Origami-Inspired Biosensors: Exploring Diverse Applications and Techniques for Shape-Changing Sensor Platforms
Abstract
1. Introduction
2. Paper-Based Origami Biosensors
2.1. Types of Paper Based Origami Biosensor
2.1.1. Single Folding
2.1.2. Double Folding
2.1.3. Multiple Folding
2.1.4. POP Up
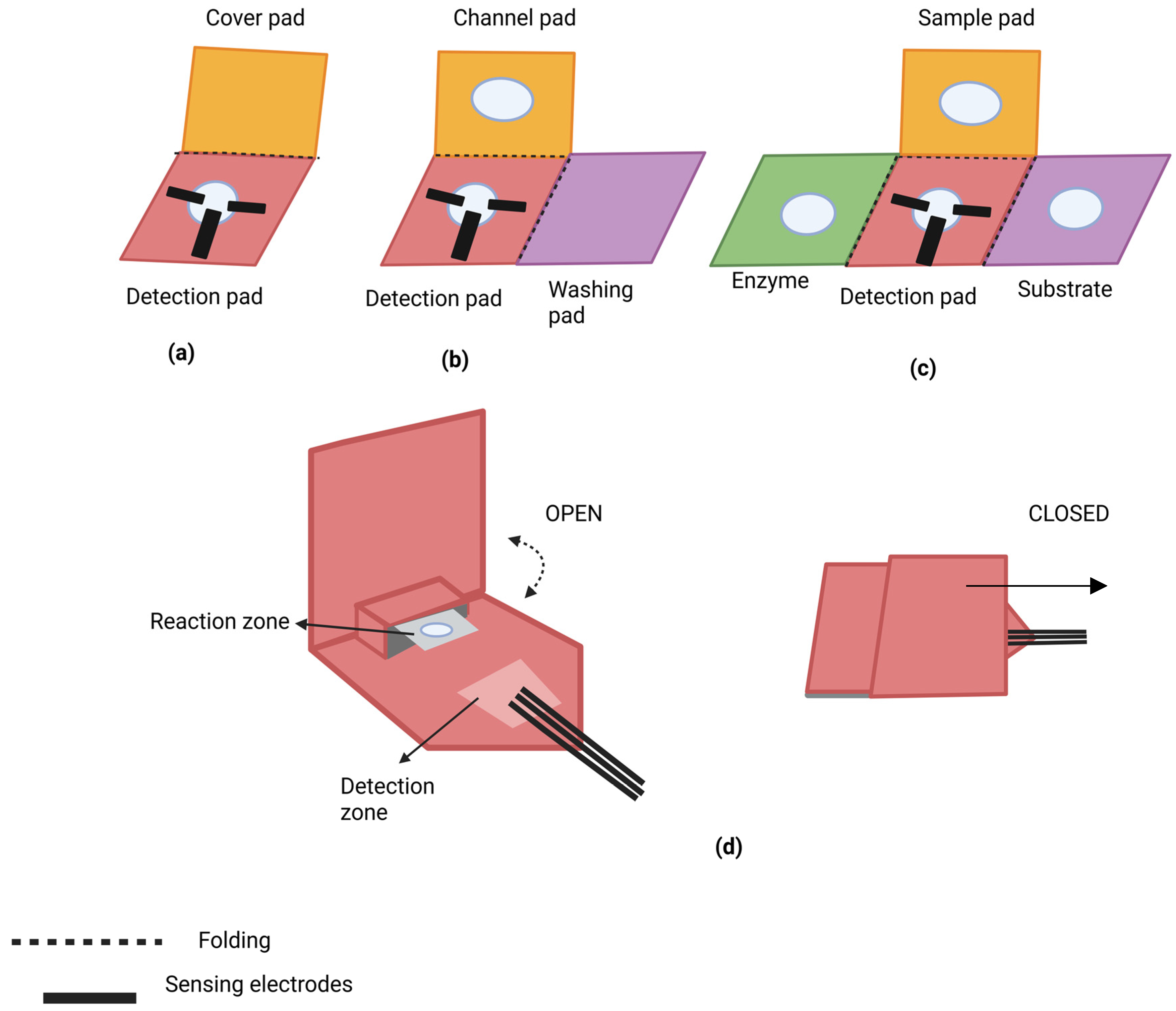
2.2. Applications of Paper Origami in Biosensing
2.2.1. Nucleic Acid Detection
2.2.2. Proteins Detection
2.2.3. Virus Detection
2.2.4. Detection of Heavy Metals
2.2.5. Agriculture and Food Safety
3. DNA Origami
3.1. Types of DNA Origami Biosensor
3.1.1. Binding-Based Biosensors
3.1.2. Nanopores
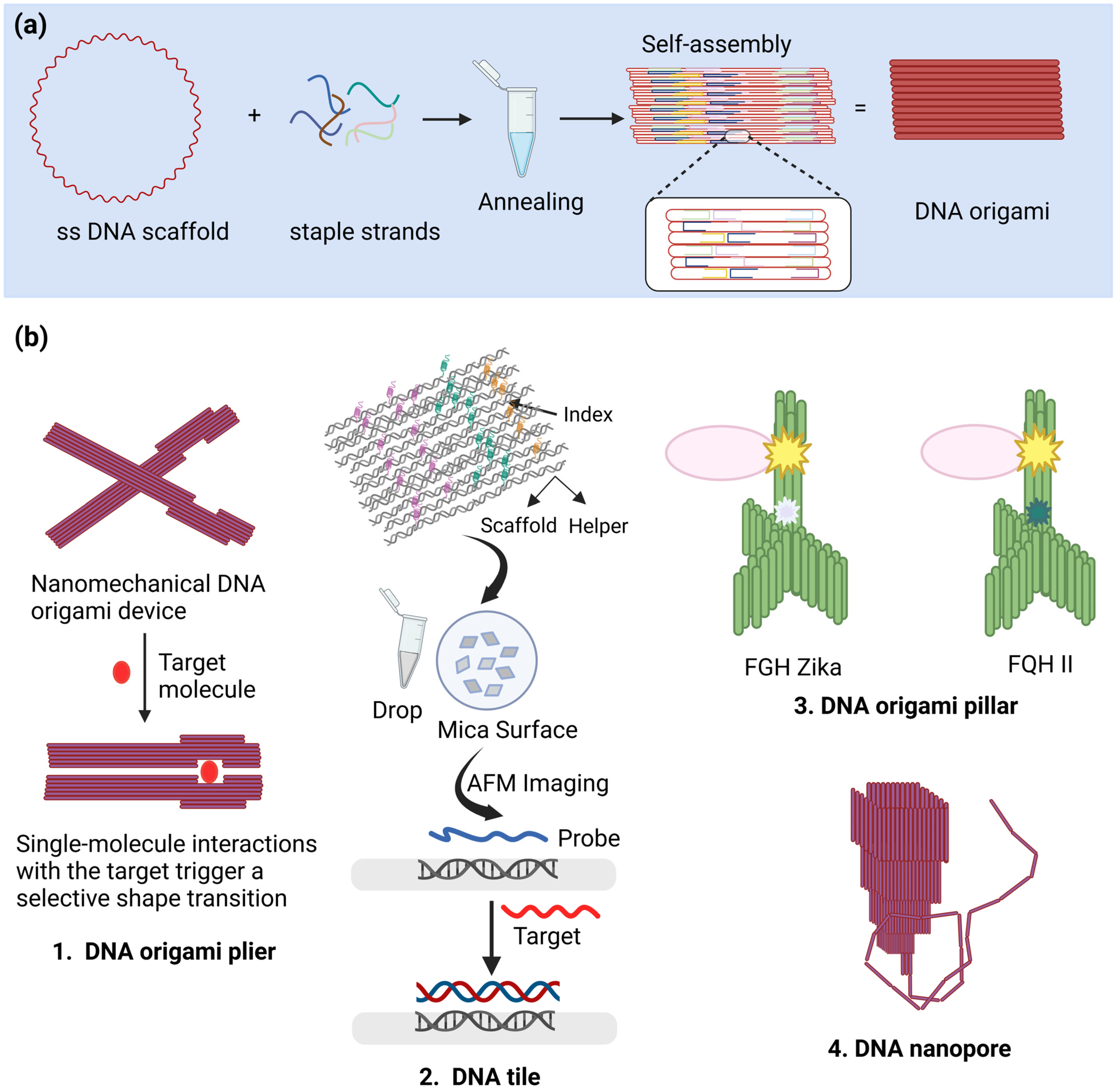
3.2. Applications of DNA Origami in Biosensing
3.2.1. Nucleic Acid/SNP Detection
3.2.2. Protein Detection
3.2.3. pH Detection
3.2.4. Enzyme Activity
3.2.5. Chemical Reactions
4. Aptamers in Biosensors
4.1. Aptamer in Paper Biosensors
4.2. Aptamers in DNA Origami Biosensors
5. Challenges in Practical Applications of Origami Biosensor
6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Peng, R.; You, Z. Origami of Thick Panels. Science (1979) 2015, 349, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Colozza, N.; Caratelli, V.; Moscone, D.; Arduini, F. Origami Paper-Based Electrochemical (Bio)Sensors: State of the Art and Perspective. Biosensors 2021, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Loretan, M.; Domljanovic, I.; Lakatos, M.; Rüegg, C.; Acuna, G.P. DNA Origami as Emerging Technology for the Engineering of Fluorescent and Plasmonic-Based Biosensors. Materials 2020, 13, 2185. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, Z.; Ma, N.; Yang, S.; Li, K.; Teng, C.; Ke, Y.; Tian, Y. Dna Origami-Enabled Biosensors. Sensors 2020, 20, 6899. [Google Scholar] [CrossRef]
- Williamson, P.; Piskunen, P.; Ijäs, H.; Butterworth, A.; Linko, V.; Corrigan, D.K. Signal Amplification in Electrochemical DNA Biosensors Using Target-Capturing DNA Origami Tiles. ACS Sens. 2023, 8, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.T.; Lantigua, D.; Meka, A.; Taing, S.; Pandher, M.; Camci-Unal, G. Paper-Based Sensors: Emerging Themes and Applications. Sensors 2018, 18, 2838. [Google Scholar] [CrossRef] [PubMed]
- Lazzarini, E.; Pace, A.; Trozzi, I.; Zangheri, M.; Guardigli, M.; Calabria, D.; Mirasoli, M. An Origami Paper-Based Biosensor for Allergen Detection by Chemiluminescence Immunoassay on Magnetic Microbeads. Biosensors 2022, 12, 825. [Google Scholar] [CrossRef] [PubMed]
- Sahraei, N.; Mazloum-Ardakani, M.; Hoseynidokht, F. Electrochemical Paper-Based Biosensors for Point-of-Care Diagnostics: Detection Methods and Applications. J. Electrochem. Sci. Eng. 2022, 12, 399–419. [Google Scholar] [CrossRef]
- Huang, Y.; Nguyen, M.K.; Natarajan, A.K.; Nguyen, V.H.; Kuzyk, A. A DNA Origami-Based Chiral Plasmonic Sensing Device. ACS Appl. Mater. Interfaces 2018, 10, 44221–44225. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R.; Punnoose, J.A.; Zhou, L.; Dey, P.; Dey, B.K.; Halvorsen, K. DNA Nanotechnology Approaches for MicroRNA Detection and Diagnosis. Nucleic Acids Res. 2019, 47, 10489–10505. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Feng, L.; Deng, R.; Wang, B.; He, Y.; Zhang, L.; Luo, D.; Chen, M.; Chang, K. Bottom-up DNA Nanostructure-Based Paper as Point-of-Care Diagnostic: From Method to Device. Interdiscip. Med. 2024, 2, e20230033. [Google Scholar] [CrossRef]
