsulate enzymes or chemicals that demonstrate high affinity, selectivity, and reactivity toward a broad spectrum of toxic molecules, reflecting significant potential for future applications [129,130,131,132].
4.1. Phosphotriesterase and Phosphotriesterase-like Lactonase (PLL) Enzymes: Structure and Function
One of the most studied OP-degrading enzymes from mesophilic bacteria is the Phosphotriesterase isolated from Brevundimonas (formerly Pseudomonas) diminuta MG (bdPTE) [133]. This enzyme encoded by opd (organophosphate degradation) gene was first isolated from bacterial cells of P. diminuta and Flavobacterium sp. (ATCC 27551), thriving in soils contaminated with parathion [134]. In vitro evolved bacterial PTEs are the most potent OP-degrading enzymes used as catalytic bioscavengers. [135]. PTE belongs to subtype I of the amidohydrolase superfamily [136] and is a homodimeric metalloprotein with a TIM-barrel structure (α/β)8 folds [137]. The active center, located at the C-terminus of the β-barrel, holds two divalent metal Zn2+ ions, that were found in the native bdPTE but the enzyme is fully active also with Cd2+, Mn2+, and Ni2+ [134]. Two divalent cations are coordinated by four histidines (H55, H57, H201, and H230), one aspartic acid (D301), and a carboxylated lysine (K169) in the enzyme bdPTE [138]. This enzyme exhibits hydrolytic activity against a broad range of organophosphate (OP) insecticides [139] as well as various G-series chemical warfare agents, including sarin, soman, tabun, cyclosarin, VX, and VR [140,141]. Notably, bdPTE displays exceptional catalytic efficiency towards paraoxon, with a kcat/KM value approaching 4.0 × 107 M⁻1s⁻1, making it the most effective enzyme for paraoxon hydrolysis among known OP-degrading enzymes [142]. PTE primarily chooses triesters as substrates, which encompass phosphonate and phosphinate compounds in addition to phosphate esters [143,144]. The hydrolysis of organophosphates by PTE occurs through a direct nucleophilic attack by a hydroxide on the phosphorus center, leading to an inversion of stereochemistry [145]. The attacking nucleophile is the bridging hydroxide in the metal center, as identified by pH-dependence studies of the manganese-substituted enzyme [146]. The reaction involves a late transition state with nearly complete bond cleavage. The phosphoryl oxygen coordinates with the metal, polarizing the phosphorus and enabling the hydroxide’s attack. The leaving group exits unprotonated, and the proton is transferred from Asp301 to His254 and then to the bulk solvent via a proton shuttle [147,148]. Moreover, crystallographic structures with such substrates were obtained [148]. A close homolog of bdPTE showing organophosphatase activity was isolated from Agrobacterium radiobacter P230 and designated as OpdA [149,150]. OpdA was evaluated for its capability to break down G- and V-series CWA. In this assessment, OpdA demonstrated strong catalytic effectiveness against soman and cyclosarin, displaying kcat/KM values of 5.8 × 102 M−1 s−1 and 3.9 × 102 M−1 s−1, respectively. However, its activity against V-type NA was notably lower. In enzyme evolution, site-saturation mutagenesis is used to create small libraries that allow all 20 amino acids to be tested at a specific site, enabling the rapid identification of the optimal amino acid. This technique was applied to the substrate binding residues of PTE, leading to the identification of the H254G/H257W/L303T (GWT) variant, which showed a 1000-fold increase in activity for a specific enantiomer compared to the wild-type enzyme. Further screenings identified other variants with enhanced activity, some up to 15,500-fold, through additional mutations and recombination [151]. Raushel and coworkers focused on mutating one of the active site loops of the enzyme PTE (loop-7) using targeted error-prone PCR [152]. This method allows for higher mutation rates in specific gene segments compared to traditional error-prone PCR. By targeting loop-7, they identified the variant L7ep3a, which showed a 9900-fold improvement in catalytic efficiency for VX, with a rate of 8.3 × 105 M⁻1 s⁻1. This variant also demonstrated enhanced activity for VR, being 20-fold more efficient than the wild-type enzyme and showing improvements for both enantiomers, especially the (SP)-VR [153]. To enhance the hydrolysis of V-agents, Tawfik and colleagues combined recombination experiments where libraries of previously identified mutants were assembled randomly with computational design to create new mutations [154]. Using a screening method involving acetylcholine esterase inactivation assays with authentic V-agents, they identified the variant G5-C23, which has an enzymatic efficiency of 8.3 × 104 M⁻1 s⁻1 for (SP)-VX and a 20-fold preference for the (SP)-enantiomer. They also found the variants G5-A53, which showed a 5800-fold improvement in catalytic efficiency for (SP)-VR compared to the wild-type enzyme. A protein engineering strategy was employed to enhance OpdA’s ability to degrade OP compounds [155,156]. Utilizing a rational design approach rooted in an in silico substrate docking, a novel mutant variant labeled W131H/F132A was created. This mutant proved a substantial increase in hydrolytic rates, showing approximately a 480-fold increase for Z-CVP and an 8-fold increase for E-CVP [155]. In a different method, a computational docking study involving malathion and the high-resolution crystal structure of OpdA indicated that malathion was too large to fit into the binding pocket of OpdA. Consequently, a technique known as combinatorial active site saturation testing (CASTing) was employed to improve the turnover rate for malathion. The most efficient mutant variant, S308L/Y309A, displayed a remarkable 5000-fold increase in catalytic efficiency towards malathion. Examination of X-ray crystal structures of this variant revealed that the substitution of Y309A facilitated improved accessibility of the substrate to the binding pocket [156].
4.2. Discovery of the PLL Family
Our group has discovered and focused on the study of an enzyme family of lactonases with promiscuous phosphotriesterase activity, dubbed Phosphotriesterase-like Lactonases (PLL) [14,47,48,157,158,159,160]. PLL family enzymes are closely related to mesophilic PTEs cited above, being classified in the same amidohydrolase superfamily. Phosphotriesterase-like lactonases, as the name suggests, share structural similarities with PTE enzymes but primarily function to catalyze the hydrolysis of lactones. This approach targets a family of enzymes exhibiting predominant lactonase activity on lactones and acyl-homoserine lactones (AHLs), along with low, promiscuous phosphotriesterase activity towards OPs [161]. Crystal structures are known for many lactonases of the PLL, and some have been biochemically characterized. The first characterized members were the hyperthermophilic paraoxonases from Sulfolobus solfataricus (SsoPox) [157,162] and Sulfolobus acidocaldarius (SacPox) [158], isolated from the hot springs of the Solfatara of Pozzuoli, in Italy. Initially studied as paraoxonases, it was later established that their primary function is lactonase activity [157,158,163]. The promiscuous phosphotriesterase activity likely evolved from lactonase functions due to environmental exposure to pesticides like paraoxon. Over time, this activity specialized in enzymes such as phosphotriesterase (PTE) and organophosphate degrading hydrolase (OPDH) in P. diminuta and A. radiobacter, respectively [47,158]. Notably, lactonases, such as SsoPox from S. solfataricus and m-PHP from Mycobacterium tuberculosis, share a similar active site structure with PTE [162,164,165,166]. Several PLL enzymes have now been identified in a diverse array of organisms, mostly from the Domains of Archaea and Bacteria. Among these, thermostable PLL enzymes have been isolated and characterized from thermophilic or extremophilic bacteria, such as GsP from Geobacillus stearothermophilus [167], GKL/GkaP from Geobacillus kaustophilus [168], and Archeae such as SisLac from Sulfolobus islandicus [169], and VmoLac/VmutPLL from Vulcanisaeta moutnovskia [170]. To this family also belong members from mesophilic microorganisms such as Mycobacterium tuberculosis (PPH), Rhodococcus erythropolis (AhlA) [14], Deinococcus radiodurans (DrOPH/Dr0930) [171,172], and Mycobacterium avium subsp. Paratuberculosis K-10 (MCP) [173]. PLL family enzymes are closely related to mesophilic PTEs from a structural and biochemical point of view [163,169]. The multi-sequence alignment between bdPTE and seven extremophilic PLL enzymes including SsoPox, SacPox, SisLac, VmoLac, Dr0930, GsP, GKL, and AhlA shows about 30% sequence identity [157]. PLLs are homodimeric enzymes belonging to the amidohydrolase superfamily, structurally similar to mesophilic PTEs [163]. Their active site, located at the C-terminus of a TIM barrel, coordinates two divalent metal cations using four conserved histidines, an aspartic acid, and a carboxylated lysine. This metal configuration, crucial for catalysis, generates a hydroxide ion essential for hydrolyzing phosphotriesters and lactones [163]. The phosphotriesterase activity of PLLs is metal-dependent, with the type of metal influencing enzyme performance, as shown in studies on bdPTE [159,174]. For instance, comparing SacPox prepared with Cd2+ (SacPox-Cd2+) to SacPox prepared with Mn2+ (SacPox-Mn2+), the latter exhibited a significant enhancement with approximately 30- and 19-fold increases in catalytic efficiencies against paraoxon and methyl paraoxon, respectively [159]. Structural analysis of bdPTE and six extremophilic PLLs revealed notable structural differences, particularly in loops 7 and 8. bdPTE has an elongated loop 7 forming a short α-helix, while loop 8 is similar in length to archaeal PLLs (SsoPox and SisLac), but shorter than in bacterial PLLs (Dr0930, GsP, and GKL/GkaP), where additional residues increase its distance from the protein core [164]. VmoLac has the shortest and most rigid loop 8, shaped into an α-helix by unique proline placement [175]. The PLL active site consists of three sub-sites: a hydrophobic channel for lactone chains, a large sub-site for N-acyl groups, and a small sub-site positioning the lactone ring over the metal center. Catalysis involves a binuclear metal center and a hydroxide ion, facilitating transition states—tetrahedral for lactones and pentacoordinate for phosphotriesters. This versatility highlights PLLs as generalists and offers insight into their rapid evolution toward specialized PTE activity [163]. The catalytic mechanisms underlying the phosphotriesterase and lactonase activities of PLL enzymes are modeled on the X-ray structure of SsoPox [163]. A catalytic hydroxide ion functions as a nucleophile, promoting the formation of intermediate species. The binuclear center and the bridging catalytic hydroxide ion are pivotal in driving the initiation of the catalytic process for both activities. The hydrolysis of lactones is inferred to follow an sp3 (tetrahedral) transition state, while the hydrolysis of phosphotriesters involves the formation of a pentacoordinate transition state. The enzyme’s ability to accommodate these distinct intermediate species underscores its classification as a generalist, shedding light on how the enzyme could have rapidly diverged into PTE in a remarkably short time [14]. PLL enzymes have also been shown to hydrolyze a diverse array of organophosphate OP pesticides, including paraoxon, methyl-paraoxon, parathion, methyl-parathion, malathion, dursban, coumaphos, and diazinon [47,128,156,157,167,168,170] as well as chemical warfare nerve agents [164]. Kinetic analyses reveal that the catalytic efficiency of SsoPox against paraoxon is 4.0 × 103 M⁻1 s⁻1, which is approximately ten times lower than that of SacPox, recorded at 2.66 × 104 M⁻1 s⁻1 at 70 °C [157,159]. Recently identified archaeal PLLs, SisLac and VmoLac demonstrate even lower catalytic efficiencies for paraoxon, measuring 6.98 × 102 M⁻1 s⁻1 and 1.86 M⁻1 s⁻1, respectively, at 70 °C [169,175]. Most analyses of thermophilic and extremophilic bacterial PLLs have been conducted at 35 °C, despite their high stability. At this temperature, GsP from Geobacillus stearothermophilus exhibits the highest kcat/KM value for paraoxon, at 54.67 M⁻1 s⁻1 [168], compared to Dr0930 (1.39 M⁻1 s⁻1 at 35 °C) [171] and GKL/GkaP (4.5 M−1 s−1 at 37 °C) [168]. Notably, GKL/GkaP shows increased catalytic efficiency at elevated temperatures, reaching 1.1 × 102 M⁻1 s⁻1 at 75 °C [165]. The phosphotriesterase activity of the enzyme AhlA toward paraoxon is so weak that only the kcat/KM ratio could be estimated (0.5 M−1 s−1) [14] (Table 2). Recently, our research has focused on the mesophilic PLL enzyme his-AhlA [14] a gene optimized for expression in E. coli [176]. Multiple sequence alignment revealed that his-AhlA shares 40% sequence identity with the thermostable protein SsoPox and 28% identity with P. diminuta PTE [14,47,158,163,177]. To further investigate the structure, we constructed a 3D model of his-AhlA, using m-PHP phosphotriesterase from Mycobacterium tuberculosis (PDB code: 4if2.1) as a template. The model displays a distorted fold (β/α)8 barrel, also known as a TIM barrel [178]. Comparison of the his-AhlA model with the 3D structure of SsoPox revealed a conserved active site. Five residues (His 55, His 57, His 201, His 230, and Asp 301) coordinate the two catalytic metal ions (Zn2⁺ in his-AhlA), along with carbamylated Lys 169, a feature present in both enzymes and in PTE [179]. Notably, his-AhlA differs by two substitutions: I168/T186 and C258/S277, and a variation in the orientation of the tryptophan side chain at position 282 (W263 in SsoPox). In SsoPox, Cys258 binds the amide group of the lactone [176]. Studies indicate that replacing cysteine with a hydrophobic residue enhances phosphotriesterase activity, while substitution with polar residues tends to decrease or abolish it [48].

