Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants
Abstract
1. Introduction
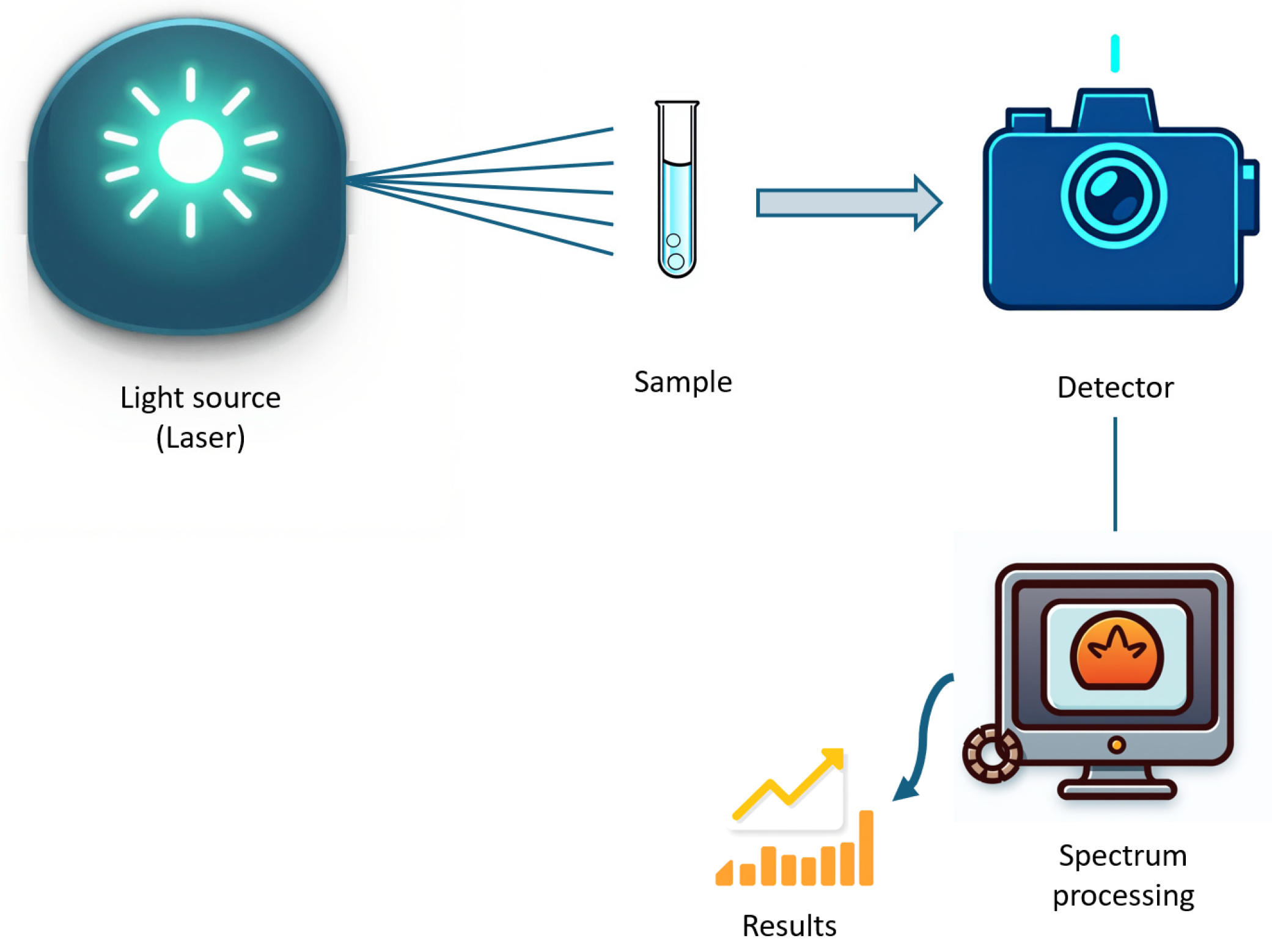
2. Spectroscopy Principles
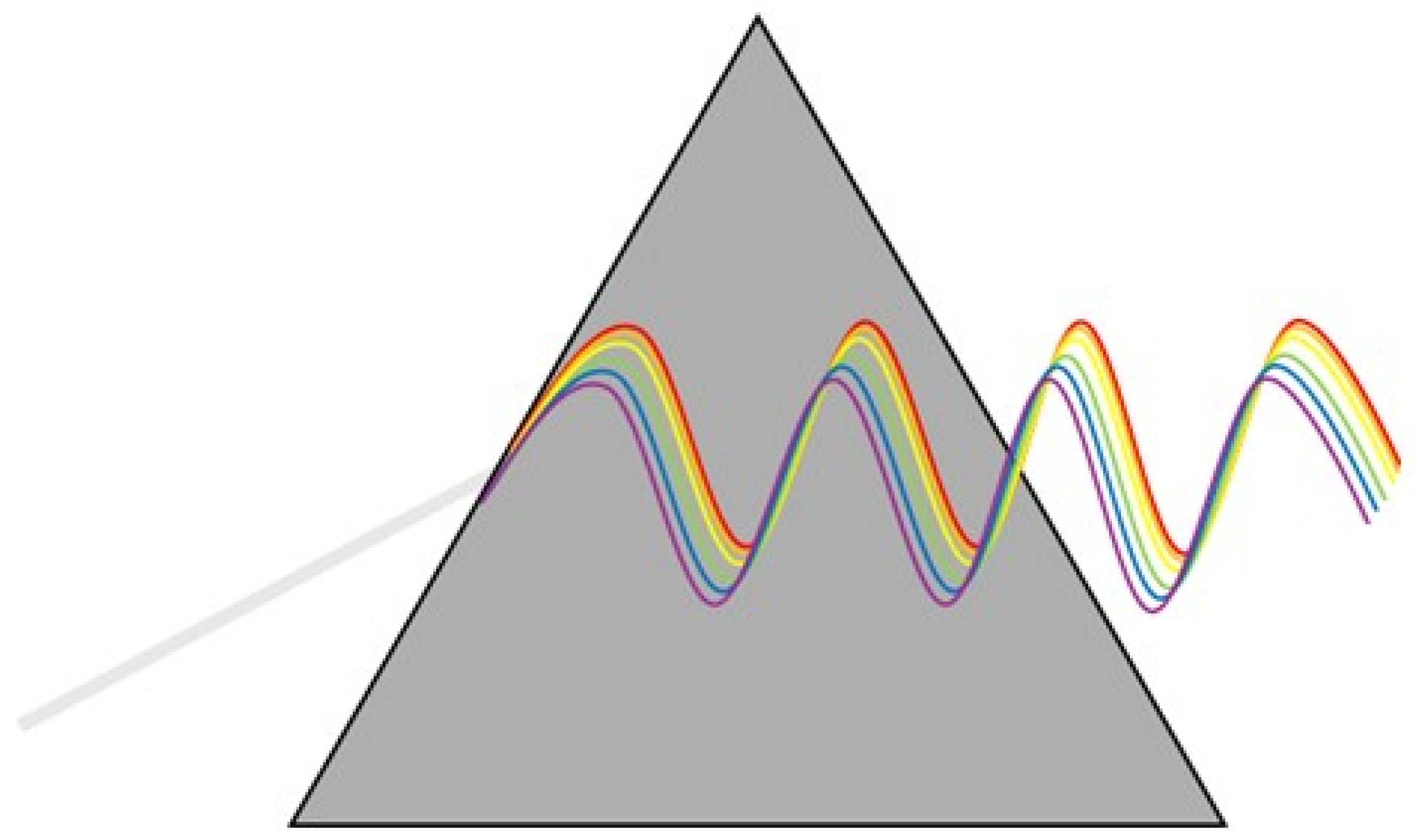
2.1. Spectroscopy
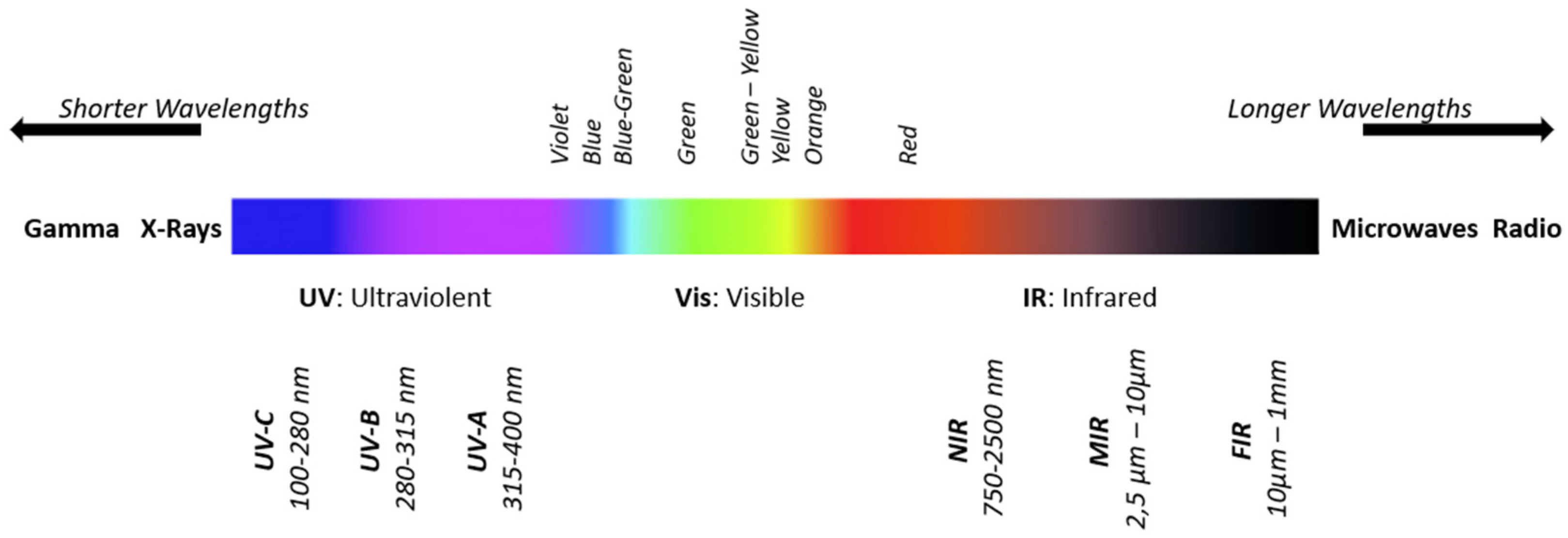
2.2. The Infrared Region of the Electromagnetic Spectrum
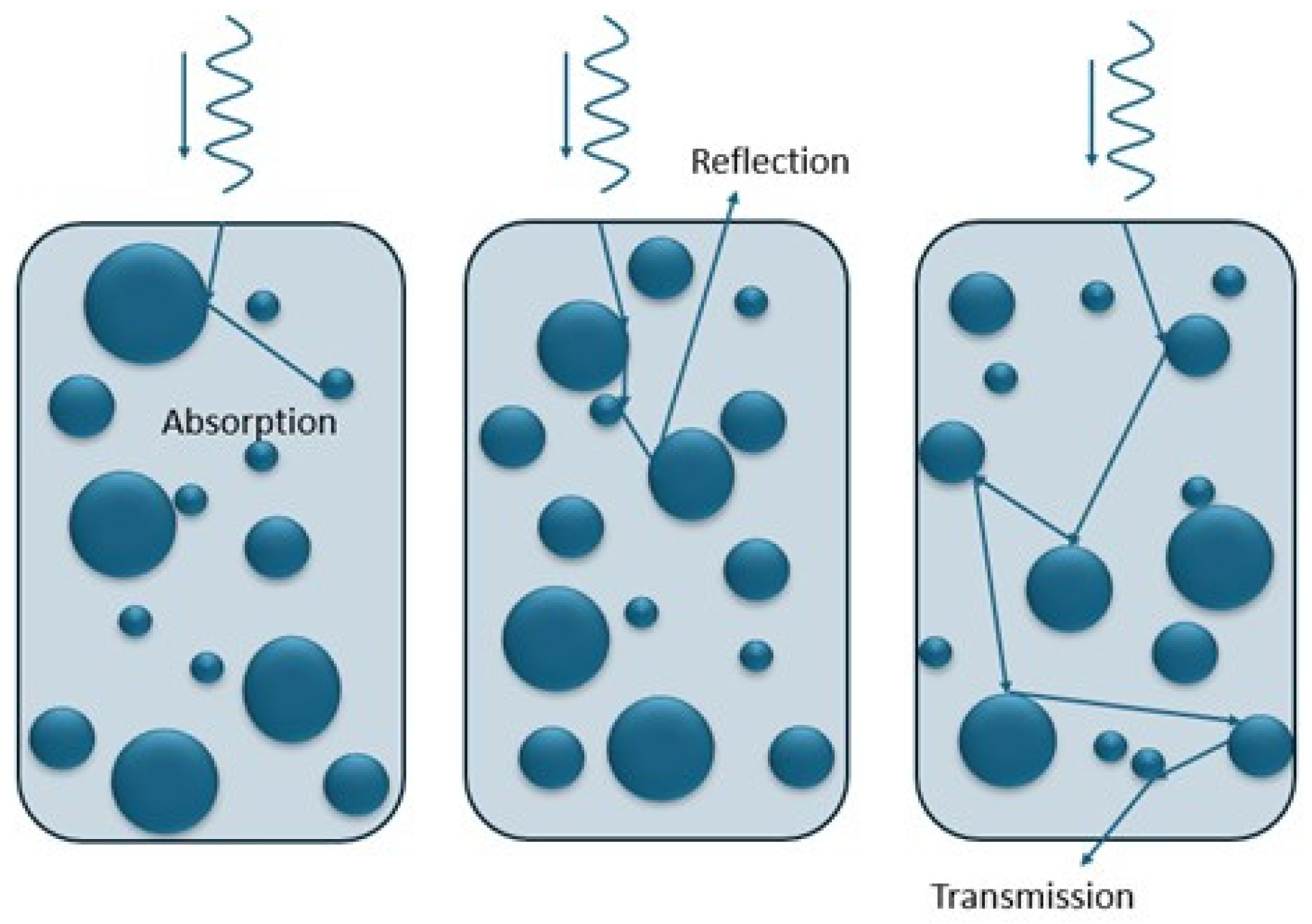
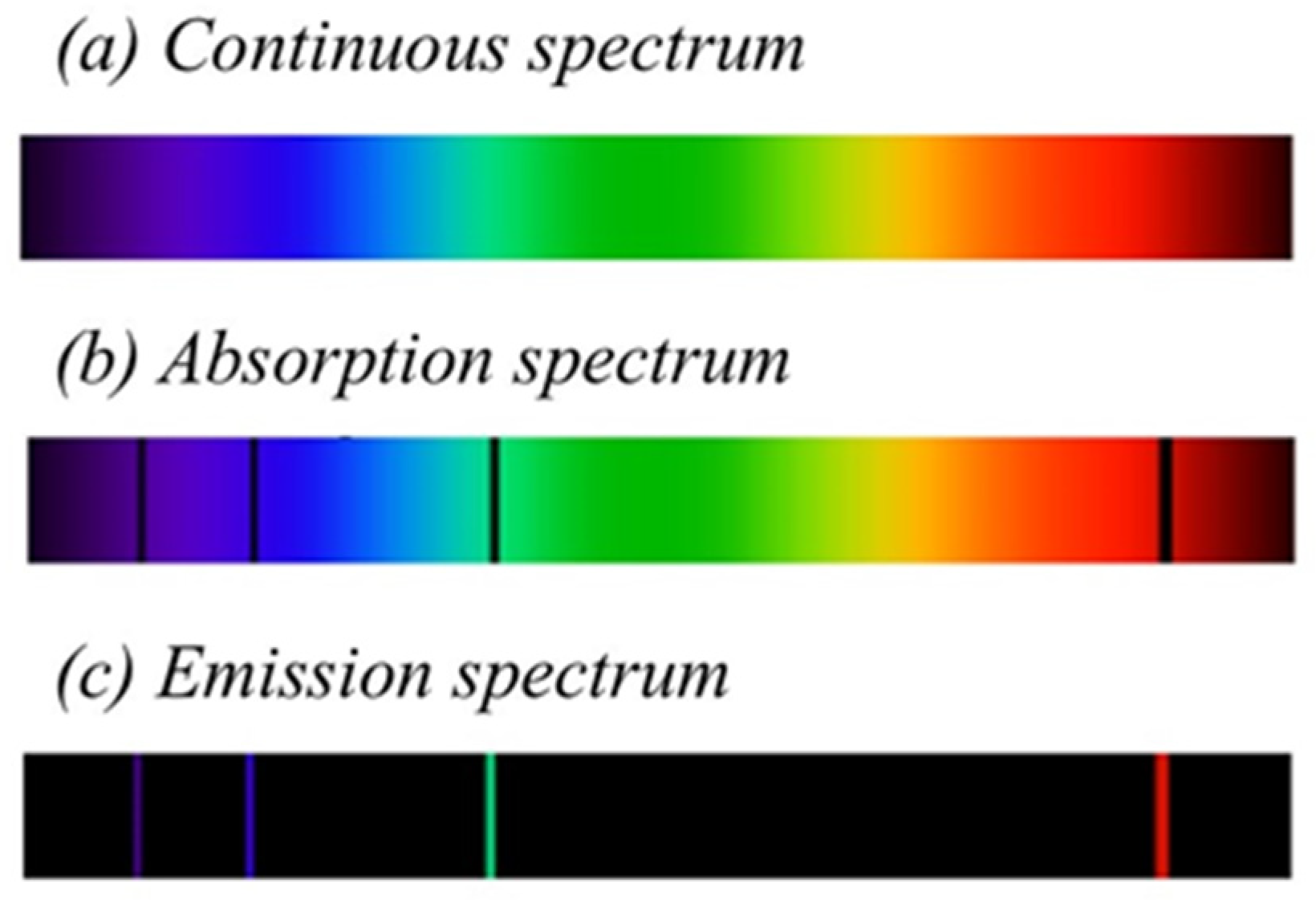
2.3. Transmittance, Reflectance, Absorption, and Emission of Light
2.4. Light Scattering
2.5. Other Optical Properties
2.6. Optical Chemosensors