- Lee, N.E.; Hong, J.H.; Lee, S.; Yoo, Y.K.; Kim, K.H.; Park, J.S.; Kim, C.; Yoon, J.; Yoon, D.S.; Lee, J.H. Integration of an Aptamer-Based Signal-On Probe and a Paper-Based Origami Preconcentrator for Small Molecule Biomarkers Detection. BioChip J. 2023, 17, 439–446. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, J.; Zheng, M.; Zhong, Y.; Yang, J.; Zhao, Y.; Wu, W.; Ye, W.; Wen, J.; Wang, Q.; et al. An Aptamer-Based Biosensor for Colorimetric Detection of Escherichia Coli O157:H7. PLoS ONE 2012, 7, e48999. [Google Scholar] [CrossRef] [PubMed]
- Sameiyan, E.; Bagheri, E.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. DNA Origami-Based Aptasensors. Biosens. Bioelectron. 2019, 143, 111662. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Butte, M.J.; Whitesides, G.M. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Asiri, A.M.; Du, D.; Wen, W.; Wang, S.; Lin, Y. Nanomaterial-Enhanced Paper-Based Biosensors. TrAC Trends Anal. Chem. 2014, 58, 31–39. [Google Scholar] [CrossRef]
- Liu, H.; Crooks, R.M. Three-Dimensional Paper Microfluidic Devices Assembled Using the Principles of Origami. J. Am. Chem. Soc. 2011, 133, 17564–17566. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiang, Y.; Lu, Y.; Crooks, R.M. Aptamer-Based Origami Paper Analytical Device for Electrochemical Detection of Adenosine. Angew. Chem. Int. Ed. 2012, 51, 6925–6928. [Google Scholar] [CrossRef] [PubMed]
- Kuswandi, B.; Hidayat, M.A.; Noviana, E. Paper-Based Sensors for Rapid Important Biomarkers Detection. Biosens. Bioelectron. X 2022, 12, 100246. [Google Scholar] [CrossRef]
- Liang, B.; Zhu, Q.; Fang, L.; Cao, Q.; Liang, X.; Ye, X. An Origami Paper Device for Complete Elimination of Interferents in Enzymatic Electrochemical Biosensors. Electrochem. Commun. 2017, 82, 43–46. [Google Scholar] [CrossRef]
- Li, W.; Qian, D.; Wang, Q.; Li, Y.; Bao, N.; Gu, H.; Yu, C. Fully-Drawn Origami Paper Analytical Device for Electrochemical Detection of Glucose. Sens. Actuators B Chem. 2016, 231, 230–238. [Google Scholar] [CrossRef]
- Li, L.; Xu, J.; Zheng, X.; Ma, C.; Song, X.; Ge, S.; Yu, J.; Yan, M. Growth of Gold-Manganese Oxide Nanostructures on a 3D Origami Device for Glucose-Oxidase Label Based Electrochemical Immunosensor. Biosens. Bioelectron. 2014, 61, 76–82. [Google Scholar] [CrossRef]
- Sun, X.; Jian, Y.; Wang, H.; Ge, S.; Yan, M.; Yu, J. Ultrasensitive Microfluidic Paper-Based Electrochemical Biosensor Based on Molecularly Imprinted Film and Boronate Affinity Sandwich Assay for Glycoprotein Detection. ACS Appl. Mater. Interfaces 2019, 11, 16198–16206. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Li, B.; Chen, L.; Qin, W. A Three-Dimensional Origami Paper-Based Device for Potentiometric Biosensing. Angew. Chem. Int. Ed. 2016, 55, 13033–13037. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Zhang, L.; Zhang, Y.; Yang, H.; Ma, C.; Ge, S.; Yan, M.; Yu, J.; Song, X. Multiplexed Enzyme-Free Electrochemical Immunosensor Based on ZnO Nanorods Modified Reduced Graphene Oxide-Paper Electrode and Silver Deposition-Induced Signal Amplification Strategy. Biosens. Bioelectron. 2015, 71, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Hennek, J.W.; Ainla, A.; Kumar, A.A.; Lan, W.J.; Im, J.; Smith, B.S.; Zhao, M.; Whitesides, G.M. A Paper-Based Pop-up Electrochemical Device for Analysis of Beta-Hydroxybutyrate. Anal. Chem. 2016, 88, 6326–6333. [Google Scholar] [CrossRef]