Table 2.
Kinetic parameters of some PLL enzymes on OP compounds.
This natural substitution in his-AhlA likely increases its affinity for lactonic substrates over phosphotriesters [14]. Additionally, the Trp 282 (263 in SsoPox) is critical for the flexibility of loop 8 and contributes to the hydrophobic channel that accommodates the substrate. In his-AhlA, this structural feature plays a significant role in substrate binding and enzymatic function [176]. An analysis of the stability of his-AhlA revealed an unusual profile characterized by high thermal stability and thermophilicity. Sequence analysis shows an increased proportion of charged residues (Asp, Glu, Lys, and Arg), which constitute 23.8% of the total residues in his-AhlA, compared to 19.7% in mesophilic PTE. Conversely, the percentage of uncharged polar residues (Gln, Asn, Thr, and Ser) is lower in SsoPox (15.9% of total residues) compared to both mesophilic enzymes his-AhlA (16.5%) and PTE (16.4%) [176,178]. Another critical distinction between his-AhlA, PTE, and SsoPox lies in the significantly higher number of salt bridges. The residues participating in salt bridges (within a 4 Å cut-off distance) were 29 per monomer for his-AhlA, compared to 25 in SsoPox, and 15 in mesophilic PTEs [176,178]. The increase in salt bridges is a fundamental determinant of a protein’s thermal stability, contributing to extensive and intricate charge-charge interaction networks [180]. These structural characteristics strongly corroborate the functional data for his-AhlA, affirming its high stability, thermophilicity, and thermostability, resembling more typical traits of a thermophilic enzyme [176]. In the case of SsoPox, the catalytic efficiencies (kcat/KM) for lactones are significantly higher (by 100-fold) than its phosphotriesterase activity against paraoxon, supporting lactone hydrolysis as its primary function [14]. AHLs, among the prime substrates for PLLs, are signaling molecules crucial in bacterial communication through a mechanism known as quorum sensing [181]. This system regulates gene expression in Gram-negative bacteria (like P. aeruginosa), impacting various biological functions. In certain pathogenic bacteria, quorum sensing governs virulence factors and the formation of biofilms [182,183]. The lactonase from R. erythropolis known as AhlA [14,176] is important for opening the γ-lactone ring. Like other PLLs, it displays relatively low phosphotriesterase activity against paraoxon [14]. More precisely, AhlA functions as a true lactonase, effectively degrading a variety of compounds, with a particular preference for relatively hydrophobic lactones, such as fragrance molecules utilized in the agri-food industry and the aromatic compound dihydrocoumarin [184]. The enzyme acts on a broad range of N-acyl-homoserine lactones (N-AHSLs), specifically those with acyl chains containing 6 to 14 carbon atoms, regardless of substitutions at carbon 3. This property not only defines the enzyme’s function but also fuels increasing interest in its potential applications [184,185,186,187] in the field of biosensing and degradation of lactones.
4.3. Molecular Evolution of PLLs to Improve Decontamination of OPs
Thermostable PLL enzymes with weak promiscuous phosphotriesterase activity have been subjected to in vitro mutagenesis to improve their efficacy as stable biocatalysts against OP compounds. [164]. Efforts to boost the hydrolytic activity of PLL enzymes towards organophosphates have been extensive, employing molecular engineering techniques such as site-directed mutagenesis and directed evolution to create novel mutants with improved phosphotriesterase activities. Earlier studies targeted specific positions (79, 97, 98, 261, and 263) in SsoPox [47], resulting in mutants with varied activity and temperature profiles, reflecting the active site’s need to balance rigidity for high-temperature stability and flexibility for catalytic activity. One notable mutant, SsoPox W263F, generated through directed evolution, exhibited a 6-fold increase in catalytic efficiency against paraoxon related to the wild type. Structural analysis revealed an expanded active site cavity in W263F to accommodate the lactone ring. Recently, our research group characterized SsoPox variants created through directed evolution and semi-rational design, including the triple mutant C258L/I261F/W263A (3Mut), which showed a catalytic efficiency of approximately 4.5 × 104 M−1 s−1 for paraoxonase activity at 65 °C. The improvement was attributed to the expanded active site from the alanine substitution at position 263 and a hydrophobic patch created by leucine and phenylalanine at positions 258 and 261, respectively. [47,48]. Additionally, site-saturated mutagenesis targeting six residues (99, 229, 255, 257, 265, 266) found through bioinformatics did not yield variants surpassing 3Mut in paraoxonase activity [48]. However, subsequent characterization by another research group confirmed an enlarged active site in these variants, aligning with our hypothesis. In the study by Suzumoto Y. et al. (2020), a novel directed evolutionary strategy was developed utilizing DNA StEP gene recombination to enhance the phosphotriesterase activity of SsoPox. The resulting mutant, V82L/C258L/I261F/W263A (4Mut), demonstrated a catalytic efficiency of 1.6 × 105 M⁻1 s⁻1 for paraoxon hydrolysis at 70 °C. [160]. In-depth structural analyses, including 3D structure and computational studies, indicated that a single mutation outside the active site can enhance dimer flexibility, improving hydrolytic activity. The 4Mut enzyme demonstrated robust activity on paraoxon, methyl-paraoxon, and methyl-parathion, with specific activities on paraoxon and methyl-paraoxon particularly high. Comprehensive kinetic analysis at optimal conditions (pH 8.0, 70 °C) showed a specific activity of 206.4 U/mg for paraoxon, a 500-fold increase compared to SsoPox. The catalytic efficiency (kcat/KM) reached 1.6 × 105 M−1 s−1, marking a significant improvement over 3Mut and SsoPox wt [164].
5. The Benefit of Thermostable Enzymes: Applications in Sensing and Bioremediation of OPs
Solid and different materials, including modified activated carbon and metal oxides, exhibit beneficial properties for the neutralization of chemical warfare agents in protective fabrics and gear, such as masks, suits, air conditioning filters, and enzyme-immobilized wipes [188,189]. Economical chemical methods, like treatments with hypochlorite and sodium hydroxide, have emerged as effective environmental options for detoxifying OPs. However, these techniques often involve harsh chemical conditions that may not be suitable for personnel safety or sensitive materials. In contrast, biocatalysts, available in either free or immobilized forms, present a promising alternative. Recent studies are increasingly concentrating on the synergistic application of OP-sensing [190] and OP-degrading enzymes [161]. Using PubMed searches (https://pubmed.ncbi.nlm.nih.gov/2024 (accessed on 7 October 2024)) with the word’s OP/nerve agents/pesticides and biosensing/detection and degradation/detoxification/decontamination, we retrieved only a few papers. Among these, in the article by Schofield, D.A. et al. [191], the authors developed a yeast-based system to detect and degrade paraoxon. The study integrates elements of genetic engineering, environmental biotechnology, and biocatalysis, aiming to create a yeast-based system capable of both detecting and breaking down paraoxon. The authors focused on engineering a strain of the yeast Saccharomyces cerevisiae expressing PTE to inactivate paraoxon. Alongside this, they designed a biosensor for pesticide detection. The biosensor was developed using a reporter gene system that was activated in the presence of paraoxon. When expressed, this reporter gene produced an easily detectable fluorescence signal, indicating the presence of the contaminant. The engineered yeast was shown to detect extremely low concentrations of paraoxon making it an overly sensitive sensor for environmental monitoring and then tested under various conditions to evaluate its ability to degrade paraoxon, confirming its effectiveness in reducing the concentration of paraoxon in the culture medium. This proved the functionality of the biocatalyst system. The degradation process was monitored using the developed biosensor, which allowed the researchers to correlate the level of degradation with the reduction in the biosensor signal. The developed system represents an innovative approach that combines detection and degradation, offering a promising solution for the treatment of hazardous contaminants such as paraoxon. In a similar vein, the work by Liu R. et al. [192] centered on the creation of a novel E. coli-based biocatalytic and biosensing system for OPs. In this study, the authors engineered E. coli cells to display heterologous proteins on their surfaces. This system was designed to both degrade toxic OPs and accurately detect their presence. The heterologous proteins were displayed on the E. coli cell surface using a surface display system, a technique where proteins of interest are anchored to the cell’s outer membrane. This enables the cells to directly interact with OP compounds in the surrounding environment. Among the displayed proteins were specific enzymes, such as PTE, which are known for their ability to degrade OPs. The engineered system proved effective in breaking down these compounds, with the modified E. coli cells converting harmful OPs into less dangerous substances, demonstrating a high level of biocatalytic activity. The surface expression of these proteins also allowed for the use of signals, such as changes in fluorescence or absorption, to indicate the presence of target compounds. The results showed that the system could operate efficiently across various temperatures and with different types of organophosphates. The authors emphasize that using whole-cell systems for displaying heterologous proteins offers advantages over traditional methods based on isolated enzymes, such as increased stability and the ability to continuously regenerate the system. The E. coli-based biocatalyst/biosensor represents a significant advancement in environmental biotechnology, combining the detection and degradation of OPs into a single cellular system. Also in the article authored by Istamboulie G. et al. [193], details of the development and application of a system for the controlled