3. Milk Composition and Quantification Techniques
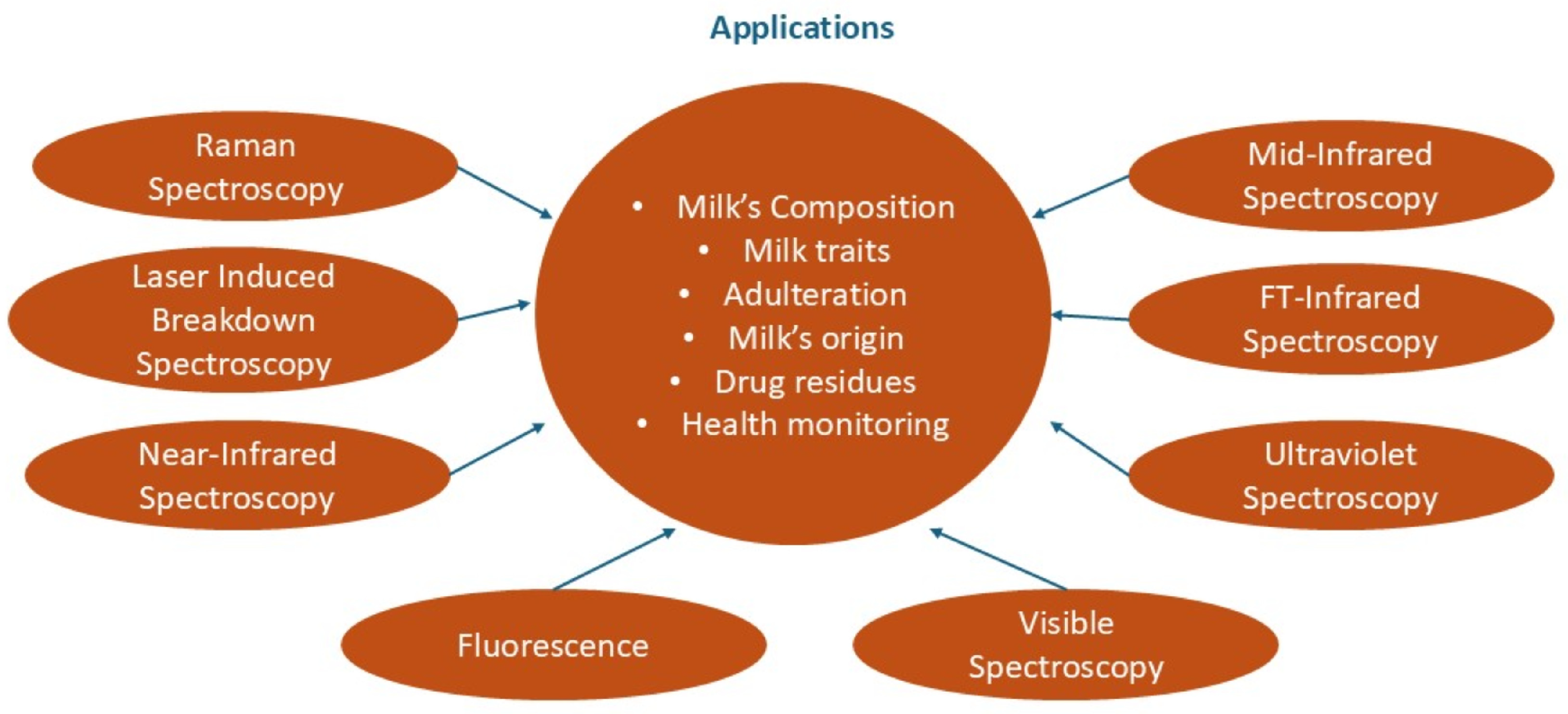
4. Spectroscopy Applications
4.1. Reflectance, Absorption, and Emission Spectroscopy
4.2. Raman Spectroscopy
4.3. Laser-Induced Breakdown Spectroscopy (LIBS)
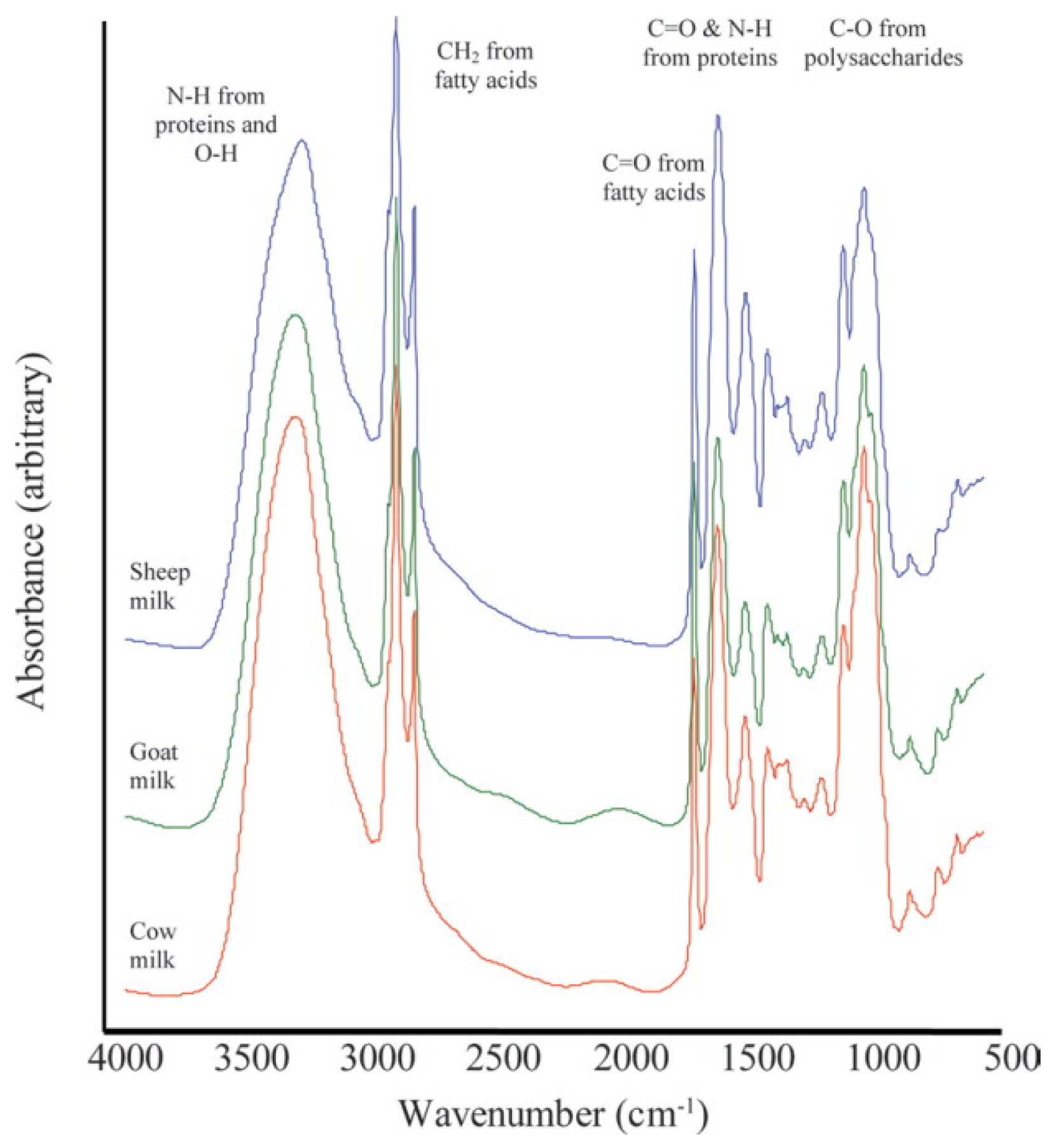
4.4. Infrared (IR) Spectroscopy
4.4.1. Near-Infrared Spectroscopy (NIRS)
Applications of Near-Infrared Spectroscopy in the Dairy Industry
- Off-line: NIRS systems are located in quality assurance/quality control (QA/QC) labs; samples are manually collected from the production line for testing;
- At-line: Samples are collected from the milk-processing line and tested using NIRS systems, which are positioned near the line;
- On-line: NIRS systems are located at the sampling point; a sample bypass is used to divert materials from the main process stream to be analyzed by the NIRS systems;
- In-line: The NIRS system is directly incorporated into the production line, utilizing various sampling techniques that allow real-time measurements.