- Srisomwat, C.; Teengam, P.; Chuaypen, N.; Tangkijvanich, P.; Vilaivan, T.; Chailapakul, O. Pop-up Paper Electrochemical Device for Label-Free Hepatitis B Virus DNA Detection. Sens. Actuators B Chem. 2020, 316, 128077. [Google Scholar] [CrossRef]
- Boonkaew, S.; Chaiyo, S.; Jampasa, S.; Rengpipat, S.; Siangproh, W.; Chailapakul, O. An Origami Paper-Based Electrochemical Immunoassay for the C-Reactive Protein Using a Screen-Printed Carbon Electrode Modified with Graphene and Gold Nanoparticles. Microchim. Acta 2019, 186, 153. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.A.; Yuan, H.; Chen, C.W.; Chien, Y.S.; Sheng, W.H.; Chen, C.F. An Electricity- And Instrument-Free Infectious Disease Sensor Based on a 3D Origami Paper-Based Analytical Device. Lab Chip 2021, 21, 1908–1915. [Google Scholar] [CrossRef]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, P.; Zha, X.; Xu, C.; Kang, S.; Zhou, M.; Nover, D.; Wang, Y. Overview Assessment of Risk Evaluation and Treatment Technologies for Heavy Metal Pollution of Water and Soil. J. Clean. Prod. 2022, 379, 134043. [Google Scholar] [CrossRef]
- Asim, M.; Nageswara Rao, K. Assessment of Heavy Metal Pollution in Yamuna River, Delhi-NCR, Using Heavy Metal Pollution Index and GIS. Environ. Monit. Assess 2021, 193, 103. [Google Scholar] [CrossRef] [PubMed]
- Malikula, R.S.; Kaonga, C.C.; Mapoma, H.W.T.; Chiipa, P.; Thulu, F.G.D. Heavy Metals and Nutrients Loads in Water, Soil, and Crops Irrigated with Effluent from WWTPs in Blantyre City, Malawi. Water 2022, 14, 121. [Google Scholar] [CrossRef]
- Xu, L.; Suo, X.-y.; Zhang, Q.; Li, X.-p.; Chen, C.; Zhang, X.Y. ELISA and Chemiluminescent Enzyme Immunoassay for Sensitive and Specific Determination of Lead (II) in Water, Food and Feed Samples. Foods 2020, 9, 305. [Google Scholar] [CrossRef]
- Fu, X.; Chen, E.; Ma, B.; Xu, Y.; Hao, P.; Zhang, M.; Ye, Z.; Yu, X.; Li, C.; Ji, Q. Establishment of an Indirect Competitive Enzyme-Linked Immunosorbent Method for the Detection of Heavy Metal Cadmium in Food Packaging Materials. Foods 2021, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Rattanarat, P.; Dungchai, W.; Cate, D.; Volckens, J.; Chailapakul, O.; Henry, C.S. Multilayer Paper-Based Device for Colorimetric and Electrochemical Quantification of Metals. Anal. Chem. 2014, 86, 3555–3562. [Google Scholar] [CrossRef]
- Xiao, W.; Gao, Y.; Zhang, Y.; Li, J.; Liu, Z.; Nie, J.; Li, J. Enhanced 3D Paper-Based Devices with a Personal Glucose Meter for Highly Sensitive and Portable Biosensing of Silver Ion. Biosens. Bioelectron. 2019, 137, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Soliman Sabra, F.; El-Deeb Mehana, E.-S. Pesticides Toxicity in Fish with Particular Reference to Insecticides. Asian J. Agric. Food Sci. 2015, 3, 1. [Google Scholar]
- Aktar, W.; Sengupta, D.; Chowdhury, A. Impact of Pesticides Use in Agriculture: Their Benefits and Hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arduini, F.; Cinti, S.; Caratelli, V.; Amendola, L.; Palleschi, G.; Moscone, D. Origami Multiple Paper-Based Electrochemical Biosensors for Pesticide Detection. Biosens. Bioelectron. 2019, 126, 346–354. [Google Scholar] [CrossRef]