degradation of chlorpyrifos and chlorfenvinfos are given. This system utilizes a detoxification column containing the enzyme PTE, which effectively breaks down the targeted pesticides. The innovative aspect of this system lies in its integration with a biosensor specifically developed to detect the presence and concentration of two OP pesticides. The biosensor is coupled with the detoxification column, facilitating real-time monitoring and control of the degradation process. When the biosensor detects chlorpyrifos or chlorfenvinfos, it triggers the detoxification column, where the PTE enzyme degrades pesticides. This approach allows for precise control over the detoxification process, maximizing the system’s efficiency and minimizing the production of unwanted by-products. The system showed high efficiency in reducing the concentrations of chlorpyrifos and chlorfenvinfos, with a high degree of specificity and rapid response. The results confirm that the integrated biosensor effectively monitors pesticide concentrations in real time, ensuring optimal control of the detoxification column and enhancing the overall effectiveness of the system. Enzyme technology is attractive due to its efficiency and specificity under mild conditions. It is non-corrosive, safe, environmentally friendly, and suitable for human therapy. [43,110,161]. In the recent review by Pashirova T. et al. [194], the use of microbial enzymes for degrading organophosphates is discussed with a focus on their potential in medical detoxification and environmental cleanup. The authors specifically highlight enzymes like PTE, which are produced by various microorganisms, including bacteria and fungi. These microbes have been isolated from contaminated environments and studied for their application potential. The article emphasizes the role of these enzymes in treating poisoning in patients exposed to OPs. Such enzymes can neutralize toxic compounds before they cause irreversible damage to the nervous system. They can be formulated into drugs or enzymatic treatments for emergency use, such as in chemical warfare scenarios, or to protect agricultural workers from pesticide exposure. Additionally, enzymes are of significant interest for environmental remediation, particularly for the decontamination of soils, water, and other polluted areas. Their application stands for an ecologically sustainable and highly effective solution for removing these contaminants from the environment. The authors describe the use of bioreactors containing microorganisms or enzymes that can process large volumes of contaminated soil or water, with the capability of being applied directly in the field for in situ remediation. Obviously, one of the primary challenges in this field is ensuring that these enzymes keep their activity and stability under varying environmental and clinical conditions. The article explores strategies to enhance enzyme stability, such as protein engineering and immobilization on solid supports. Another challenge is the large-scale production of these enzymes in a cost-effective manner, for which the authors discuss microbial fermentation techniques and genetic engineering as potential solutions. These studies underscore the importance of stable microbial enzymes in addressing both health-related and environmental issues caused by OP contamination. Naturally stable enzymes, including SsoPox, SacPox, and mutants SsoW263F and Sso3Mut, were tested for their detoxification capabilities against paraoxon applied to a cotton strip. When used individually at specific concentrations, these enzymes demonstrated impressive detoxification performance in the presence of detergents (such as SDS or commercial soap) and organic solvents (ethanol or methanol) [48]. Each of the four enzymes was monitored at two different concentrations (25 and 50 µg/mL) with or without SDS (0.025% w/v) on nerve agent solution (NAS) substrate (a mixture of paraoxon, parathion, and methylparaoxon). While SsoPox3Mut demonstrated the highest performance, it was still unable to fully degrade the NAS substrate, achieving approximately 80% efficiency [96]. Carboxylesterases may be utilized at the end of the decontamination process to eliminate any residual agents that remain after hydrolysis, following the principle that “clean is clean”. These experiments provide a proof-of-principle of the potential development of enzyme formulations that combine stable enzymes for the efficient degradation of organophosphate mixtures in aqueous solutions and on various surface types and materials [96]. An important experiment was performed, when it was tested, first on a small box and then in a large room, with the same pool of aerosolized enzymes to prove activity against nebulized nerve agent models [164]. The stability of thermophilic PLLs allowed us to use them under simulated real working conditions. A mixture of three enzymes (SsoPox263F, SsoPox3Mut, and SacPox) has been nebulized in a sealed chamber and found to be active even on the surfaces of varied materials placed inside the chamber during the experiment. The materials tested were cotton, aluminum, glass, and linoleum. The results show that enzymes are active already in the “air” and the enzyme triplet decontaminates 99% of the nebulized simulants mixture (100 µM each paraoxon and m-paraoxon, 25 µM m-parathion) [96]. To strengthen the case for utilizing Sso3Mut as a bioscavenger and to evaluate the impact of the introduced mutations, an analysis of its activity was conducted against “live” nerve agents belonging to the “G series,” namely cyclosarin, sarin, soman, tabun, and VX, at 37 °C. It compared the performance of 4Mut with that of other enzymes. The process involved incubating the enzyme with each nerve agent and recording acetylcholinesterase (AChE) inhibition at different time points, enabling us to calculate the detoxification kinetics (Table 3).

Table 3.
Kinetic parameters of extremophilic PLL enzymes against G-series nerve agents.
From the results obtained, it has been shown that of the PLLs used, both SsoPox 3Mut and SsoPox 4Mut show comparable specificity on OP nerve agents tested. Some of us have also reported the development of an integrated system based on a highly responsive biosensor (mutant of the EST2) with a limit of detection (LOD) of 0.4 nM for paraoxon, after 5 s of exposure time (Table 1, [98]). The “detection and reaction” strategy, based on the prototypes discussed, could serve as a deterrent against terrorist acts involving deadly nerve agents, especially given that our enzymes have shown remarkable activity compared to PTE, even at room temperature. Additionally, since we utilized OP pesticides as model compounds, this study suggests that the system is well suited for detecting and identifying low concentrations of pesticides in the environment, such as those resulting from runoff from cotton farms [94].
Recent Advancements and Applications of BIOSENSORS: A Further Example of Integrated System
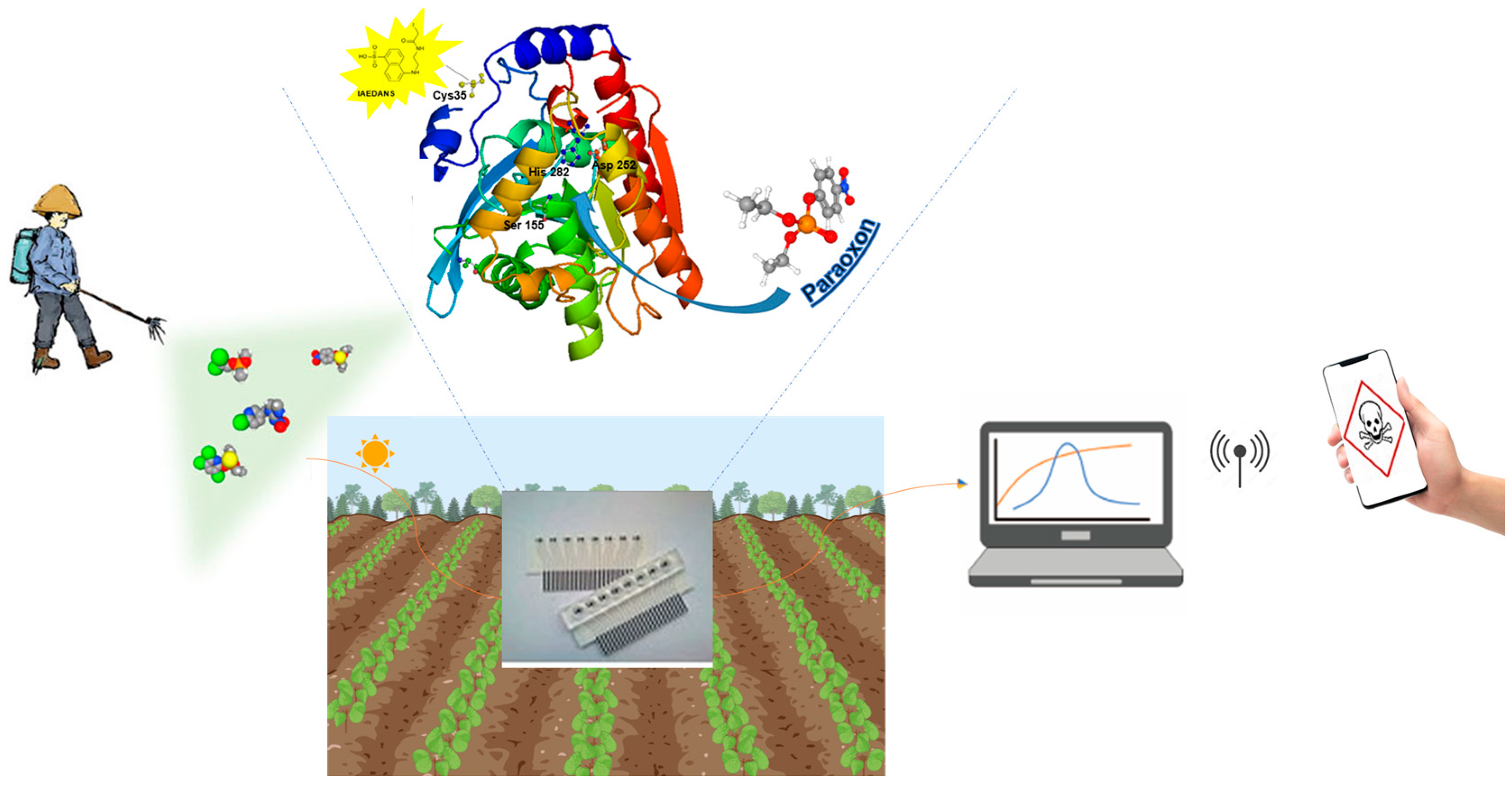
Recent improvements in biosensor technology for healthcare have led to the development of three primary categories of biosensors: those designed for in vitro diagnosis using blood, saliva, or urine samples; continuous monitoring biosensors (CMBs); and wearable biosensors. The in vitro diagnostic biosensors field has seen remarkable advancements, fueled by recent breakthroughs such as clustered regularly interspaced short palindromic repeats (CRISPR)/Cas technologies and enhancements in established platforms like lateral flow assays (LFAs) and microfluidic or electrochemical paper-based analytical devices (μPADs/ePADs) [195]. Additionally, biosensors hold great promise for environmental monitoring, providing critical insights into air and water quality, pollution levels, and the presence of hazardous substances. Deploying networks of biosensors enables the detection of contaminants and biohazards, facilitating timely interventions to mitigate risks. Recent studies have demonstrated the ability of biosensors to detect chemical warfare agents simulants, such as an optical array device capable of efficiently detecting DMMP gas (a simulant for G-Type nerve agents) at concentrations as low as 0.1 ppm, utilizing a simple smartphone as a detector. Also, enzymes can be used in similar prototypes. The signal could then be processed and transmitted via a smartphone, which would control let to say a high-performance drone for the rapid and safe dispersal of an enzyme solution to decontaminate the area (Figure 4) Furthermore, advances in sensor technologies for nerve agents have focused on developing innovative sprayable sensors based on polymers, organic or inorganic dyes, or nanoparticles [196].
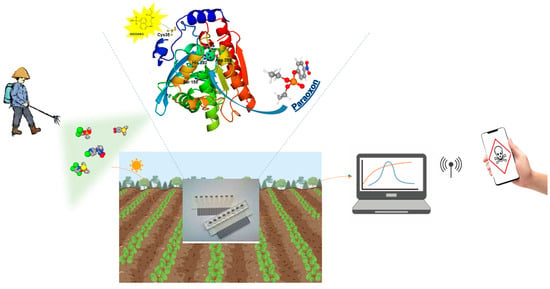
Figure 4.
Example of a portable biosensor for field detection of pesticides. Enhanced detection with a mutated EST2 enzyme in the biosensor, paired with a smartphone controlled signal.
These chemosensors offer convenience and versatility despite some limitations. Enzymes, unlike other materials, bind specifically and irreversibly to nerve agents, neutralizing them and making them safe for humans. As a result, enzyme-based sprayable colorimetric sensors can not only identify contaminated areas but also decontaminate them, providing a major advantage over other chemical methods [196]. Although enzyme-based sensors tend to be more costly and harder to provide a direct signal their excellent selectivity, rapid response, and ability to eventually detect and detoxify contaminants represent significant improvements over traditional systems. Moreover, they can be adapted through engineering methodologies. A promising approach involves computational redesign (using molecular modeling, transition state simulations, and QM/MM methods) of known enzymes. As artificial intelligence continues to advance, in silico methods are expected to produce new, mutated enzymes with enhanced activity across a broader spectrum of OPs. In the future, aerial and autonomous systems such as drones may provide even more significant benefits, as these platforms have the capability to quickly spray and capture images of vast regions. Collaboration across multiple disciplines could transform the future of NA detection. The participation of experts in engineering, materials science, chemistry, and data science is expected to result in advancements in sensor design, development methods, and data analysis.