Near-Infrared Spectroscopy Systems for Milk Analysis
Handheld and Portable Near-Infrared Spectroscopy Systems
4.4.2. Mid-Infrared Spectroscopy (MIRS)
4.5. Other Spectroscopy Methods
| Spectroscopy Method | Wavelength (nm) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE | Accuracy (%) | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FT-IR | 600–4000 (cm−1) | oven-dried | 63 | cow R, goat R, sheep R | mixed samples | composition | 0.92 0.93 0.96 | 6.40 *p 5.61 * p 3.98 * p | - | [35] | ||
| FT-IR | 400–4000 (cm−1) | liquid | 23 | cow R, goat R, sheep R | untreated samples | fat content animal of origin | - | - | 78.0 74.0 | [96] | ||
| Ultraviolent | 220–400 | liquid | 23 | cow R, goat R, sheep R | diluted samples | fat content animal of origin | - | - | 96.0 91.0 | [96] | ||
| Fluorescence | 240–500 exc 290–750 em | liquid | 23 | cow R, goat R, sheep R | untreated samples | fat content animal of origin | - | - | 70.0 91.0 | [96] | ||
| Fluorescence | 250–380 exc 280–640 em | liquid | 40 | cow | untreated samples | milk origin clss. | - | - | 76.9 † 70.4 †† | [102] | ||
| Fluorescence | 250–550 exc | liquid | 242 | cow | homogenized samples | carotenoid vitamins FAs | 0.01–0.54 0.03–0.17 0.01–0.50 | 0.01–0.17 μg/mL SEP 0.17 μg/mL–918.32 pg/mL SEP 0.15–13.76 g/100 g SEP | - | [78] | ||
| Fluorescence | 240–260 exc 320–440 exc | liquid | 12 | retail | spiked, diluted samples | melamine A | 0.97 ††† 0.95 ††† | PARAFAC: 68.6 ppm p U-PLS/RBL: 81.9 ppm p | - | [99] | ||
| Fluorescence | 330 exc 420 em | liquid | 23 | ND | skimmed, mixed, heated, homogenized samples | heat treatment discrimination | >0.95 | - | - | [103] | ||
| Fluorescence | 250–350 exc 260–500 em | liquid | 30 | cow | pasteurized samples | characterization of pasteurized milk | - | - | - | [104] | ||
| Visible | 400–1000 refl 400–1000 trans | liquid | 300 | cow | untreated samples | fat crude protein lactose urea | refl 0.978 0.861 0.557 - | trans 0.395 0.687 0.111 - | refl 0.11% p 0.18% p 0.22% p - | trans 0.629% p 0.274% p 0.317% p - | - | [61] |
| Visible light scatter | 400–1000 | liquid | 21 | retail | mixed, spiked, diluted samples | fat protein | 0.973 0.964 | 0.047% 0.032% | - | [100] | ||
| UV/Vis | 183–667 | liquid FR liquid HPH | 240 240 | cow | heated, homogenized, diluted or untreated samples | fat, protein, lactose, TSC | - | Liquid FR 0.13% p–0.46% p HPH FR 0.09% p–0.27% p | - | [101] | ||
| Fusion NIRS-LIBS | ≈185–2500 | powder | 50 | vetch root | pelleted samples | milk origin | - | - | 95.8 | [55] | ||
4.6. Benchmarking of Spectroscopy Methods
5. Machine Learning Principles
5.1. Logistic Regression (LR)
5.2. Decision Trees (DTs)
5.3. Random Forest (RF)
5.4. Support Vector Machine (SVM)
5.5. k-Nearest Neighbor (k-NN)
5.6. Naïve Bayes (NB)
5.7. Linear Regression
5.8. Linear Discriminant Analysis (LDA)
5.9. Boosting
Adaptive Boosting/Adaboost
5.10. Gradient Boosting Machine (GBM)
5.11. Neural Networks (NN)
5.12. Partial Least Square (PLS)
5.13. Partial Least Square Regression (PLSR)
6. Application of Machine Learning Methods in Milk Quality Assessment
6.1. Milk Quality and Composition Assessment
| ML | Tools | No and Type of Milk Samples | Application | R2 | RMSE | Acc | Se | Sp | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| NN | MIRS | 730 b | RCT k20 heat stability κ-CN | 0.50 0.36 0.45 0.42 | (1) 6.397 min (1) 2.770 min (1) 5.464 min (1) 1.095 g/L | - | - | - | [90] |
| MFFANN | NIRS | 385 b | blood metabolites | - | - | - | - | - | [150] |
| ANN | NIRS | 499 b | milk technological properties (CFp, CYcurd, Recprotein, etc.) | 0.45 to 0.71 | (2) 0.02% to 0.84 mm | - | - | - | [58] |
| FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.09 to 0.81 | 0.03 L/L to 80.59 U/L | - | - | - | [151] | |
| k-NN | sensors | 1059 ND | milk quality | - | - | 98.58% | - | - | [9] |
| PLS | FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.08 to 0.83 | 0.03 L/L to 106.37 U/L | - | - | - | [151] |
| FTIR | 471 b | κ-casein BCS BHB | (3) 0.90 tr 0.77 v (3) 0.95 tr 0.57 v (3) 0.88 tr 0.76 v | (1) 1.41 g/L (1) 0.35 (1) 0.10 | - | - | - | [148] | |
| PLS-DA | MIRS | 730 b | technological and protein properties of milk | - | - | 0.40–0.80 | 0.44 | - | [90] |
| MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.968 | 0.977 | 0.962 | [149] | |
| LDA | MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.968 | 0.980 | 0.961 | [149] |
| SVM | MIRS | 730 b | technological and protein properties of milk | - | - | 0.43–0.80 | 0.44 (overall) | 1.00 (overall) | [86] |
| MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.947 | 0.962 | 0.938 | [149] | |
| Boosting | MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.754 | 0.587 | 0.842 | [149] |
| Boosting DT | MIRS | 730 b | coagulation | - | - | - | 0.50 | 0.98 | [90] |
| MB-DA | MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.964 | 0.972 | 0.959 | [149] |
| GBM | NIRS | 499 b | milk technological properties (CFp, CYcurd, Recprotein, etc.) | 0.45 to 0.70 | (2) 0.02% to 0.87 mm | - | - | - | [58] |
| FTIR | 471 b | κ-casein BCS BHB | (4) 0.97 tr 0.81 v (4) 0.91 tr 0.63 v (4) 0.90 tr 0.77 v | (1) 1.08 (1) 0.25 (1) 0.09 | - | - | - | [148] | |
| FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.10 to 0.83 | 0.03 L/L to 75.69 U/L | - | - | - | [151] | |
| XGB | NIRS | 499 b | milk technological properties (CFp, CYcurd, Recprotein, etc.) | 0.43 to 0.63 | (2) 0.02% to 0.90 mm | - | - | - | [58] |
| FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.08 to 0.78 | 0.03 L/L to 80.23 U/L | - | - | - | [151] | |
| RF | MIRS | 730 b | αS1-CN, κ-CN | - | - | 0.48 0.45 | 0.44 | - | [90] |
| FTIR | 471 b | κ-casein BCS BHB | (3) 0.96 tr 0.80 v (3) 0.95 tr 0.61 v (3) 0.90 tr 0.79 v | (1) 1.18 (1) 0.26 (1) 0.10 | - | - | - | [148] | |
| MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.696 | 0.447 | 0.827 | [149] | |
| DRF | FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.09 to 0.79 | 0.03 L/L to 82.49 U/L | - | - | - | [151] |
| EN | NIRS | 499 b | milk technological properties (CFp, CYcurd, Recprotein, etc.) | 0.46 to 0.71 | (2) 0.02% to 0.78 mm | - | - | - | [58] |
| FTIR | 471 b | κ-casein BCS BHB | (3) 0.96 tr 0.79 v (3) 0.92 tr 0.59 v (3) 0.89 tr 0.78 v | (1) 1.25 (1) 0.27 (1) 0.10 | - | - | - | [148] | |