- Jabeen, N.; Shahzad, K. Studies on Identification of the Food Born Fungal Pathogens and Their Pathogenic Effects on Human Health. Acta Sci. Agric. 2019, 3, 186–191. [Google Scholar] [CrossRef]
- Khiyami, M.; Al-Faris, N.; Busaeed, B.; Sher, H. Food Borne Pathogen Contamination in Minimally Processed Vegetable Salads in Riyadh, Saudi Arabia. J. Med. Plant Res. 2011, 5, 444–451. [Google Scholar]
- Wang, K.; Lin, X.; Zhang, M.; Li, Y.; Luo, C.; Wu, J. Review of Electrochemical Biosensors for Food Safety Detection. Biosensors 2022, 12, 959. [Google Scholar] [CrossRef] [PubMed]
- Ivanišević, I.; Milardović, S.; Kassal, P. Recent Advances in (Bio)Chemical Sensors for Food Safety and Quality Based on Silver Nanomaterials. Food Technol. Biotechnol. 2021, 59, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Kuswandi, B.; Hidayat, M.A.; Noviana, E. Paper-Based Electrochemical Biosensors for Food Safety Analysis. Biosensors 2022, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Li, J.; Liu, L.; Ge, S.; Yan, M.; Liu, H.; Yu, J. Origami-Based “Book” Shaped Three-Dimensional Electrochemical Paper Microdevice for Sample-to-Answer Detection of Pathogens. RSC Adv. 2020, 10, 25808–25816. [Google Scholar] [CrossRef]
- Lu, J.; Ge, S.; Ge, L.; Yan, M.; Yu, J. Electrochemical DNA Sensor Based on Three-Dimensional Folding Paper Device for Specific and Sensitive Point-of-Care Testing. Electrochim. Acta 2012, 80, 334–341. [Google Scholar] [CrossRef]
- Scida, K.; Li, B.; Ellington, A.D.; Crooks, R.M. DNA Detection Using Origami Paper Analytical Devices. Anal. Chem. 2013, 85, 9713–9720. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Seeman, N.C. DNA in a Material World. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Zadegan, R.M.; Norton, M.L. Structural DNA Nanotechnology: From Design to Applications. Int. J. Mol. Sci. 2012, 13, 7149–7162. [Google Scholar] [CrossRef]
- Ramm, B.; Khmelinskaia, A.; Franquelim, H.G.; Schwille, P. Patterning DNA Origami on Membranes Through Protein Self-Organization. In Natural Computing Series; Springer: Singapore, 2023; Volume Part F821. [Google Scholar]
- Hartl, C.; Frank, K.; Amenitsch, H.; Fischer, S.; Liedl, T.; Nickel, B. Position Accuracy of Gold Nanoparticles on DNA Origami Structures Studied with Small-Angle X-Ray Scattering. Nano Lett. 2018, 18, 2609–2615. [Google Scholar] [CrossRef] [PubMed]
- Dietz, H. Toward Applications for DNA Nanotechnology—More Bricks To Build With. ChemBioChem 2016, 17, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The Beauty and Utility of DNA Origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Anderson, N.; Kizer, M.; Halvorsen, K.; Wang, X. Beyond the Fold: Emerging Biological Applications of DNA Origami. ChemBioChem 2016, 17, 1081–1089. [Google Scholar] [CrossRef]
- Ke, Y.; Nangreave, J.; Yan, H.; Lindsay, S.; Liu, Y. Developing DNA Tiles for Oligonucleotide Hybridization Assay with Higher Accuracy and Efficiency. Chem. Commun. 2008, 43, 5622–5624. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Lindsay, S.; Chang, Y.; Liu, Y.; Yan, H. Self-Assembled Water-Soluble Nucleic Acid Probe Tiles for Label-Free RNA Hybridization Assays. Science (1979) 2008, 319, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA Origami “single-Molecule Beacons” Directly Imaged by Atomic Force Microscopy. Nat. Commun. 2011, 2, 449. [Google Scholar] [CrossRef]