6. Conclusions
The increasing need for sustainable and efficient methods to detect, detoxify, and decontaminate OP compounds highlights the promise of enzyme-based approaches. Enzymatic biosensors, particularly those using thermostable carboxylesterases and PLLs, offer significant advantages over traditional chemical methods, including specificity, eco-friendliness, and adaptability to challenging environments. Advances in protein engineering have further improved enzyme activity, stability, and specificity, enhancing their application potential for sensing and degrading OPs in diverse contexts, such as environmental remediation, food safety, and defense against chemical warfare agents. Furthermore, integrating these enzymes into biosensor platforms, coupled with innovative technologies like smartphone applications and drones for rapid deployment, paves the way for real-time monitoring and decontamination strategies. These systems can effectively address critical challenges in detecting and neutralizing OP contamination, with implications for public health, environmental protection, and global security. While challenges such as enzyme production costs and stability remain, ongoing research in nanotechnology, protein engineering, and immobilization techniques continues to expand the feasibility of these solutions. The integration of enzymatic sensing and degradation represents a pivotal step forward in developing comprehensive, scalable, and sustainable systems for managing the risks associated with OP compounds.
Author Contributions
G.M. and E.P. conceived the project, E.A.L., N.S.K.A. and G.C. wrote and corrected parts of the paper, M.M. and E.P. coordinated the work. All authors have read and agreed to the published version of the manuscript.
Funding
NutrAge FOE 2021 DM MUR n. 844/2021 IBBC 2B and Project: “Nuove strategie per la diagnostica medica e molecolare e per la tracciabilità ed il monitoraggio dei prodotti alimentari” DD n. 350 of 25/05/2017 to GM.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, H.; Wang, C.; Chang, W.-Y.; Liu, H. Factors Affecting Chinese Farmers’ Environment-Friendly Pesticide Application Behavior: A Meta-Analysis. J. Clean. Prod. 2023, 409, 137277. [Google Scholar] [CrossRef]
- He, Y.; Guo, C.; Lv, J.; Deng, Y.; Xu, J. Occurrence, Sources, and Ecological Risks of Three Classes of Insecticides in Sediments of the Liaohe River Basin, China. Environ. Sci. Pollut. Res. 2021, 28, 62726–62735. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Zhu, J.; Pan, Y.; Wu, S.; Dang, Y.; Xu, B.; Yang, H. The Positive Impacts of Landscape Fragmentation on the Diversification of Agricultural Production in Zhejiang Province, China. J. Clean. Prod. 2020, 251, 119722. [Google Scholar] [CrossRef]
- Zhang, D.; Xiao, Y.; Xu, P.; Yang, X.; Wu, Q.; Wu, K. Insecticide Resistance Monitoring for the Invasive Populations of Fall Armyworm, Spodoptera Frugiperda in China. J. Integr. Agric. 2021, 20, 783–791. [Google Scholar] [CrossRef]
- Voorhees, J.R.; Rohlman, D.S.; Lein, P.J.; Pieper, A.A. Neurotoxicity in Preclinical Models of Occupational Exposure to Organophosphorus Compounds. Front. Neurosci. 2017, 10, 590. [Google Scholar] [CrossRef]
- Simonelli, A.; Carfora, A.; Basilicata, P.; Liguori, B.; Mascolo, P.; Policino, F.; Niola, M.; Campobasso, C.P. Suicide by Pesticide (Phorate) Ingestion: Case Report and Review of Literature. Toxics 2022, 10, 205. [Google Scholar] [CrossRef]
- World Health Organization; Food and Agriculture Organization of the United Nations. Preventing Suicide: A Resource for Pesticide Registrars and Regulators; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Eddleston, M. Novel Clinical Toxicology and Pharmacology of Organophosphorus Insecticide Self-Poisoning. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 341–360. [Google Scholar] [CrossRef]
- Enserink, M.U.N. Taps Special Labs to Investigate Syrian Attack. Science 2013, 341, 1050–1051. [Google Scholar] [CrossRef]
- Sprague, R.M.; Ladd, M.; Ashurst, J.V. EMS Resuscitation During Contamination While Wearing PPE. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Picard, B.; Chataigner, I.; Maddaluno, J.; Legros, J. Introduction to Chemical Warfare Agents, Relevant Simulants and Modern Neutralisation Methods. Org. Biomol. Chem. 2019, 17, 6528–6537. [Google Scholar] [CrossRef]
- Vale, J.A.; Marrs, T.C.; Maynard, R.L. Novichok: A Murderous Nerve Agent Attack in the UK. Clin. Toxicol. 2018, 56, 1093–1097. [Google Scholar] [CrossRef]
- Plaza, P.I.; Martínez-López, E.; Lambertucci, S.A. The Perfect Threat: Pesticides and Vultures. Sci. Total Environ. 2019, 687, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Afriat, L.; Roodveldt, C.; Manco, G.; Tawfik, D.S. The Latent Promiscuity of Newly Identified Microbial Lactonases Is Linked to a Recently Diverged Phosphotriesterase. Biochemistry 2006, 45, 13677–13686. [Google Scholar] [CrossRef] [PubMed]
- Manco, G.; Merone, L.; Mandrich, L. Structural and Kinetic Overview of the Carboxylesterase EST2 from Alicyclobacillus Acidocaldarius: A Comparison with the Other Members Ofthe HSL Family. Protein Pept. Lett. 2009, 16, 1189–1200. [Google Scholar] [CrossRef]
- Mukherjee, S.; Gupta, R.D. Organophosphorus Nerve Agents: Types, Toxicity, and Treatments. J. Toxicol. 2020, 2020, 1–16. [Google Scholar] [CrossRef]
- Balali-Mood, B. Chemistry and Classification of OP Compounds. In Basic and Clinical Toxicology of Organophosphorus Compounds; Balali-Mood, M., Abdollahi, M., Eds.; Springer: London, UK, 2014; pp. 1–23. ISBN 978-1-4471-5624-6. [Google Scholar]
- Kumar, S.; Kaushik, G.; Villarreal-Chiu, J.F. Scenario of Organophosphate Pollution and Toxicity in India: A Review. Environ. Sci. Pollut. Res. 2016, 23, 9480–9491. [Google Scholar] [CrossRef]
- Gupta, R.C. Classification and Uses of Organophosphates and Carbamates. In Toxicology of Organophosphate & Carbamate Compounds; Elsevier: Amsterdam, The Netherlands, 2006; pp. 5–24. ISBN 978-0-12-088523-7. [Google Scholar]
- Organisation for the Prohibition of Chemical Weapons. 2019. Available online: https://www.opcw.org/ (accessed on 7 October 2024).
- Kranawetvogl, A.; Küppers, J.; Siegert, M.; Gütschow, M.; Worek, F.; Thiermann, H.; Elsinghorst, P.W.; John, H. Bioanalytical Verification of V-Type Nerve Agent Exposure: Simultaneous Detection of Phosphonylated Tyrosines and Cysteine-Containing Disulfide-Adducts Derived from Human Albumin. Anal. Bioanal. Chem. 2018, 410, 1463–1474. [Google Scholar] [CrossRef]
- Moshiri, M.; Darchini-Maragheh, E.; Balali-Mood, M. Advances in Toxicology and Medical Treatment of Chemical Warfare Nerve Agents. DARU J. Pharm. Sci. 2012, 20, 81. [Google Scholar] [CrossRef]
- Bajgar, J. Nerve Agents Poisoning and Its Treatment in Schematic Figures and Tables; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Hulse, E.J.; Haslam, J.D.; Emmett, S.R.; Woolley, T. Organophosphorus Nerve Agent Poisoning: Managing the Poisoned Patient. Br. J. Anaesth. 2019, 123, 457–463. [Google Scholar] [CrossRef]
- Holmgaard, R.; Nielsen, J.B. Dermal Absorption of Pesticides—Valuation of Variability and Prevention; DANISH Ministry of the Environment, Environment Protection Agency: Odense, Denmark, 2009. [Google Scholar]
- Chai, P.R.; Hayes, B.D.; Erickson, T.B.; Boyer, E.W. Novichok Agents: A Historical, Current, and Toxicological Perspective. Toxicol. Commun. 2018, 2, 45–48. [Google Scholar] [CrossRef]
- Franca, T.; Kitagawa, D.; Cavalcante, S.; Da Silva, J.; Nepovimova, E.; Kuca, K. Novichoks: The Dangerous Fourth Generation of Chemical Weapons. Int. J. Mol. Sci. 2019, 20, 1222. [Google Scholar] [CrossRef]
- Charejoo, A.; Arabfard, M.; Jafari, A.; Nourian, Y.H. A Complete, Evidence-Based Review on Novichok Poisoning Based on Epidemiological Aspects and Clinical Management. Front. Toxicol. 2023, 4, 1004705. [Google Scholar] [CrossRef] [PubMed]
- Smithson, A.E.; Mirrzayanov, V.S.; Lajoie, R. Chemical Weapons Disarmament in Russia: Problems and Prospects; Stimson Center: Washington, DC, USA, 1995. [Google Scholar]
- Harvey, S.P.; McMahon, L.R.; Berg, F.J. Hydrolysis and Enzymatic Degradation of Novichok Nerve Agents. Heliyon 2020, 6, e03153. [Google Scholar] [CrossRef]
- Mousavi, M.; Hellström-Lindahl, E.; Guan, Z.-Z.; Bednar, I.; Nordberg, A. Expression of Nicotinic Acetylcholine Receptors in Human and Rat Adrenal Medulla. Life Sci. 2001, 70, 577–590. [Google Scholar] [CrossRef]
- Aroniadou-Anderjaska, V.; Apland, J.P.; Figueiredo, T.H.; De Araujo Furtado, M.; Braga, M.F. Acetylcholinesterase Inhibitors (Nerve Agents) as Weapons of Mass Destruction: History, Mechanisms of Action, and Medical Countermeasures. Neuropharmacology 2020, 181, 108298. [Google Scholar] [CrossRef]
- Mew, E.J.; Padmanathan, P.; Konradsen, F.; Eddleston, M.; Chang, S.-S.; Phillips, M.R.; Gunnell, D. The Global Burden of Fatal Self-Poisoning with Pesticides 2006-15: Systematic Review. J. Affect. Disord. 2017, 219, 93–104. [Google Scholar] [CrossRef]
- Ojha, A.; Gupta, Y. Evaluation of Genotoxic Potential of Commonly Used Organophosphate Pesticides in Peripheral Blood Lymphocytes of Rats. Hum. Exp. Toxicol. 2015, 34, 390–400. [Google Scholar] [CrossRef]
- Salazar-Arredondo, E.; Solís-Heredia, M.D.J.; Rojas-García, E.; Hernández-Ochoa, I.; Quintanilla-Vega, B. Sperm Chromatin Alteration and DNA Damage by Methyl-Parathion, Chlorpyrifos and Diazinon and Their Oxon Metabolites in Human Spermatozoa. Reprod. Toxicol. 2008, 25, 455–460. [Google Scholar] [CrossRef]