| MIRS | 4320 b | grass-fed/ non-grass-fed milk classification | - | - | 0.951 | 0.960 | 0.946 | [149] | |
| FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.12 to 0.87 | 0.03 L/L to 82.99 U/L | - | - | - | [151] | |
| LASSO | MIRS | 730 n | CMS, κ-CN | 0.08 0.42 | (1) 25.286 mm (1) 1.095 g/L | - | - | - | [90] |
| MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.959 | 0.970 | 0.953 | [149] | |
| PC-LR | MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.667 | 0.117 | 0.956 | [149] |
| RR | MIRS | 730 b | a30, β-CN, β-LG A | 0.37 0.35 0.19 | 12.495 mm 1.759 g/L 1.050 g/L | - | - | - | [90] |
| MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.880 | 0.779 | 0.933 | [149] | |
| Stacking Ensemble | NIRS | 385 b | blood metabolites | - | - | - | - | - | [149] |
| FTIR | 2701 b | blood metabolites (hematocrit, myeloperoxidase, globulins, etc.) | 0.13 to 0.87 | 0.03 L/L to 76.33 U/L | - | - | - | [151] | |
| VarSel-DA | MIRS | 4320 b | grass-fed/non-grass-fed milk classification | - | - | 0.890 | 0.845 | 0.913 | [149] |
| PLS + ANN | MIRS | 6619 b | LF in milk | 0.60 c 0.55 cv 0.60 v | 130.59 c mg/L 139.01 cv mg/L 162.17 v mg/L | - | - | - | [152] |
| PLSR | MIRS | 6619 b | LF in milk | 0.53 c 0.51 cv 0.61 v | 140.94 c mg/L 144.31 cv mg/L 163.76 v mg/L | - | - | - | [152] |
| PLS + SVM | MIRS | 6619 b | LF in milk | 0.53 c 0.53 cv 0.63 v | 144.32 c mg/L 144.60 cv mg/L 174.92 v mg/L | - | - | - | [152] |
| PLS + Polynomial SVM | MIRS | 6619 b | LF in milk | 0.64 c 0.56 cv 0.62 v | 125.89 c mg/L 138.40 cv mg/L 166.75 v mg/L | - | - | - | [152] |
6.2. Fraud Detection and Adulteration Identification
| ML | Tools | No and Type of Milk Samples | Application | R2 | RMSE | Se (%) | Sp (%) | Accuracy | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| NN | LIBS | 22 b, c, o | melamine in toddler milk powder | 0.999 | - | - | - | Acc: 100% | [50] |
| UV, Vis, IR | ND | adulterants in milk | - | - | - | - | Acc: 100% | [155] | |
| CNN | LIBS | 25 r | protein adulteration in milk powder | - | - | - | - | Acc: 97.8% | [57] |
| PLS-DA | NIRS | 600 b, c | fraud in goat milk: water urea bovine whey milk authentic | - | - | 100 in all cases | 100 in all cases | - | [153] |
| PLSR | Fluorescence | 40 b | adulteration in milk | 0.99 | (1) 1.16 (2) 6.24 | - | - | - | [154] |
| NB | UV, Vis, IR | ND | adulterants in milk | - | - | - | - | 90% | [155] |
| DT | UV, Vis, IR | ND | adulterants in milk | - | - | - | - | 91.7% | [155] |
| LDA | UV, Vis, IR | ND | adulterants in milk | - | - | - | - | 88.1% | [155] |
| FTIR | ND | heat treatment to milk | - | - | - | - | 0.84 | [157] | |
| RF | FTIR | ND | heat treatment to milk | - | - | - | - | 0.92 | [157] |
| LIBS | 25 r | protein adulteration in milk powder | - | - | 0.886 (train) 0.871 (test) | [57] | |||
| k-NN | NIRS | 600 b, c | fraud in goat milk: water urea bovine whey milk authentic | - | - | 76.0 80.0 96.0 80.0 99.0 | 96.6 95.4 100 100 88.0 | - | [153] |
| FTIR | ND | heat treatment to milk | - | - | - | - | 0.86 | [157] | |
| LIBS | 25 r | protein adulteration in milk powder | - | - | - | - | 0.884 (train) 0.867 (test) | [57] | |
| SVM | UV, Vis, IR | ND | adulterants in milk | - | - | - | - | 90% | [155] |
| LIBS | 25 r | protein adulteration in milk powder | - | - | - | - | 0.961 (train) 0.938 (test) | [57] | |
| FTIR | ND | heat treatment to milk | - | - | - | - | 0.90 | [157] | |
| CART | FTIR | 520 b | fraud of cheese whey to milk | - | - | - | - | 96.2% (train), 97.2% (test) | [156] |
| MLP | FTIR | 520 b | fraud of cheese whey to milk | - | - | - | - | 97.8% | [156] |
6.3. Milk Source and Origin Classification
7. Future Research
8. Conclusions
Author Contributions
Funding

Acknowledgments
Conflicts of Interest
References
- McLeod, A. World Livestock 2011: Livestock in Food Security; FAO: Rome, Italy, 2011; ISBN 978-92-5-107013-0. [Google Scholar]
- UN (United Nations) Department of Economic and Social Affairs, Population Division. World Population Prospects. 2019. Available online: https://population.un.org/wpp/publications/files/wpp2019_highlights.pdf (accessed on 29 April 2024).
- Neethirajan, S.; Kemp, B. Digital Livestock Farming. Sens. Bio-Sens. Res. 2021, 32, 100408. [Google Scholar] [CrossRef]
- Halachmi, I.; Guarino, M.; Bewley, J.; Pastell, M. Smart Animal Agriculture: Application of Real-Time Sensors to Improve Animal Well-Being and Production. Annu. Rev. Anim. Biosci. 2019, 7, 403–425. [Google Scholar] [CrossRef] [PubMed]
- Ochs, D.S.; Wolf, C.A.; Widmar, N.J.O.; Bir, C. Consumer Perceptions of Egg-Laying Hen Housing Systems. Poult. Sci. 2018, 97, 3390–3396. [Google Scholar] [CrossRef]
- Anestis, V.; Bartzanas, T.; Kittas, C. Life Cycle Inventory Analyis for the Milk Produced in a Greek Commercial Dairy Farm—The Link to Precision Livestock Farming. ResearchGate. Available online: https://www.researchgate.net/publication/310005326_Life_cycle_inventory_analysis_for_the_milk_produced_by_a_Greek_commercial_dairy_farm (accessed on 15 September 2024).
- Pereira, P.C. Milk Nutritional Composition and Its Role in Human Health. Nutrition 2014, 30, 619–627. [Google Scholar] [CrossRef]
- Evangelista, C.; Basiricò, L.; Bernabucci, U. An Overview on the Use of Near Infrared Spectroscopy (NIRS) on Farms for the Management of Dairy Cows. Agriculture 2021, 11, 296. [Google Scholar] [CrossRef]
- Samad, A.; Taze, S.; Kürsad Uçar, M. Enhancing Milk Quality Detection with Machine Learning: A Comparative Analysis of KNN and Distance-Weighted KNN Algorithms. Int. J. Innov. Sci. Res. Technol. (IJISRT) 2024, 9, 2021–2029. [Google Scholar] [CrossRef]
- Tullo, E.; Finzi, A.; Guarino, M. Review: Environmental Impact of Livestock Farming and Precision Livestock Farming as a Mitigation Strategy. Sci. Total Environ. 2019, 650, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- Helwatkar, A.; Riordan, D.; Walsh, J. Sensor Technology for Animal Health Monitoring. Int. J. Smart Sens. Intell. Syst. 2014, 7, 1–6. [Google Scholar] [CrossRef]
- Neethirajan, S. Recent Advances in Wearable Sensors for Animal Health Management. Sens. Bio-Sens. Res. 2017, 12, 15–29. [Google Scholar] [CrossRef]
- Spectroscopy | Definition, Types, & Facts | Britannica. Available online: https://www.britannica.com/science/spectroscopy (accessed on 12 September 2024).