- Hernández-Ainsa, S.; Keyser, U.F. DNA Origami Nanopores: Developments, Challenges and Perspectives. Nanoscale 2014, 6, 14121–14132. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J. Nanometer-Scale Pores: Potential Applications for Analyte Detection and DNA Characterization. Dis. Markers 2002, 18, 185–191. [Google Scholar] [CrossRef]
- Storm, A.J.; Chen, J.H.; Ling, X.S.; Zandbergen, H.W.; Dekker, C. Fabrication of Solid-State Nanopores with Single-Nanometre Precision. Nat. Mater. 2003, 2, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.A.W.; Keyser, U.F. Nanopores Formed by DNA Origami: A Review. FEBS Lett. 2014, 588, 3564–3570. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Scott, A.; Rotem, D.; Mehta, K.K.; Bayley, H.; Dekker, C. Hybrid Pore Formation by Directed Insertion of α-Haemolysin into Solid-State Nanopores. Nat. Nanotechnol. 2010, 5, 874–877. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Engst, C.R.; Ablay, M.; Divitini, G.; Ducati, C.; Liedl, T.; Keyser, U.F. DNA Origami Nanopores. Nano Lett. 2012, 12, 512–517. [Google Scholar] [CrossRef]
- Wei, R.; Martin, T.G.; Rant, U.; Dietz, H. DNA Origami Gatekeepers for Solid-State Nanopores. Angew. Chem. Int. Ed. 2012, 51, 4864–4867. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ainsa, S.; Bell, N.A.W.; Thacker, V.V.; Göpfrich, K.; Misiunas, K.; Fuentes-Perez, M.E.; Moreno-Herrero, F.; Keyser, U.F. DNA Origami Nanopores for Controlling DNA Translocation. ACS Nano 2013, 7, 6024–6030. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ainsa, S.; Keyser, U.F. DNA Origami Nanopores: An Emerging Tool in Biomedicine. Nanomedicine 2013, 8, 1551–1554. [Google Scholar] [CrossRef] [PubMed]
- Ochmann, S.E.; Vietz, C.; Trofymchuk, K.; Acuna, G.P.; Lalkens, B.; Tinnefeld, P. Optical Nanoantenna for Single Molecule-Based Detection of Zika Virus Nucleic Acids without Molecular Multiplication. Anal. Chem. 2017, 89, 13000–13007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Y.; Fan, C.; Li, C.; Li, Y.; Qian, L.; Fu, Y.; Shi, Y.; Hu, J.; He, L. Asymmetric DNA Origami for Spatially Addressable and Index-Free Solution-Phase DNA Chips. Adv. Mater. 2010, 22, 2672–2675. [Google Scholar] [CrossRef]
- Zhang, Z.; Zeng, D.; Ma, H.; Feng, G.; Hu, J.; He, L.; Li, C.; Fan, C. A DNA-Origami Chip Platform for Label-Free SNP Genotyping Using Toehold-Mediated Strand Displacement. Small 2010, 6, 1854–1858. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, H.K.K.; Chakraborty, B.; Sha, R.; Seeman, N.C. The Label-Free Unambiguous Detection and Symbolic Display of Single Nucleotide Polymorphisms on DNA Origami. Nano Lett. 2011, 11, 910–913. [Google Scholar] [CrossRef]
- Panyutin, I.G.; Hsieh, P. The Kinetics of Spontaneous DNA Branch Migration. Proc. Natl. Acad. Sci. USA 1994, 91, 2021–2025. [Google Scholar] [CrossRef] [PubMed]