- Guyton, K.Z.; Loomis, D.; Grosse, Y.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Scoccianti, C.; Mattock, H.; Straif, K. Carcinogenicity of Tetrachlorvinphos, Parathion, Malathion, Diazinon, and Glyphosate. Lancet Oncol. 2015, 16, 490–491. [Google Scholar] [CrossRef]
- Pszczolińska, K.; Michel, M. The QuEChERS Approach for the Determination of Pesticide Residues in Soil Samples: An Overview. J. AOAC Int. 2016, 99, 1403–1414. [Google Scholar] [CrossRef]
- Pasupuleti, R.R.; Tsai, P.; Lin, P.D.; Wu, M.; Ponnusamy, V.K. Rapid and Sensitive Analytical Procedure for Biomonitoring of Organophosphate Pesticide Metabolites in Human Urine Samples Using a Vortex-assisted Salt-induced Liquid–Liquid Microextraction Technique Coupled with Ultra-high-performance Liquid Chromatography/Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8565. [Google Scholar] [CrossRef]
- Nasiri, M.; Ahmadzadeh, H.; Amiri, A. Organophosphorus Pesticides Extraction with Polyvinyl Alcohol Coated Magnetic Graphene Oxide Particles and Analysis by Gas Chromatography-Mass Spectrometry: Application to Apple Juice and Environmental Water. Talanta 2021, 227, 122078. [Google Scholar] [CrossRef] [PubMed]
- Febbraio, F. Biochemical Strategies for the Detection and Detoxification of Toxic Chemicals in the Environment. World J. Biol. Chem. 2017, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, M.; Yu, H.; She, Y.; Cao, Z.; Ye, J.; Abd El-Aty, A.M.; Hacımüftüoğlu, A.; Wang, J.; Lao, S. An Overview on the Mechanisms and Applications of Enzyme Inhibition-Based Methods for Determination of Organophosphate and Carbamate Pesticides. J. Agric. Food Chem. 2020, 68, 7298–7315. [Google Scholar] [CrossRef] [PubMed]
- Lyagin, I.V.; Efremenko, E.N.; Varfolomeev, S.D. Enzymatic Biosensors for Determination of Pesticides. Russ. Chem. Rev. 2017, 86, 339–355. [Google Scholar] [CrossRef]
- Raushel, F.M. Catalytic Detoxification. Nature 2011, 469, 310–311. [Google Scholar] [CrossRef]
- Wille, T.; Kaltenbach, L.; Thiermann, H.; Worek, F. Investigation of Kinetic Interactions between Approved Oximes and Human Acetylcholinesterase Inhibited by Pesticide Carbamates. Chem.-Biol. Interact. 2013, 206, 569–572. [Google Scholar] [CrossRef]
- Gupta, R.D.; Goldsmith, M.; Ashani, Y.; Simo, Y.; Mullokandov, G.; Bar, H.; Ben-David, M.; Leader, H.; Margalit, R.; Silman, I.; et al. Directed Evolution of Hydrolases for Prevention of G-Type Nerve Agent Intoxication. Nat. Chem. Biol. 2011, 7, 120–125. [Google Scholar] [CrossRef]
- Masson, P.; Nachon, F. Cholinesterase Reactivators and Bioscavengers for Pre- and Post-exposure Treatments of Organophosphorus Poisoning. J. Neurochem. 2017, 142, 26–40. [Google Scholar] [CrossRef]
- Merone, L.; Mandrich, L.; Porzio, E.; Rossi, M.; Müller, S.; Reiter, G.; Worek, F.; Manco, G. Improving the Promiscuous Nerve Agent Hydrolase Activity of a Thermostable Archaeal Lactonase. Bioresour. Technol. 2010, 101, 9204–9212. [Google Scholar] [CrossRef]
- Del Giudice, I.; Coppolecchia, R.; Merone, L.; Porzio, E.; Carusone, T.M.; Mandrich, L.; Worek, F.; Manco, G. An Efficient Thermostable Organophosphate Hydrolase and Its Application in Pesticide Decontamination. Biotechnol. Bioeng. 2016, 113, 724–734. [Google Scholar] [CrossRef]
- LeJeune, K.E.; Wild, J.R.; Russell, A.J. Nerve Agents Degraded by Enzymatic Foams. Nature 1998, 395, 27–28. [Google Scholar] [CrossRef] [PubMed]
- LeJeune, K.E.; Russell, A.J. Covalent Binding of a Nerve Agent Hydrolyzing Enzyme within Polyurethane Foams. Biotechnol. Bioeng. 2000, 51, 450–457. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Pittman, T.W.; Zhang, X.; Punyadeera, C.; Henry, C.S. Electrochemical Immunosensor for the Quantification of Galectin-3 in Saliva. Sens. Actuators B Chem. 2024, 400, 134811. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, Z.; Khalife, M.; Costache, V.; Camacho, M.J.; Cardoso, S.; Martins, V.; Gadjanski, I.; Radovic, M.; Vidic, J. Rapid Detection and Identification of Vancomycin-Sensitive Bacteria Using an Electrochemical Apta-Sensor. ACS Omega 2024, 9, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-Care Diagnostics for Infectious Diseases: From Methods to Devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- Morrison, D.W.G.; Dokmeci, M.R.; Demirci, U.; Khademhosseini, A. Clinical Applications of Micro- and Nanoscale Biosensors. In Biomedical Nanostructures; Gonsalves, K.E., Halberstadt, C.R., Laurencin, C.T., Nair, L.S., Eds.; Wiley: Hoboken, NJ, USA, 2007; pp. 439–460. ISBN 978-0-471-92552-1. [Google Scholar]
- Nguyen, H.H.; Lee, S.H.; Lee, U.J.; Fermin, C.D.; Kim, M. Immobilized Enzymes in Biosensor Applications. Materials 2019, 12, 121. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Freitas, A.C.; Pereira, R.; Duarte, A.C.; Rocha Santos, T.A.P. Recent Developments in Recognition Elements for Chemical Sensors and Biosensors. TrAC Trends Anal. Chem. 2015, 68, 2–17. [Google Scholar] [CrossRef]
- Lim, S.A.; Ahmed, M.U. Introduction to Food Biosensors. In Food Biosensors; Ahmed, M.U., Zourob, M., Tamiya, E., Eds.; The Royal Society of Chemistry: Cambridge, 2016; pp. 1–21. ISBN 978-1-78262-361-8. [Google Scholar]
- Perumal, V.; Hashim, U. Advances in Biosensors: Principle, Architecture and Applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Liu, H.; Ge, J.; Ma, E.; Yang, L. Advanced Biomaterials for Biosensor and Theranostics. In Biomaterials in Translational Medicine; Elsevier: Amsterdam, The Netherlands, 2019; pp. 213–255. ISBN 978-0-12-813477-1. [Google Scholar]
- Öndeş, B.; Akpınar, F.; Uygun, M.; Muti, M.; Aktaş Uygun, D. High Stability Potentiometric Urea Biosensor Based on Enzyme Attached Nanoparticles. Microchem. J. 2021, 160, 105667. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Cavacini, L. Structure and Function of Immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef] [PubMed]
- Facure, M.H.M.; Mercante, L.A.; Mattoso, L.H.C.; Correa, D.S. Detection of Trace Levels of Organophosphate Pesticides Using an Electronic Tongue Based on Graphene Hybrid Nanocomposites. Talanta 2017, 167, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Fan, C.; Chen, M.; Zhang, X.; Yan, Q.; Wang, Y.; Zhang, S.; Gong, Z.; Shi, L.; Li, X.; et al. Recent Advances of Fluorescent Biosensors Based on Cyclic Signal Amplification Technology in Biomedical Detection. J. Nanobiotechnol. 2021, 19, 403. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Noumani, A.; Yadav, A.K.; Solanki, P.R. FRET Based Biosensor: Principle Applications Recent Advances and Challenges. Diagnostics 2023, 13, 1375. [Google Scholar] [CrossRef] [PubMed]
- Mu, F.; Zhou, X.; Fan, F.; Chen, Z.; Shi, G. A Fluorescence Biosensor for Therapeutic Drug Monitoring of Vancomycin Using in Vivo Microdialysis. Anal. Chim. Acta 2021, 1151, 338250. [Google Scholar] [CrossRef]
- Levine, M. Fluorescence-Based Sensing of Pesticides Using Supramolecular Chemistry. Front. Chem. 2021, 9, 616815. [Google Scholar] [CrossRef]
- Choi, H.K.; Yoon, J. Enzymatic Electrochemical/Fluorescent Nanobiosensor for Detection of Small Chemicals. Biosensors 2023, 13, 492. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Li, Q.; Tian, Y.; Gao, X.; Zhang, W.; Sun, Z.; Mou, Y.; Sun, X.; Guo, Y.; et al. Development of a Fluorescent Sensor Based on TPE-Fc and GSH-AuNCs for the Detection of Organophosphorus Pesticide Residues in Vegetables. Food Chem. 2024, 431, 137067. [Google Scholar] [CrossRef]
- Krejci, E.; Duval, N.; Chatonnet, A.; Vincens, P.; Massoulié, J. Cholinesterase-like Domains in Enzymes and Structural Proteins: Functional and Evolutionary Relationships and Identification of a Catalytically Essential Aspartic Acid. Proc. Natl. Acad. Sci. USA 1991, 88, 6647–6651. [Google Scholar] [CrossRef]
- Langin, D.; Laurell, H.; Holst, L.S.; Belfrage, P.; Holm, C. Gene Organization and Primary Structure of Human Hormone-Sensitive Lipase: Possible Significance of a Sequence Homology with a Lipase of Moraxella TA144, an Antarctic Bacterium. Proc. Natl. Acad. Sci. USA 1993, 90, 4897–4901. [Google Scholar] [CrossRef]
- Hemilä, H.; Koivula, T.T.; Palva, I. Hormone-Sensitive Lipase Is Closely Related to Several Bacterial Proteins, and Distantly Related to Acetylcholinesterase and Lipoprotein Lipase: Identification of a Superfamily of Esterases and Lipases. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1994, 1210, 249–253. [Google Scholar] [CrossRef]
- Lenfant, N.; Hotelier, T.; Velluet, E.; Bourne, Y.; Marchot, P.; Chatonnet, A. ESTHER, the Database of the α/β-Hydrolase Fold Superfamily of Proteins: Tools to Explore Diversity of Functions. Nucleic Acids Res. 2012, 41, D423–D429. [Google Scholar] [CrossRef] [PubMed]
- Chatonnet, A.; Perochon, M.; Velluet, E.; Marchot, P. The ESTHER Database on Alpha/Beta Hydrolase Fold Proteins—An Overview of Recent Developments. Chem.-Biol. Interact. 2023, 383, 110671. [Google Scholar] [CrossRef]
- Holm, C.; Kirchgessner, T.G.; Svenson, K.L.; Fredrikson, G.; Nilsson, S.; Miller, C.G.; Shively, J.E.; Heinzmann, C.; Sparkes, R.S.; Mohandas, T.; et al. Hormone-Sensitive Lipase: Sequence, Expression, and Chromosomal Localization to 19 Cent-Q13.3. Science 1988, 241, 1503–1506. [Google Scholar] [CrossRef]
- Pohanka, M. Cholinesterases, a target of pharmacology and toxicology. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2011, 155, 219–223. [Google Scholar] [CrossRef]
- Pohanka, M. Butyrylcholinesterase as a Biochemical Marker. Bratisl. Lek. Listy 2013, 114, 726–734. [Google Scholar] [CrossRef]