- Pu, Y.-Y.; O’Donnell, C.; Tobin, J.T.; O’Shea, N. Review of Near-Infrared Spectroscopy as a Process Analytical Technology for Real-Time Product Monitoring in Dairy Processing. Int. Dairy J. 2020, 103, 104623. [Google Scholar] [CrossRef]
- Agelet, L.E.; Hurburgh, C.R. A Tutorial on Near Infrared Spectroscopy and Its Calibration. Crit. Rev. Anal. Chem. 2010, 40, 246–260. [Google Scholar] [CrossRef]
- Herschel, W. Investigation of the Powers of the Prismatic Colours to Heat and Illuminate Objects; With Remarks, That Prove the Different Refrangibility of Radiant Heat. To Which Is Added, an Inquiry into the Method of Viewing the Sun Advantageously, with Telescopes of Large Apertures and High Magnifying Powers. Philos. Trans. R. Soc. Lond. 1800, 90, 255–283. [Google Scholar]
- Gastélum-Barrios, A.; Soto-Zarazúa, G.M.; Escamilla-García, A.; Toledano-Ayala, M.; Macías-Bobadilla, G.; Jauregui-Vazquez, D. Optical Methods Based on Ultraviolet, Visible, and Near-Infrared Spectra to Estimate Fat and Protein in Raw Milk: A Review. Sensors 2020, 20, 3356. [Google Scholar] [CrossRef]
- Dispersion (Optics). Wikipedia. 2024. Available online: https://en.wikipedia.org/wiki/Dispersion_(optics) (accessed on 9 December 2024).
- Ma, W.; Ji, X.; Ding, L.; Yang, S.X.; Guo, K.; Li, Q. Automatic Monitoring Methods for Greenhouse and Hazardous Gases Emitted from Ruminant Production Systems: A Review. Sensors 2024, 24, 4423. [Google Scholar] [CrossRef] [PubMed]
- Fazio, E.; Spadaro, S.; Corsaro, C.; Neri, G.; Leonardi, S.G.; Neri, F.; Lavanya, N.; Sekar, C.; Donato, N.; Neri, G. Metal-Oxide Based Nanomaterials: Synthesis, Characterization and Their Applications in Electrical and Electrochemical Sensors. Sensors 2021, 21, 2494. [Google Scholar] [CrossRef] [PubMed]
- Hulanicki’, A.; Geab, S.; Ingman, F.O.L.K.E. Chemical sensors: Definitions and classification. Pure Appl. Chem. 1991, 63, 1247–1250. [Google Scholar] [CrossRef]
- Kunes, R.; Bartos, P.; Iwasaka, G.K.; Lang, A.; Hankovec, T.; Smutny, L.; Cerny, P.; Poborska, A.; Smetana, P.; Kriz, P.; et al. In-Line Technologies for the Analysis of Important Milk Parameters during the Milking Process: A Review. Agriculture 2021, 11, 239. [Google Scholar] [CrossRef]
- Vaskova, H.; Buckova, M. Measuring the Lactose Content in Milk. MATEC Web Conf. 2016, 76, 05011. [Google Scholar] [CrossRef]
- He, H.; Sun, D.-W.; Pu, H.; Chen, L.; Lin, L. Applications of Raman Spectroscopic Techniques for Quality and Safety Evaluation of Milk: A Review of Recent Developments. Crit. Rev. Food Sci. Nutr. 2019, 59, 770–793. [Google Scholar] [CrossRef]
- Pellegrino, L.; Cattaneo, S.; De Noni, I. Nutrition and Health: Effects of Processing on Protein Quality of Milk and Milk Products. In Encyclopedia of Dairy Sciences, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1067–1074. ISBN 978-0-08-100596-5. [Google Scholar]
- Reference Material for Somatic Cell Counting—European Commission. Available online: https://joint-research-centre.ec.europa.eu/jrc-news-and-updates/reference-material-somatic-cell-counting-2020-02-11_en (accessed on 1 October 2024).
- Gelasakis, A.I.; Mavrogianni, V.S.; Petridis, I.G.; Vasileiou, N.G.C.; Fthenakis, G.C. Mastitis in Sheep—The Last 10 Years and the Future of Research. Vet. Microbiol. 2015, 181, 136–146. [Google Scholar] [CrossRef]
- Melfsen, A.; Hartung, E.; Haeussermann, A. Accuracy of In-Line Milk Composition Analysis with Diffuse Reflectance near-Infrared Spectroscopy. J. Dairy Sci. 2012, 95, 6465–6476. [Google Scholar] [CrossRef] [PubMed]
- Numthuam, S.; Hongpathong, J.; Charoensook, R.; Rungchang, S. Method Development for the Analysis of Total Bacterial Count in Raw Milk Using Near-infrared Spectroscopy. J. Food Saf. 2017, 37, e12335. [Google Scholar] [CrossRef]
- Nicolaou, N.; Goodacre, R. Rapid and Quantitative Detection of the Microbial Spoilage in Milk Using Fourier Transform Infrared Spectroscopy and Chemometrics. Analyst 2008, 133, 1424. [Google Scholar] [CrossRef] [PubMed]
- Pampoukis, G.; Lytou, A.E.; Argyri, A.A.; Panagou, E.Z.; Nychas, G.-J.E. Recent Advances and Applications of Rapid Microbial Assessment from a Food Safety Perspective. Sensors 2022, 22, 2800. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.; Villarroya, I. NIR Spectroscopy: A Rapid-Response Analytical Tool. TrAC Trends Anal. Chem. 2002, 21, 240–250. [Google Scholar] [CrossRef]
- Wikimedia Commons. Available online: https://commons.wikimedia.org/wiki/File:Spectral_lines_en.PNG?uselang=en-gb (accessed on 30 August 2024).
- Chen, H.; Tan, C.; Lin, Z.; Wu, T. Classification of Different Liquid Milk by Near-Infrared Spectroscopy and Ensemble Modeling. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 251, 119460. [Google Scholar] [CrossRef]
- Nicolaou, N.; Xu, Y.; Goodacre, R. Fourier Transform Infrared Spectroscopy and Multivariate Analysis for the Detection and Quantification of Different Milk Species. J. Dairy Sci. 2010, 93, 5651–5660. [Google Scholar] [CrossRef]
- Nanou, E.; Stefas, D.; Couris, S. Milk’s Inorganic Content Analysis via Laser Induced Breakdown Spectroscopy. Food Chem. 2023, 407, 135169. [Google Scholar] [CrossRef] [PubMed]
- Raman, C.V.; Krishnan, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Mazurek, S.; Szostak, R.; Czaja, T.; Zachwieja, A. Analysis of Milk by FT-Raman Spectroscopy. Talanta 2015, 138, 285–289. [Google Scholar] [CrossRef] [PubMed]
- El-Abassy, R.M.; Eravuchira, P.J.; Donfack, P.; Von Der Kammer, B.; Materny, A. Fast Determination of Milk Fat Content Using Raman Spectroscopy. Vib. Spectrosc. 2011, 56, 3–8. [Google Scholar] [CrossRef]
- Rodrigues Júnior, P.H.; De Sá Oliveira, K.; Almeida, C.E.R.D.; De Oliveira, L.F.C.; Stephani, R.; Pinto, M.D.S.; Carvalho, A.F.D.; Perrone, Í.T. FT-Raman and Chemometric Tools for Rapid Determination of Quality Parameters in Milk Powder: Classification of Samples for the Presence of Lactose and Fraud Detection by Addition of Maltodextrin. Food Chem. 2016, 196, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.M.; Krishna, H.; Majumder, S.K.; Gupta, P.K. Detection of Urea Adulteration in Milk Using Near-Infrared Raman Spectroscopy. Food Anal. Methods 2015, 8, 93–102. [Google Scholar] [CrossRef]
- McGoverin, C.M.; Clark, A.S.S.; Holroyd, S.E.; Gordon, K.C. Raman Spectroscopic Quantification of Milk Powder Constituents. Anal. Chim. Acta 2010, 673, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Amjad, A.; Ullah, R.; Khan, S.; Bilal, M.; Khan, A. Raman Spectroscopy Based Analysis of Milk Using Random Forest Classification. Vib. Spectrosc. 2018, 99, 124–129. [Google Scholar] [CrossRef]
- Noll, R. Laser-Induced Breakdown Spectroscopy: Fundamentals and Applications; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2012; ISBN 978-3-642-20667-2. [Google Scholar]
- What Is LIBS? | SciAps. Available online: https://www.sciaps.com/products/libs/what-is-libs (accessed on 12 September 2024).