- Raveendran, M.; Lee, A.J.; Sharma, R.; Wälti, C.; Actis, P. Rational Design of DNA Nanostructures for Single Molecule Biosensing. Nat. Commun. 2020, 11, 4384. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Hu, C.; Wang, P.; Liu, R.; Zuo, X.; Liu, X.; Song, S.; Fan, C.; He, D.; Sun, G. Novel Rolling Circle Amplification and DNA Origami-Based DNA Belt-Involved Signal Amplification Assay for Highly Sensitive Detection of Prostate-Specific Antigen (PSA). ACS Appl. Mater. Interfaces 2014, 6, 20372–20377. [Google Scholar] [CrossRef]
- Kuzuya, A.; Watanabe, R.; Yamanaka, Y.; Tamaki, T.; Kaino, M.; Ohya, Y. Nanomechanical DNA Origami PH Sensors. Sensors 2014, 14, 19329–19335. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.V.; Tørring, T.; Rotaru, A.; Jacobsen, M.F.; Ravnsbæk, J.B.; Subramani, R.; Mamdouh, W.; Kjems, J.; Mokhir, A.; Besenbacher, F.; et al. Single-Molecule Chemical Reactions on DNA Origami. Nat. Nanotechnol. 2010, 5, 200–203. [Google Scholar] [CrossRef]
- Tintoré, M.; Gállego, I.; Manning, B.; Eritja, R.; Fàbrega, C. DNA Origami as a DNA Repair Nanosensor at the Single-Molecule Level. Angew. Chem. Int. Ed. 2013, 52, 7747–7750. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, L.; Guo, R.; Zhang, Y.; Li, F.; Li, M.; Li, J.; Shi, J.; Qu, F.; Zuo, X.; et al. DNA Origami Nanocalipers for PH Sensing at the Nanoscale. Chem. Commun. 2022, 58, 3673–3676. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef]
- Sequeira-Antunes, B.; Ferreira, H.A. Nucleic Acid Aptamer-Based Biosensors: A Review. Biomedicines 2023, 11, 3201. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Xie, T.; Kang, B.; Yu, X.; Schaffter, S.W.; Schulman, R. Plug-and-Play Protein Biosensors Using Aptamer-Regulated in Vitro Transcription. Nat. Commun. 2024, 15, 7973. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Jo, H.; Oh, S.S. Detection and beyond: Challenges and Advances in Aptamer-Based Biosensors. Mater. Adv. 2020, 1, 2663–2687. [Google Scholar] [CrossRef]
- Sakai, Y.; Islam, M.S.; Adamiak, M.; Shiu, S.C.C.; Tanner, J.A.; Heddle, J.G. DNA Aptamers for the Functionalisation of DNA Origami Nanostructures. Genes 2018, 9, 571. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, D.; Fan, J.; Nie, J.; Le, S.; Zhu, W.; Yang, J.; Li, J. Naked-Eye Quantitative Aptamer-Based Assay on Paper Device. Biosens. Bioelectron. 2016, 78, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, W.; Chen, W.; Liu, L.; Ma, W.; Zhu, Y.; Xu, L.; Kuang, H.; Xu, C. An Aptamer-Based Chromatographic Strip Assay for Sensitive Toxin Semi-Quantitative Detection. Biosens. Bioelectron. 2011, 26, 3059–3062. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Guo, Q.; Zhang, C.; Sun, Z.; Weng, X. Microfluidic Origami Nano-Aptasensor for Peanut Allergen Ara H1 Detection. Food Chem. 2021, 365, 130511. [Google Scholar] [CrossRef]
- Godonoga, M.; Lin, T.Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.; Kuzuya, A.; et al. A DNA Aptamer Recognising a Malaria Protein Biomarker Can Function as Part of a DNA Origami Assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef] [PubMed]
- Walter, H.K.; Bauer, J.; Steinmeyer, J.; Kuzuya, A.; Niemeyer, C.M.; Wagenknecht, H.A. DNA Origami Traffic Lights with a Split Aptamer Sensor for a Bicolor Fluorescence Readout. Nano Lett. 2017, 17, 2467–2472. [Google Scholar] [CrossRef]
- Hosseini, S.; Vázquez-Villegas, P.; Martínez-Chapa, S.O. Paper and Fiber-Based Bio-Diagnostic Platforms: Current Challenges and Future Needs. Appl. Sci. 2017, 7, 863. [Google Scholar] [CrossRef]