- Pohanka, M. Acetylcholinesterase Inhibitors: A Patent Review (2008–Present). Expert. Opin. Ther. Pat. 2012, 22, 871–886. [Google Scholar] [CrossRef]
- Julian, P.L.; Pikl, J. Studies in the Indole Series. III. On the Synthesis of Physostigmine. J. Am. Chem. Soc. 1935, 57, 539–544. [Google Scholar] [CrossRef]
- Lilienfeld, S. Galantamine—A Novel Cholinergic Drug with a Unique Dual Mode of Action for the Treatment of Patients with Alzheimer’s Disease. CNS Drug Rev. 2002, 8, 159–176. [Google Scholar] [CrossRef]
- Silver, A. The Biology of Cholinesterases; Front. Biol.; North-Holland Publishing Company (Elsevier): Amsterdam, The Netherlands, 1974. [Google Scholar]
- Massoulié, J.; Pezzementi, L.; Bon, S.; Krejci, E.; Vallette, F.-M. Molecular and Cellular Biology of Cholinesterases. Prog. Neurobiol. 1993, 41, 31–91. [Google Scholar] [CrossRef]
- Cohen, J.A.; Oosterbaan, R.A. Cholinesterases and Anticholinesterase Agents; Koelle, G.B., Ed.; Springer: Berlin/Heidelberg, Germany, 1963; pp. 299–373. [Google Scholar]
- Millard, C.B.; Broomfield, C.A. A Computer Model of Glycosylated Human Butyrylcholinesterase. Biochem. Biophys. Res. Commun. 1992, 189, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Tsigelny, I.; Taylor, P.; Bourne, P.E.; Shindyalov, I.N.; Südhof, T.C. Common EF-hand Motifs in Cholinesterases and Neuroligins Suggest a Role for Ca2+ Binding in Cell Surface Associations. Protein Sci. 2000, 9, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Mesulam, M.; Guillozet, A.; Shaw, P.; Quinn, B. Widely Spread Butyrylcholinesterase Can Hydrolyze Acetylcholine in the Normal and Alzheimer Brain. Neurobiol. Dis. 2002, 9, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Froment, M.-T.; Fortier, P.-L.; Visicchio, J.-E.; Bartels, C.F.; Lockridge, O. Butyrylcholinesterase-Catalysed Hydrolysis of Aspirin, a Negatively Charged Ester, and Aspirin-Related Neutral Esters. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1998, 1387, 41–52. [Google Scholar] [CrossRef]
- Darvesh, S.; Hopkins, D.A.; Geula, C. Neurobiology of Butyrylcholinesterase. Nat. Rev. Neurosci. 2003, 4, 131–138. [Google Scholar] [CrossRef]
- Manco, G.; Adinolfi, E.; Pisani, F.M.; Ottolina, G.; Carrea, G.; Rossi, M. Overexpression and Properties of a New Thermophilic and Thermostable Esterase from Bacillus Acidocaldarius with Sequence Similarity to Hormone-Sensitive Lipase Subfamily. Biochem. J. 1998, 332 Pt 1, 203–212. [Google Scholar] [CrossRef]
- Manco, G.; Febbraio, F.; Adinolfi, E.; Rossi, M. Homology Modeling and Active-site Residues Probing of the Thermophilic Alicyclobacillus Acidocaldarius Esterase 2. Protein Sci. 1999, 8, 1789–1796. [Google Scholar] [CrossRef]
- Mandrich, L.; De Santi, C.; De Pascale, D.; Manco, G. Effect of Low Organic Solvents Concentration on the Stability and Catalytic Activity of HSL-like Carboxylesterases: Analysis from Psychrophiles to (Hyper)Thermophiles. J. Mol. Catal. B Enzym. 2012, 82, 46–52. [Google Scholar] [CrossRef]
- Pennacchio, A.; Mandrich, L.; Manco, G.; Trincone, A. Enlarging the Substrate Portfolio of the Thermophilic Esterase EST2 from Alicyclobacillus Acidocaldarius. Extremophiles 2015, 19, 1001–1011. [Google Scholar] [CrossRef]
- De Simone, G.; Galdiero, S.; Manco, G.; Lang, D.; Rossi, M.; Pedone, C. A Snapshot of a Transition State Analogue of a Novel Thermophilic Esterase Belonging to the Subfamily of Mammalian Hormone-Sensitive Lipase 1 1Edited by D. Rees. J. Mol. Biol. 2000, 303, 761–771. [Google Scholar] [CrossRef]
- Febbraio, F.; D’Andrea, S.E.; Mandrich, L.; Merone, L.; Rossi, M.; Nucci, R.; Manco, G. Irreversible Inhibition of the Thermophilic Esterase EST2 from Alicyclobacillus Acidocaldarius. Extremophiles 2008, 12, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Pezzullo, M.; Del Vecchio, P.; Mandrich, L.; Nucci, R.; Rossi, M.; Manco, G. Comprehensive Analysis of Surface Charged Residues Involved in Thermal Stability in Alicyclobacillus Acidocaldarius Esterase 2. Protein Eng. Des. Sel. 2013, 26, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Porzio, E.; Bettazzi, F.; Mandrich, L.; Del Giudice, I.; Restaino, O.F.; Laschi, S.; Febbraio, F.; De Luca, V.; Borzacchiello, M.G.; Carusone, T.M.; et al. Innovative Biocatalysts as Tools to Detect and Inactivate Nerve Agents. Sci. Rep. 2018, 8, 13773. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Singh, P.K. Enzyme-Based Optical Biosensors for Organophosphate Class of Pesticide Detection. Phys. Chem. Chem. Phys. 2020, 22, 15105–15119. [Google Scholar] [CrossRef]
- Kim, M.; Kwon, J.E.; Lee, K.; Koh, W.-G. Signal-Amplifying Nanoparticle/Hydrogel Hybrid Microarray Biosensor for Metal-Enhanced Fluorescence Detection of Organophosphorus Compounds. Biofabrication 2018, 10, 035002. [Google Scholar] [CrossRef]
- Wu, X.; Song, Y.; Yan, X.; Zhu, C.; Ma, Y.; Du, D.; Lin, Y. Carbon Quantum Dots as Fluorescence Resonance Energy Transfer Sensors for Organophosphate Pesticides Determination. Biosens. Bioelectron. 2017, 94, 292–297. [Google Scholar] [CrossRef]
- Yan, X.; Li, H.; Han, X.; Su, X. A Ratiometric Fluorescent Quantum Dots Based Biosensor for Organophosphorus Pesticides Detection by Inner-Filter Effect. Biosens. Bioelectron. 2015, 74, 277–283. [Google Scholar] [CrossRef]
- Kamelipour, N.; Mohsenifar, A.; Tabatabaei, M.; Rahmani-Cherati, T.; Khoshnevisan, K.; Allameh, A.; Milani, M.M.; Najavand, S.; Etemadikia, B. Fluorometric Determination of Paraoxon in Human Serum Using a Gold Nanoparticle-Immobilized Organophosphorus Hydrolase and Coumarin 1 as a Competitive Inhibitor. Microchim. Acta 2014, 181, 239–248. [Google Scholar] [CrossRef]
- Rodrigues, A.C.M.; Barbieri, M.V.; Chino, M.; Manco, G.; Febbraio, F. A 3D Printable Adapter for Solid-State Fluorescence Measurements: The Case of an Immobilized Enzymatic Bioreceptor for Organophosphate Pesticides Detection. Anal. Bioanal. Chem. 2022, 414, 1999–2008. [Google Scholar] [CrossRef]
- De Luca, V.; Mandrich, L.; Manco, G. Development of a Qualitative Test to Detect the Presence of Organophosphate Pesticides on Fruits and Vegetables. Life 2023, 13, 490. [Google Scholar] [CrossRef]
- Febbraio, F.; Merone, L.; Cetrangolo, G.P.; Rossi, M.; Nucci, R.; Manco, G. Thermostable Esterase 2 from Alicyclobacillus Acidocaldarius as Biosensor for the Detection of Organophosphate Pesticides. Anal. Chem. 2011, 83, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Purcarea, C.; Ruginescu, R.; Banciu, R.M.; Vasilescu, A. Extremozyme-Based Biosensors for Environmental Pollution Monitoring: Recent Developments. Biosensors 2024, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Carullo, P.; Chino, M.; Cetrangolo, G.P.; Terreri, S.; Lombardi, A.; Manco, G.; Cimmino, A.; Febbraio, F. Direct Detection of Organophosphate Compounds in Water by a Fluorescence-Based Biosensing Device. Sens. Actuators B Chem. 2018, 255, 3257–3266. [Google Scholar] [CrossRef]
- Barbieri, M.V.; Rodrigues, A.C.; Febbraio, F. Monitoring of Pesticide Amount in Water and Drinkable Food by a Fluorescence-based Biosensor. EFSA J. 2022, 20, e200403. [Google Scholar] [CrossRef]
- Cetrangolo, G.P.; Gori, C.; Rusko, J.; Terreri, S.; Manco, G.; Cimmino, A.; Febbraio, F. Determination of Picomolar Concentrations of Paraoxon in Human Urine by Fluorescence-Based Enzymatic Assay. Sensors 2019, 19, 4852. [Google Scholar] [CrossRef]
- Garefalaki, V.; Manco, G.; Porzio, E.; Institute of Biochemistry and Cell Biology-National Research Council (IBBC-CNR), Italy. Use of Biosensors for Rapid and Sensitive Detection of Pesticides in Food Samples for Food Safety Chemical Risk Assessment. EFSA J. 2022, 20, e200922. [Google Scholar] [CrossRef]
- Carullo, P.; Cetrangolo, G.P.; Mandrich, L.; Manco, G.; Febbraio, F. Fluorescence Spectroscopy Approaches for the Development of a Real-Time Organophosphate Detection System Using an Enzymatic Sensor. Sensors 2015, 15, 3932–3951. [Google Scholar] [CrossRef]
- Tortora, F.; Porzio, E.; Galoppo, C.; Febbraio, F.; Manco, G. Development of a Fluorescence-Based Biosensor for Coumaphos. Sens. Transducers 2021, 253, 18–22. [Google Scholar]
- Doctor, B.P.; Saxena, A. Bioscavengers for the Protection of Humans against Organophosphate Toxicity. Chem.-Biol. Interact. 2005, 157–158, 167–171. [Google Scholar] [CrossRef]
- Braue, E.H., Jr.; Hobson, S.T. Active Topical Skin Protectants Containing OPAA Enzymes and Clecs. US6410604B1, 2001. [Google Scholar]
- Rosenberg, Y.J.; Laube, B.; Mao, L.; Jiang, X.; Hernandez-Abanto, S.; Lee, K.D.; Adams, R. Pulmonary Delivery of an Aerosolized Recombinant Human Butyrylcholinesterase Pretreatment Protects against Aerosolized Paraoxon in Macaques. Chem.-Biol. Interact. 2013, 203, 167–171. [Google Scholar] [CrossRef]
- Rosenberg, Y.J.; Adams, R.J.; Hernandez-Abanto, S.; Jiang, X.; Sun, W.; Mao, L.; Lee, K.D. Pharmacokinetics and Immunogenicity of a Recombinant Human Butyrylcholinesterase Bioscavenger in Macaques Following Intravenous and Pulmonary Delivery. Chem.-Biol. Interact. 2015, 242, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Lushchekina, S.V.; Schopfer, L.M.; Grigorenko, B.L.; Nemukhin, A.V.; Varfolomeev, S.D.; Lockridge, O.; Masson, P. Optimization of Cholinesterase-Based Catalytic Bioscavengers Against Organophosphorus Agents. Front. Pharmacol. 2018, 9, 211. [Google Scholar] [CrossRef]
- Yang, Y.-C. ChemInform Abstract: Chemical Detoxification of Nerve Agent VX. ChemInform 2010, 30. [Google Scholar] [CrossRef]
- Pearson, G.S.; Magee, R.S. Critical Evaluation of Proven Chemical Weapon Destruction Technologies (IUPAC Technical Report). Pure Appl. Chem. 2002, 74, 187–316. [Google Scholar] [CrossRef]