- Musazzi, S.; Perini, U. Laser-Induced Breakdown Spectroscopy: Theory and Applications; Springer: Berlin, Germany, 2014; ISBN 978-3-642-45085-3. [Google Scholar]
- Dos Santos Augusto, A.; Barsanelli, P.L.; Pereira, F.M.V.; Pereira-Filho, E.R. Calibration Strategies for the Direct Determination of Ca, K, and Mg in Commercial Samples of Powdered Milk and Solid Dietary Supplements Using Laser-Induced Breakdown Spectroscopy (LIBS). Food Res. Int. 2017, 94, 72–78. [Google Scholar] [CrossRef]
- Markiewicz-Keszycka, M.; Cama-Moncunill, X.; Casado-Gavalda, M.P.; Dixit, Y.; Cama-Moncunill, R.; Cullen, P.J.; Sullivan, C. Laser-Induced Breakdown Spectroscopy (LIBS) for Food Analysis: A Review. Trends Food Sci. Technol. 2017, 65, 80–93. [Google Scholar] [CrossRef]
- Nanou, E.; Pliatsika, N.; Stefas, D.; Couris, S. Identification of the Animal Origin of Milk via Laser-Induced Breakdown Spectroscopy. Food Control 2023, 154, 110007. [Google Scholar] [CrossRef]
- Moncayo, S.; Manzoor, S.; Rosales, J.D.; Anzano, J.; Caceres, J.O. Qualitative and Quantitative Analysis of Milk for the Detection of Adulteration by Laser Induced Breakdown Spectroscopy (LIBS). Food Chem. 2017, 232, 322–328. [Google Scholar] [CrossRef]
- Bilge, G.; Sezer, B.; Eseller, K.E.; Berberoglu, H.; Topcu, A.; Boyaci, I.H. Determination of Whey Adulteration in Milk Powder by Using Laser Induced Breakdown Spectroscopy. Food Chem. 2016, 212, 183–188. [Google Scholar] [CrossRef]
- Abdel-Salam, Z.; Al Sharnoubi, J.; Harith, M.A. Qualitative Evaluation of Maternal Milk and Commercial Infant Formulas via LIBS. Talanta 2013, 115, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Salam, Z.; El-Saeid, R.; Abdelghany, S.; Abdel-Salam, S.; Radwan, M. Assessment of Milk Quality at Farm Level Using Laser Techniques. Egypt. J. Chem. 2022, 66, 273–278. [Google Scholar] [CrossRef]
- Cama-Moncunill, X.; Markiewicz-Keszycka, M.; Dixit, Y.; Cama-Moncunill, R.; Casado-Gavalda, M.P.; Cullen, P.J.; Sullivan, C. Feasibility of Laser-Induced Breakdown Spectroscopy (LIBS) as an at-Line Validation Tool for Calcium Determination in Infant Formula. Food Control 2017, 78, 304–310. [Google Scholar] [CrossRef]
- Eum, C.; Jang, D.; Kim, J.; Choi, S.; Cha, K.; Chung, H. Improving the Accuracy of Spectroscopic Identification of Geographical Origins of Agricultural Samples through Cooperative Combination of Near-Infrared and Laser-Induced Breakdown Spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2018, 149, 281–287. [Google Scholar] [CrossRef]
- Sezer, B.; Durna, S.; Bilge, G.; Berkkan, A.; Yetisemiyen, A.; Boyaci, I.H. Identification of Milk Fraud Using Laser-Induced Breakdown Spectroscopy (LIBS). Int. Dairy J. 2018, 81, 1–7. [Google Scholar] [CrossRef]
- Huang, W.; Guo, L.; Kou, W.; Zhang, D.; Hu, Z.; Chen, F.; Chu, Y.; Cheng, W. Identification of Adulterated Milk Powder Based on Convolutional Neural Network and Laser-Induced Breakdown Spectroscopy. Microchem. J. 2022, 176, 107190. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Giannuzzi, D.; Bisutti, V.; Pegolo, S.; Trevisi, E.; Schiavon, S.; Gallo, L.; Fineboym, D.; Katz, G.; Cecchinato, A. Real-Time Milk Analysis Integrated with Stacking Ensemble Learning as a Tool for the Daily Prediction of Cheese-Making Traits in Holstein Cattle. J. Dairy Sci. 2022, 105, 4237–4255. [Google Scholar] [CrossRef] [PubMed]
- Brandão, M.C.M.P.; Carmo, A.P.; Bell, M.J.V.; Anjos, V.C. Characterization of milk by infrared spectroscopy. Rev. Do Inst. De Laticínios Cândido Tostes 2010, 65, 30–33. [Google Scholar]
- Aernouts, B.; Van Beers, R.; Watté, R.; Huybrechts, T.; Lammertyn, J.; Saeys, W. Visible and Near-Infrared Bulk Optical Properties of Raw Milk. J. Dairy Sci. 2015, 98, 6727–6738. [Google Scholar] [CrossRef] [PubMed]
- Aernouts, B.; Polshin, E.; Lammertyn, J.; Saeys, W. Visible and Near-Infrared Spectroscopic Analysis of Raw Milk for Cow Health Monitoring: Reflectance or Transmittance? J. Dairy Sci. 2011, 94, 5315–5329. [Google Scholar] [CrossRef]
- Korelidou, V.; Simitzis, P.; Massouras, T.; Gelasakis, A.I. Infrared Thermography as a Diagnostic Tool for the Assessment of Mastitis in Dairy Ruminants. Animals 2024, 14, 2691. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, D.F. The beer-lambert law. J. Chem. Educ. 1962, 39, 333. [Google Scholar] [CrossRef]
- Givens, D.I.; De Boever, J.L.; Deaville, E.R. The Principles, Practices and Some Future Applications of near Infrared Spectroscopy for Predicting the Nutritive Value of Foods for Animals and Humans. Nutr. Res. Rev. 1997, 10, 83–114. [Google Scholar] [CrossRef] [PubMed]
- Yakubu, H.G.; Kovacs, Z.; Toth, T.; Bazar, G. The Recent Advances of Near-Infrared Spectroscopy in Dairy Production—A Review. Crit. Rev. Food Sci. Nutr. 2022, 62, 810–831. [Google Scholar] [CrossRef] [PubMed]
- 14:00-17:00 ISO 21543:2006; Milk Products—Guidelines for the Application of Near Infrared Spectrometry. ISO: Geneva, Switzerland, 2006. Available online: https://www.iso.org/standard/40318.html (accessed on 12 September 2024).
- 14:00-17:00 ISO 21543:2020; Milk and Milk Products—Guidelines for the Application of Near Infrared Spectrometry. ISO: Geneva, Switzerland, 2020. Available online: https://www.iso.org/standard/77606.html (accessed on 13 September 2024).