- Linko, V.; Keller, A. Stability of DNA Origami Nanostructures in Physiological Media: The Role of Molecular Interactions. Small 2023, 19, e2301935. [Google Scholar] [CrossRef]
- Yan, W.; Li, S.; Deguchi, M.; Zheng, Z.; Rus, D.; Mehta, A. Origami-Based Integration of Robots That Sense, Decide, and Respond. Nat. Commun. 2023, 14, 1553. [Google Scholar] [CrossRef] [PubMed]
- Jeon, B.; Guareschi, M.M.; Stewart, J.M.; Wu, E.; Gopinath, A.; Arroyo-Currás, N.; Dauphin-Ducharme, P.; Plaxco, K.W.; Lukeman, P.S.; Rothemund, P.W.K. Modular DNA Origami-Based Electrochemical Detection of DNA and Proteins. arXiv 2023, arXiv:2312.06554. [Google Scholar]
- Chang, K.; Zhao, Y.; Wang, M.; Xu, Z.; Zhu, L.; Xu, L.; Wang, Q. Advances in Metal-Organic Framework-Plasmonic Metal Composites Based SERS Platforms: Engineering Strategies in Chemical Sensing, Practical Applications and Future Perspectives in Food Safety. Chem. Eng. J. 2023, 459, 141539. [Google Scholar] [CrossRef]
- Wang, Q.; Chang, K.; Yang, Q.; Wu, W. Semiconductor-Based Surface-Enhanced Raman Scattering Sensing Platforms: State of the Art, Applications and Prospects in Food Safety. Trends Food Sci. Technol. 2024, 147, 104460. [Google Scholar] [CrossRef]

| Paper Origami Biosensors | DNA Origami Biosensors | Aptamer Origami Biosensor | |
|---|---|---|---|
| Material Composition | Cellulose-based paper | DNA strands | Single stranded oligonucleotides (DNA/RNA) |
| Fabrication Techniques | Cutting, folding, and assembling 3D structures with paper | DNA origami technique (scaffold DNA and staple strands for folding) | Aptamer-based folding (using aptamers for target binding and folding) |
| Detection Mechanisms | Colorimetric, electrochemical, fluorescence-based | Fluorescence, FRET, SPR, electrochemical | Fluorescence, FRET, SPR, electrochemical |
| Target Analytes | Pathogens, environmental toxins, small molecules | Proteins, nucleic acids, metal ions | Small molecules, proteins, ions, pathogens |
| Sensitivity | Moderate, depending on design and readout method | High sensitivity due to precise molecular design | Moderate to high, depending on aptamer specificity |
| Applications | POC diagnostics, Food safety, environmental monitoring | Medical diagnostics, drug delivery, nanoscale sensing | Medical diagnostics, biosensing, therapeutic monitoring |
| Advantages | Cost-effective, simple, disposable, portable | High precision, customizable structures | High specificity, versatility in target binding |
| Limitations | Lower sensitivity compared to DNA origami | Complex fabrication, higher cost | Limited by aptamer selection and stability |
| Biosensor Type | Target | Sensing Substrate | Linear Range | LOD | Ref. |
|---|---|---|---|---|---|
| Single folding | Glucose | Glucose preloaded on paper | 0–24 mM | - | [20] |
| Glucose | Glucose oxidase + ferrocenecarboxylic acid | 1–12 mM | 0.05 mM | [21] | |
| Prostate protein antigen | Carbon nanosperes-glucose oxidase-monoclonal antibody label | 0.005–100 ng/mL | 0.0012 ng/mL | [22] | |
| ssDNA | AuNPs/GS modified SPWPE | 0.0008–500 pM | 0.2 fM | [47] | |
| Cd2+, Ni2+, Pb2+, Fe2+, Cu2+and Cr2+ | Bi and potassium ferricyanide | 0.25–7.5 ng | 0.25 ng | [36] | |