- Kim, K.; Tsay, O.G.; Atwood, D.A.; Churchill, D.G. Destruction and Detection of Chemical Warfare Agents. Chem. Rev. 2011, 111, 5345–5403. [Google Scholar] [CrossRef]
- Tucker, M. Reduced Weight Decontamination Formulation for Neutralization of Chemical and Biological Warfare Agents. U.S. Patent 8741174 B1, 3 June 2014. [Google Scholar]
- Poirier, L.; Jacquet, P.; Elias, M.; Daudé, D.; Chabrière, E. Decontamination of organophosphorus compounds: Towards new alternatives. Ann. Pharm. Fr. 2017, 75, 209–226. [Google Scholar] [CrossRef]
- Pearson, J.P.; Gray, K.M.; Passador, L.; Tucker, K.D.; Eberhard, A.; Iglewski, B.H.; Greenberg, E.P. Structure of the Autoinducer Required for Expression of Pseudomonas Aeruginosa Virulence Genes. Proc. Natl. Acad. Sci. USA 1994, 91, 197–201. [Google Scholar] [CrossRef]
- Reddy, P.V.L.; Kim, K.-H. A Review of Photochemical Approaches for the Treatment of a Wide Range of Pesticides. J. Hazard. Mater. 2015, 285, 325–335. [Google Scholar] [CrossRef]
- Pashirova, T.; Shaihutdinova, Z.; Mansurova, M.; Kazakova, R.; Shambazova, D.; Bogdanov, A.; Tatarinov, D.; Daudé, D.; Jacquet, P.; Chabrière, E.; et al. Enzyme Nanoreactor for In Vivo Detoxification of Organophosphates. ACS Appl. Mater. Interfaces 2022, 14, 19241–19252. [Google Scholar] [CrossRef]
- Xu, F.; Kang, T.; Deng, J.; Liu, J.; Chen, X.; Wang, Y.; Ouyang, L.; Du, T.; Tang, H.; Xu, X.; et al. Functional Nanoparticles Activate a Decellularized Liver Scaffold for Blood Detoxification. Small 2016, 12, 2067–2076. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Huang, J.; Li, W.; Luo, Y.; Sui, X.; Li, J.; Wang, Y.; Yang, J. A Protein Nanocomposite for Ultra-Fast, Efficient and Non-Irritating Skin Decontamination of Nerve Agents. Nanoscale 2020, 12, 4400–4409. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, M.; Zhang, Y.; Lee, J.H.; Escajadillo, T.; Gong, H.; Fang, R.H.; Gao, W.; Nizet, V.; Zhang, L. Antivirulence Therapy: Broad-Spectrum Neutralization of Pore-Forming Toxins with Human Erythrocyte Membrane-Coated Nanosponges (Adv. Healthcare Mater. 13/2018). Adv. Healthc. Mater. 2018, 7, 1870049. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Yan, M.; Lau, M.Y.; Hu, J.; Han, H.; Yang, O.O.; Liang, S.; Wei, W.; Wang, H.; et al. Biomimetic Enzyme Nanocomplexes and Their Use as Antidotes and Preventive Measures for Alcohol Intoxication. Nat. Nanotechnol. 2013, 8, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Xing, R.; Guan, J.; Yang, B.; Zhao, X.; Yang, J.; Zhan, C.; Zhang, S. A Nanoantidote Alleviates Glioblastoma Chemotoxicity without Efficacy Compromise. Nano Lett. 2021, 21, 5158–5166. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Lu, Y. Enzyme Therapeutics for Systemic Detoxification. Adv. Drug Deliv. Rev. 2015, 90, 24–39. [Google Scholar] [CrossRef]
- Forster, V.; Leroux, J.-C. Nano-Antidotes for Drug Overdose and Poisoning. Sci. Transl. Med. 2015, 7, 290ps14. [Google Scholar] [CrossRef]
- Rabanel, J.-M.; Delbreil, P.; Banquy, X.; Brambilla, D.; Ramassamy, C. Periphery-Confined Particulate Systems for the Management of Neurodegenerative Diseases and Toxicity: Avoiding the Blood-Brain-Barrier Challenge. J. Control. Release 2020, 322, 286–299. [Google Scholar] [CrossRef]
- Serdar, C.M.; Gibson, D.T.; Munnecke, D.M.; Lancaster, J.H. Plasmid Involvement in Parathion Hydrolysis by Pseudomonas Diminuta. Appl. Environ. Microbiol. 1982, 44, 246–249. [Google Scholar] [CrossRef]
- Efremenko, E.N.; Lyagin, I.V.; Klyachko, N.L.; Bronich, T.; Zavyalova, N.V.; Jiang, Y.; Kabanov, A.V. A Simple and Highly Effective Catalytic Nanozyme Scavenger for Organophosphorus Neurotoxins. J. Control. Release 2017, 247, 175–181. [Google Scholar] [CrossRef]
- Bigley, A.N.; Harvey, S.P.; Narindoshvili, T.; Raushel, F.M. Substrate Analogues for the Enzyme-Catalyzed Detoxification of the Organophosphate Nerve Agents—Sarin, Soman, and Cyclosarin. Biochemistry 2021, 60, 2875–2887. [Google Scholar] [CrossRef]
- Seibert, C.M.; Raushel, F.M. Structural and Catalytic Diversity within the Amidohydrolase Superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.-C.; Bigley, A.; Li, Y.; Ghanem, E.; Cadieux, C.L.; Kasten, S.A.; Reeves, T.E.; Cerasoli, D.M.; Raushel, F.M. Stereoselective Hydrolysis of Organophosphate Nerve Agents by the Bacterial Phosphotriesterase. Biochemistry 2010, 49, 7978–7987. [Google Scholar] [CrossRef] [PubMed]
- Holden, H.M.; Raushel, F.M. From the Three-Dimensional Structure of Phosphotriesterase. Biochemistry 2021, 60, 3413–3415. [Google Scholar] [CrossRef]
- Dumas, D.P.; Caldwell, S.R.; Wild, J.R.; Raushel, F.M. Purification and Properties of the Phosphotriesterase from Pseudomonas Diminuta. J. Biol. Chem. 1989, 264, 19659–19665. [Google Scholar] [CrossRef]
- Dumas, D.P.; Durst, H.D.; Landis, W.G.; Raushel, F.M.; Wild, J.R. Inactivation of Organophosphorus Nerve Agents by the Phosphotriesterase from Pseudomonas Diminuta. Arch. Biochem. Biophys. 1990, 277, 155–159. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, D. Transient Expression of Organophosphorus Hydrolase to Enhance the Degrading Activity of Tomato Fruit on Coumaphos. J. Zhejiang Univ. Sci. B 2009, 10, 142–146. [Google Scholar] [CrossRef]
- Caldwell, S.R.; Newcomb, J.R.; Schlecht, K.A.; Raushel, F.M. Limits of Diffusion in the Hydrolysis of Substrates by the Phosphotriesterase from Pseudomonas Diminuta. Biochemistry 1991, 30, 7438–7444. [Google Scholar] [CrossRef]
- Lyagin, I.V.; Andrianova, M.S.; Efremenko, E.N. Extensive Hydrolysis of Phosphonates as Unexpected Behaviour of the Known His6-Organophosphorus Hydrolase. Appl. Microbiol. Biotechnol. 2016, 100, 5829–5838. [Google Scholar] [CrossRef]
- Nowlan, C.; Li, Y.; Hermann, J.C.; Evans, T.; Carpenter, J.; Ghanem, E.; Shoichet, B.K.; Raushel, F.M. Resolution of Chiral Phosphate, Phosphonate, and Phosphinate Esters by an Enantioselective Enzyme Library. J. Am. Chem. Soc. 2006, 128, 15892–15902. [Google Scholar] [CrossRef]
- Lewis, V.E.; Donarski, W.J.; Wild, J.R.; Raushel, F.M. Mechanism and Stereochemical Course at Phosphorus of the Reaction Catalyzed by a Bacterial Phosphotriesterase. Biochemistry 1988, 27, 1591–1597. [Google Scholar] [CrossRef]
- Samples, C.R.; Howard, T.; Raushel, F.M.; DeRose, V.J. Protonation of the Binuclear Metal Center within the Active Site of Phosphotriesterase. Biochemistry 2005, 44, 11005–11013. [Google Scholar] [CrossRef] [PubMed]
- Benning, M.M.; Hong, S.-B.; Raushel, F.M.; Holden, H.M. The Binding of Substrate Analogs to Phosphotriesterase. J. Biol. Chem. 2000, 275, 30556–30560. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Tsai, P.-C.; Chen, S.-L.; Himo, F.; Almo, S.C.; Raushel, F.M. Structure of Diethyl Phosphate Bound to the Binuclear Metal Center of Phosphotriesterase. Biochemistry 2008, 47, 9497–9504. [Google Scholar] [CrossRef] [PubMed]
- Horne, I.; Sutherland, T.D.; Harcourt, R.L.; Russell, R.J.; Oakeshott, J.G. Identification of an Opd (Organophosphate Degradation) Gene in an Agrobacterium Isolate. Appl. Environ. Microbiol. 2002, 68, 3371–3376. [Google Scholar] [CrossRef]
- Horne, I.; Qiu, X.; Russell, R.J.; Oakeshott, J.G. The Phosphotriesterase Gene opdA in Agrobacterium Radiobacter P230 Is Transposable. FEMS Microbiol. Lett. 2003, 222, 1–8. [Google Scholar] [CrossRef]
- Bigley, A.N.; Raushel, F.M. The Evolution of Phosphotriesterase for Decontamination and Detoxification of Organophosphorus Chemical Warfare Agents. Chem.-Biol. Interact. 2019, 308, 80–88. [Google Scholar] [CrossRef]
- Bigley, A.N.; Xu, C.; Henderson, T.J.; Harvey, S.P.; Raushel, F.M. Enzymatic Neutralization of the Chemical Warfare Agent VX: Evolution of Phosphotriesterase for Phosphorothiolate Hydrolysis. J. Am. Chem. Soc. 2013, 135, 10426–10432. [Google Scholar] [CrossRef]
- Bigley, A.N.; Mabanglo, M.F.; Harvey, S.P.; Raushel, F.M. Variants of Phosphotriesterase for the Enhanced Detoxification of the Chemical Warfare Agent VR. Biochemistry 2015, 54, 5502–5512. [Google Scholar] [CrossRef]
- Cherny, I.; Greisen, P.; Ashani, Y.; Khare, S.D.; Oberdorfer, G.; Leader, H.; Baker, D.; Tawfik, D.S. Engineering V-Type Nerve Agents Detoxifying Enzymes Using Computationally Focused Libraries. ACS Chem. Biol. 2013, 8, 2394–2403. [Google Scholar] [CrossRef]
- Jackson, C.J.; Weir, K.; Herlt, A.; Khurana, J.; Sutherland, T.D.; Horne, I.; Easton, C.; Russell, R.J.; Scott, C.; Oakeshott, J.G. Structure-Based Rational Design of a Phosphotriesterase. Appl. Environ. Microbiol. 2009, 75, 5153–5156. [Google Scholar] [CrossRef]
- Naqvi, T.; Warden, A.C.; French, N.; Sugrue, E.; Carr, P.D.; Jackson, C.J.; Scott, C. A 5000-Fold Increase in the Specificity of a Bacterial Phosphotriesterase for Malathion through Combinatorial Active Site Mutagenesis. PLoS ONE 2014, 9, e94177. [Google Scholar] [CrossRef] [PubMed]
- Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A Thermostable Phosphotriesterase from the Archaeon Sulfolobus Solfataricus: Cloning, Overexpression and Properties. Extremophiles 2005, 9, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Porzio, E.; Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A New Phosphotriesterase from Sulfolobus Acidocaldarius and Its Comparison with the Homologue from Sulfolobus Solfataricus. Biochimie 2007, 89, 625–636. [Google Scholar] [CrossRef]
- Porzio, E.; Di Gennaro, S.; Palma, A.; Manco, G. Mn2+ Modulates the Kinetic Properties of an Archaeal Member of the PLL Family. Chem. Biol. Interact. 2013, 203, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Suzumoto, Y.; Dym, O.; Roviello, G.N.; Worek, F.; Sussman, J.L.; Manco, G. Structural and Functional Characterization of New SsoPox Variant Points to the Dimer Interface as a Driver for the Increase in Promiscuous Paraoxonase Activity. Int. J. Mol. Sci. 2020, 21, 1683. [Google Scholar] [CrossRef]
- Porzio, E.; Del Giudice, I.; Manco, G. Engineering of Extremophilic Phosphotriesterase-Like Lactonases for Biotechnological Applications. In Biotechnology of Extremophiles; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Hiblot, J.; Gotthard, G.; Chabriere, E.; Elias, M. Characterisation of the Organophosphate Hydrolase Catalytic Activity of SsoPox. Sci. Rep. 2012, 2, 779. [Google Scholar] [CrossRef]
- Elias, M.; Dupuy, J.; Merone, L.; Mandrich, L.; Porzio, E.; Moniot, S.; Rochu, D.; Lecomte, C.; Rossi, M.; Masson, P.; et al. Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities. J. Mol. Biol. 2008, 379, 1017–1028. [Google Scholar] [CrossRef]
- Manco, G.; Porzio, E.; Suzumoto, Y. Enzymatic Detoxification: A Sustainable Means of Degrading Toxic Organophosphate Pesticides and Chemical Warfare Nerve Agents. J. Chem. Technol. Biotechnol. 2018, 93, 2064–2082. [Google Scholar] [CrossRef]
- Zhang, Y.; An, J.; Yang, G.-Y.; Bai, A.; Zheng, B.; Lou, Z.; Wu, G.; Ye, W.; Chen, H.-F.; Feng, Y.; et al. Active Site Loop Conformation Regulates Promiscuous Activity in a Lactonase from Geobacillus Kaustophilus HTA426. PLoS ONE 2015, 10, e0115130. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.; Liu, X.; Zhou, W.; Rao, Z. The Crystal Structure of the Phosphotriesterase from M. tuberculosis, Another Member of Phosphotriesterase-like Lactonase Family. Biochem. Biophys. Res. Commun. 2019, 510, 224–229. [Google Scholar] [CrossRef]
- Hawwa, R.; Aikens, J.; Turner, R.J.; Santarsiero, B.D.; Mesecar, A.D. Structural Basis for Thermostability Revealed through the Identification and Characterization of a Highly Thermostable Phosphotriesterase-like Lactonase from Geobacillus Stearothermophilus. Arch. Biochem. Biophys. 2009, 488, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.Y.; Xue, B.; Lee, K.H.; Tung, A.; Wu, L.; Robinson, R.C.; Yew, W.S. Directed Evolution of a Thermostable Quorum-Quenching Lactonase from the Amidohydrolase Superfamily. J. Biol. Chem. 2010, 285, 40911–40920. [Google Scholar] [CrossRef] [PubMed]
- Hiblot, J.; Gotthard, G.; Chabriere, E.; Elias, M. Structural and Enzymatic Characterization of the Lactonase SisLac from Sulfolobus Islandicus. PLoS ONE 2012, 7, e47028. [Google Scholar] [CrossRef]
- Kallnik, V.; Bunescu, A.; Sayer, C.; Bräsen, C.; Wohlgemuth, R.; Littlechild, J.; Siebers, B. Characterization of a Phosphotriesterase-like Lactonase from the Hyperthermoacidophilic Crenarchaeon Vulcanisaeta Moutnovskia. J. Biotechnol. 2014, 190, 11–17. [Google Scholar] [CrossRef]
- Hawwa, R.; Larsen, S.D.; Ratia, K.; Mesecar, A.D. Structure-Based and Random Mutagenesis Approaches Increase the Organophosphate-Degrading Activity of a Phosphotriesterase Homologue from Deinococcus Radiodurans. J. Mol. Biol. 2009, 393, 36–57. [Google Scholar] [CrossRef]
- Mandrich, L.; Di Gennaro, S.; Palma, A.; Manco, G. A Further Biochemical Characterization of DrPLL the Thermophilic Lactonase from Deinococcus Radiodurans. Protein Pept. Lett. 2013, 20, 36–44. [Google Scholar] [CrossRef]
- Chow, J.Y.; Wu, L.; Yew, W.S. Directed Evolution of a Quorum-Quenching Lactonase from Mycobacterium Avium Subsp. Paratuberculosis K-10 in the Amidohydrolase Superfamily. Biochemistry 2009, 48, 4344–4353. [Google Scholar] [CrossRef]
- Rochu, D.; Viguié, N.; Renault, F.; Crouzier, D.; Froment, M.-T.; Masson, P. Contribution of the Active-Site Metal Cation to the Catalytic Activity and to the Conformational Stability of Phosphotriesterase: Temperature- and pH-Dependence. Biochem. J. 2004, 380, 627–633. [Google Scholar] [CrossRef]
- Hiblot, J.; Bzdrenga, J.; Champion, C.; Chabriere, E.; Elias, M. Crystal Structure of VmoLac, a Tentative Quorum Quenching Lactonase from the Extremophilic Crenarchaeon Vulcanisaeta Moutnovskia. Sci. Rep. 2015, 5, 8372. [Google Scholar] [CrossRef]
- Marone, M.; Porzio, E.; Lampitella, E.A.; Manco, G. A Mesophilic Phosphotriesterase-like Lactonase Shows High Stability and Proficiency as Quorum Quenching Enzyme. Chem. Biol. Interact. 2023, 383, 110657. [Google Scholar] [CrossRef]
- Roodveldt, C.; Tawfik, D.S. Shared Promiscuous Activities and Evolutionary Features in Various Members of the Amidohydrolase Superfamily. Biochemistry 2005, 44, 12728–12736. [Google Scholar] [CrossRef] [PubMed]
- Farber, G.K.; Petsko, G.A. The Evolution of α/β Barrel Enzymes. Trends Biochem. Sci. 1990, 15, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Mandrich, L.; Merone, L.; Manco, G. Hyperthermophilic Phosphotriesterases/Lactonases for the Environment and Human Health. Environ. Technol. 2010, 31, 1115–1127. [Google Scholar] [CrossRef]
- Del Vecchio, P.; Elias, M.; Merone, L.; Graziano, G.; Dupuy, J.; Mandrich, L.; Carullo, P.; Fournier, B.; Rochu, D.; Rossi, M.; et al. Structural Determinants of the High Thermal Stability of SsoPox from the Hyperthermophilic Archaeon Sulfolobus Solfataricus. Extremophiles 2009, 13, 461–470. [Google Scholar] [CrossRef]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum Sensing and Chromobacterium Violaceum: Exploitation of Violacein Production and Inhibition for the Detection of N-Acylhomoserine Lactones. Microbiology 1997, 143 Pt 12, 3703–3711. [Google Scholar] [CrossRef]
- Reimmann, C.; Ginet, N.; Michel, L.; Keel, C.; Michaux, P.; Krishnapillai, V.; Zala, M.; Heurlier, K.; Triandafillu, K.; Harms, H.; et al. Genetically Programmed Autoinducer Destruction Reduces Virulence Gene Expression and Swarming Motility in Pseudomonas Aeruginosa PAO1 The GenBank Accession Number for the aiiA Nucleotide Sequence Is AF397400. The GenBank Accession Numbers for the Nucleotide Sequences of the 16S rRNA Genes of Strains A23 and A24 Are AF397398 and AF397399. Microbiology 2002, 148, 923–932. [Google Scholar] [CrossRef]
- Wagner, V.E.; Bushnell, D.; Passador, L.; Brooks, A.I.; Iglewski, B.H. Microarray Analysis of Pseudomonas Aeruginosa Quorum-Sensing Regulons: Effects of Growth Phase and Environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef]
- Barbey, C.; Crépin, A.; Bergeau, D.; Ouchiha, A.; Mijouin, L.; Taupin, L.; Orange, N.; Feuilloley, M.; Dufour, A.; Burini, J.-F.; et al. In Planta Biocontrol of Pectobacterium Atrosepticum by Rhodococcus Erythropolis Involves Silencing of Pathogen Communication by the Rhodococcal Gamma-Lactone Catabolic Pathway. PLoS ONE 2013, 8, e66642. [Google Scholar] [CrossRef]
- Crépin, A.; Beury-Cirou, A.; Barbey, C.; Farmer, C.; Hélias, V.; Burini, J.-F.; Faure, D.; Latour, X. N-Acyl Homoserine Lactones in Diverse Pectobacterium and Dickeya Plant Pathogens: Diversity, Abundance, and Involvement in Virulence. Sensors 2012, 12, 3484–3497. [Google Scholar] [CrossRef]
- Singh, B.K. Organophosphorus-Degrading Bacteria: Ecology and Industrial Applications. Nat. Rev. Microbiol. 2009, 7, 156–164. [Google Scholar] [CrossRef]
- Uroz, S.; Oger, P.M.; Chapelle, E.; Adeline, M.-T.; Faure, D.; Dessaux, Y. A Rhodococcus qsdA-Encoded Enzyme Defines a Novel Class of Large-Spectrum Quorum-Quenching Lactonases. Appl. Environ. Microbiol. 2008, 74, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Han, X.; Jia, H.; Cai, J.; Zhou, Y.; Zheng, Z. AuPt Alloy Nanostructures with Tunable Composition and Enzyme-like Activities for Colorimetric Detection of Bisulfide. Sci. Rep. 2017, 7, 40103. [Google Scholar] [CrossRef] [PubMed]
- Mondloch, J.E.; Katz, M.J.; Isley Iii, W.C.; Ghosh, P.; Liao, P.; Bury, W.; Wagner, G.W.; Hall, M.G.; DeCoste, J.B.; Peterson, G.W.; et al. Destruction of Chemical Warfare Agents Using Metal–Organic Frameworks. Nat. Mater. 2015, 14, 512–516. [Google Scholar] [CrossRef]
- Mishra, R.K.; Dominguez, R.B.; Bhand, S.; Muñoz, R.; Marty, J.-L. A Novel Automated Flow-Based Biosensor for the Determination of Organophosphate Pesticides in Milk. Biosens. Bioelectron. 2012, 32, 56–61. [Google Scholar] [CrossRef]
- Schofield, D.A.; Westwater, C.; Barth, J.L.; DiNovo, A.A. Development of a Yeast Biosensor–Biocatalyst for the Detection and Biodegradation of the Organophosphate Paraoxon. Appl. Microbiol. Biotechnol. 2007, 76, 1383–1394. [Google Scholar] [CrossRef]
- Liu, R.; Yang, C.; Xu, Y.; Xu, P.; Jiang, H.; Qiao, C. Development of a Whole-Cell Biocatalyst/Biosensor by Display of Multiple Heterologous Proteins on the Escherichia Coli Cell Surface for the Detoxification and Detection of Organophosphates. J. Agric. Food Chem. 2013, 61, 7810–7816. [Google Scholar] [CrossRef]
- Istamboulie, G.; Durbiano, R.; Fournier, D.; Marty, J.-L.; Noguer, T. Biosensor-Controlled Degradation of Chlorpyrifos and Chlorfenvinfos Using a Phosphotriesterase-Based Detoxification Column. Chemosphere 2010, 78, 1–6. [Google Scholar] [CrossRef]
- Pashirova, T.; Salah-Tazdaït, R.; Tazdaït, D.; Masson, P. Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation. Int. J. Mol. Sci. 2024, 25, 7822. [Google Scholar] [CrossRef]
- Kim, E.R.; Joe, C.; Mitchell, R.J.; Gu, M.B. Biosensors for Healthcare: Current and Future Perspectives. Trends Biotechnol. 2023, 41, 374–395. [Google Scholar] [CrossRef]
- Safarkhani, M.; Kim, H.; Han, S.; Taghavimandi, F.; Park, Y.; Umapathi, R.; Jeong, Y.-S.; Shin, K.; Huh, Y.S. Advances in Sprayable Sensors for Nerve Agent Detection. Coord. Chem. Rev. 2024, 509, 215804. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).