| Double folding | Glycoproteins | SiO2@Au/dsDNA/CeO2 nanocomposite | 1–107 pg/mL | 0.87 pg/mL | [23] |
| ssDNA | fluorophore-labeled ssDNA | - | <5 nM | [48] | |
| SARS-CoV-2 antibodies | spike protein receptor-binding domain (SP RBD) of SARS-CoV-2 | - | 1 ng/ml | [30] | |
| Multiple folding | Organophosphorus pesticides | Inhibition of Butyrylcholinesterase, with butyrylcholine-sensitive membrane | 0.1 and 1.0 nM | 0.06 nM | [24] |
| (a) HCG (b) PSA(c) carcinoembryonic antigen | Reduced graphene oxide/Ag@BSA/2° antibody as signal label. | (a) 0.002–120 mIU/mL (b) 0.001–110 pg/mL (c) 0.001–100 pg/mL | (a) 0.0007 mIU/mL (b) 0.35 pg/mL (c) 0.33 pg/mL | [25] | |
| HIV p24 | anti-HIV-1 p24 | 0.03 ng/mL to 3 ng/mL | 0.01 ng/mL | [29] | |
| (i) paraoxon, (ii) 2,4-dichlorophenoxyacetic acid (iii) atrazine | (i) butyrylcholinesterase, (ii) alkaline phosphatase, (iii) tyrosinase |
| (i) 2 ppb (ii) 50 ppb (iii) - | [39] | |
| Pop up | β-hydroxybutyrate | 3-hydrozybutyrate dehydrogenase | 0.1–6.0 mM | 0.3 mM | [26] |
| Applications | Target | Sensing Component | Detection/Visualization Technique | Efficiency/ LOD | Ref. |
|---|---|---|---|---|---|
| Nucleic acid detection | Rag-1, c-myc and β-actin | DNA origami tile having complementary probe | Atomic Force Microscopy | ~1000 molecules | [58] |
| SNP detection | SM (single-base mismatch) DNA | - | “toehold”-mediated strand-displacement reaction/Atomic Force Microscopy | - | [71] |
| Protein detection | Streptavidin | Biotinylated DNA plier | Atomic Force Microscopy | 2 molecules | [59] |
| Prostate-specific antigen (PSA) | RCA based DNA belts | Magnetic bead-based ELISA/AFM | 50 aM | [75] | |
| C-reactive protein (CRP) | CRP-specific aptamer | AFM | 3 nM | [74] | |
| pH detection | HEPES buffer | DNA origami plier | AFM | - | [76] |
| pH buffer | DNA nanocaliper | Transmission electron microscope | - | [79] | |
| Enzyme activity | human O6-alkylguanine-DNA alkyltransferase (hAGT) | DNA quadruplex | AFM | - | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, S.; Suleman, S.; Anzar, N.; Narang, J.; Pilloton, R.; Timur, S.; Guler Celik, E.; Pundir, C.S.; Shukla, S.K. Origami-Inspired Biosensors: Exploring Diverse Applications and Techniques for Shape-Changing Sensor Platforms. Chemosensors 2024, 12, 276. https://doi.org/10.3390/chemosensors12120276
Patil S, Suleman S, Anzar N, Narang J, Pilloton R, Timur S, Guler Celik E, Pundir CS, Shukla SK. Origami-Inspired Biosensors: Exploring Diverse Applications and Techniques for Shape-Changing Sensor Platforms. Chemosensors. 2024; 12(12):276. https://doi.org/10.3390/chemosensors12120276
Chicago/Turabian StylePatil, Shikha, Shariq Suleman, Nigar Anzar, Jagriti Narang, Roberto Pilloton, Suna Timur, Emine Guler Celik, Chandra S. Pundir, and Sudheesh K. Shukla. 2024. "Origami-Inspired Biosensors: Exploring Diverse Applications and Techniques for Shape-Changing Sensor Platforms" Chemosensors 12, no. 12: 276. https://doi.org/10.3390/chemosensors12120276
APA StylePatil, S., Suleman, S., Anzar, N., Narang, J., Pilloton, R., Timur, S., Guler Celik, E., Pundir, C. S., & Shukla, S. K. (2024). Origami-Inspired Biosensors: Exploring Diverse Applications and Techniques for Shape-Changing Sensor Platforms. Chemosensors, 12(12), 276. https://doi.org/10.3390/chemosensors12120276