- Albanell, E.; Caja, G.; Such, X.; Rovai, M.; Salama, A.A.K.; Casals, R. Determination of Fat, Protein, Casein, Total Solids, and Somatic Cell Count in Goat’s Milk by Near-Infrared Reflectance Spectroscopy. J. AOAC Int. 2003, 86, 746–752. [Google Scholar] [CrossRef]
- Revilla, I.; Escuredo, O.; González-Martín, M.I.; Palacios, C. Fatty Acids and Fat-Soluble Vitamins in Ewe’s Milk Predicted by near Infrared Reflectance Spectroscopy. Determination of Seasonality. Food Chem. 2017, 214, 468–477. [Google Scholar] [CrossRef]
- Holroyd, S.E. The Use of near Infrared Spectroscopy on Milk and Milk Products. J. Near Infrared Spectrosc. 2013, 21, 311–322. [Google Scholar] [CrossRef]
- Coppa, M.; Martin, B.; Agabriel, C.; Chassaing, C.; Sibra, C.; Constant, I.; Graulet, B.; Andueza, D. Authentication of Cow Feeding and Geographic Origin on Milk Using Visible and Near-Infrared Spectroscopy. J. Dairy Sci. 2012, 95, 5544–5551. [Google Scholar] [CrossRef]
- Al-Qadiri, H.M.; Lin, M.; Al-Holy, M.A.; Cavinato, A.G.; Rasco, B.A. Monitoring Quality Loss of Pasteurized Skim Milk Using Visible and Short Wavelength Near-Infrared Spectroscopy and Multivariate Analysis. J. Dairy Sci. 2008, 91, 950–958. [Google Scholar] [CrossRef]
- Cattaneo, T.M.P.; Cabassi, G.; Profaizer, M.; Giangiacomo, R. Contribution of Light Scattering to near Infrared Absorption in Milk. J. Near Infrared Spectrosc. 2009, 17, 337–343. [Google Scholar] [CrossRef]
- Tsenkova, R.; Meilina, H.; Kuroki, S.; Burns, D.H. Near Infrared Spectroscopy Using Short Wavelengths and Leave-One-Cow-Out Cross-Validation for Quantification of Somatic Cells in Milk. J. Near Infrared Spectrosc. 2009, 17, 345–351. [Google Scholar] [CrossRef]
- Coppa, M.; Ferlay, A.; Leroux, C.; Jestin, M.; Chilliard, Y.; Martin, B.; Andueza, D. Prediction of Milk Fatty Acid Composition by near Infrared Reflectance Spectroscopy. Int. Dairy J. 2010, 20, 182–189. [Google Scholar] [CrossRef]
- Núñez-Sánchez, N.; Martínez-Marín, A.L.; Polvillo, O.; Fernández-Cabanás, V.M.; Carrizosa, J.; Urrutia, B.; Serradilla, J.M. Near Infrared Spectroscopy (NIRS) for the Determination of the Milk Fat Fatty Acid Profile of Goats. Food Chem. 2016, 190, 244–252. [Google Scholar] [CrossRef]
- Wu, D.; He, Y.; Feng, S.; Sun, D.-W. Study on Infrared Spectroscopy Technique for Fast Measurement of Protein Content in Milk Powder Based on LS-SVM. J. Food Eng. 2008, 84, 124–131. [Google Scholar] [CrossRef]
- Soulat, J.; Andueza, D.; Graulet, B.; Girard, C.L.; Labonne, C.; Aït-Kaddour, A.; Martin, B.; Ferlay, A. Comparison of the Potential Abilities of Three Spectroscopy Methods: Near-Infrared, Mid-Infrared, and Molecular Fluorescence, to Predict Carotenoid, Vitamin and Fatty Acid Contents in Cow Milk. Foods 2020, 9, 592. [Google Scholar] [CrossRef] [PubMed]
- Coppa, M.; Revello-Chion, A.; Giaccone, D.; Ferlay, A.; Tabacco, E.; Borreani, G. Comparison of near and Medium Infrared Spectroscopy to Predict Fatty Acid Composition on Fresh and Thawed Milk. Food Chem. 2014, 150, 49–57. [Google Scholar] [CrossRef]
- Balabin, R.M.; Smirnov, S.V. Melamine Detection by Mid- and near-Infrared (MIR/NIR) Spectroscopy: A Quick and Sensitive Method for Dairy Products Analysis Including Liquid Milk, Infant Formula, and Milk Powder. Talanta 2011, 85, 562–568. [Google Scholar] [CrossRef]
- Henn, R.; Kirchler, C.G.; Grossgut, M.-E.; Huck, C.W. Comparison of Sensitivity to Artificial Spectral Errors and Multivariate LOD in NIR Spectroscopy—Determining the Performance of Miniaturizations on Melamine in Milk Powder. Talanta 2017, 166, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Llano Suárez, P.; Soldado, A.; González-Arrojo, A.; Vicente, F.; De La Roza-Delgado, B. Rapid On-Site Monitoring of Fatty Acid Profile in Raw Milk Using a Handheld near Infrared Sensor. J. Food Compos. Anal. 2018, 70, 1–8. [Google Scholar] [CrossRef]
- Liu, N.; Parra, H.A.; Pustjens, A.; Hettinga, K.; Mongondry, P.; Van Ruth, S.M. Evaluation of Portable Near-Infrared Spectroscopy for Organic Milk Authentication. Talanta 2018, 184, 128–135. [Google Scholar] [CrossRef]
- De La Roza-Delgado, B.; Garrido-Varo, A.; Soldado, A.; González Arrojo, A.; Cuevas Valdés, M.; Maroto, F.; Pérez-Marín, D. Matching Portable NIRS Instruments for in Situ Monitoring Indicators of Milk Composition. Food Control 2017, 76, 74–81. [Google Scholar] [CrossRef]
- Diaz-Olivares, J.A.; Adriaens, I.; Stevens, E.; Saeys, W.; Aernouts, B. Online Milk Composition Analysis with an On-Farm near-Infrared Sensor. Comput. Electron. Agric. 2020, 178, 105734. [Google Scholar] [CrossRef]
- Kalinin, A.; Krasheninnikov, V.; Sadovskiy, S.; Yurova, E. Determining the Composition of Proteins in Milk Using a Portable near Infrared Spectrometer. J. Near Infrared Spectrosc. 2013, 21, 409–415. [Google Scholar] [CrossRef]
- Santos, P.M.; Pereira-Filho, E.R.; Rodriguez-Saona, L.E. Rapid Detection and Quantification of Milk Adulteration Using Infrared Microspectroscopy and Chemometrics Analysis. Food Chem. 2013, 138, 19–24. [Google Scholar] [CrossRef]
- Etzion, Y.; Linker, R.; Cogan, U.; Shmulevich, I. Determination of Protein Concentration in Raw Milk by Mid-Infrared Fourier Transform Infrared/Attenuated Total Reflectance Spectroscopy. J. Dairy Sci. 2004, 87, 2779–2788. [Google Scholar] [CrossRef]
- Dabrowska, A.; David, M.; Freitag, S.; Andrews, A.M.; Strasser, G.; Hinkov, B.; Schwaighofer, A.; Lendl, B. Broadband Laser-Based Mid-Infrared Spectroscopy Employing a Quantum Cascade Detector for Milk Protein Analysis. Sens. Actuators B Chem. 2022, 350, 130873. [Google Scholar] [CrossRef]
- Frizzarin, M.; Gormley, I.C.; Berry, D.P.; Murphy, T.B.; Casa, A.; Lynch, A.; McParland, S. Predicting Cow Milk Quality Traits from Routinely Available Milk Spectra Using Statistical Machine Learning Methods. J. Dairy Sci. 2021, 104, 7438–7447. [Google Scholar] [CrossRef]
- De Marchi, M.; Fagan, C.C.; O’Donnell, C.P.; Cecchinato, A.; Dal Zotto, R.; Cassandro, M.; Penasa, M.; Bittante, G. Prediction of Coagulation Properties, Titratable Acidity, and pH of Bovine Milk Using Mid-Infrared Spectroscopy. J. Dairy Sci. 2009, 92, 423–432. [Google Scholar] [CrossRef]
- De Marchi, M.; Toffanin, V.; Cassandro, M.; Penasa, M. Invited Review: Mid-Infrared Spectroscopy as Phenotyping Tool for Milk Traits. J. Dairy Sci. 2014, 97, 1171–1186. [Google Scholar] [CrossRef]
- Ceniti, C.; Spina, A.A.; Piras, C.; Oppedisano, F.; Tilocca, B.; Roncada, P.; Britti, D.; Morittu, V.M. Recent Advances in the Determination of Milk Adulterants and Contaminants by Mid-Infrared Spectroscopy. Foods 2023, 12, 2917. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.F.; Uniacke-Lowe, T.; McSweeney, P.L.H.; O’Mahony, J.A. Dairy Chemistry and Biochemistry; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-14891-5. [Google Scholar]
- Loudiyi, M.; Temiz, H.T.; Sahar, A.; Haseeb Ahmad, M.; Boukria, O.; Hassoun, A.; Aït-Kaddour, A. Spectroscopic Techniques for Monitoring Changes in the Quality of Milk and Other Dairy Products during Processing and Storage. Crit. Rev. Food Sci. Nutr. 2022, 62, 3063–3087. [Google Scholar] [CrossRef] [PubMed]
- Fragkoulis, N.; Samartzis, P.C.; Velegrakis, M. Commercial Milk Discrimination by Fat Content and Animal Origin Using Optical Absorption and Fluorescence Spectroscopy. Int. Dairy J. 2021, 123, 105181. [Google Scholar] [CrossRef]
- Andersen, C.M.; Mortensen, G. Fluorescence Spectroscopy: A Rapid Tool for Analyzing Dairy Products. J. Agric. Food Chem. 2008, 56, 720–729. [Google Scholar] [CrossRef]
- Shaikh, S.; O’Donnell, C. Applications of Fluorescence Spectroscopy in Dairy Processing: A Review. Curr. Opin. Food Sci. 2017, 17, 16–24. [Google Scholar] [CrossRef]
- Barreto, M.C.; Braga, R.G.; Lemos, S.G.; Fragoso, W.D. Determination of Melamine in Milk by Fluorescence Spectroscopy and Second-Order Calibration. Food Chem. 2021, 364, 130407. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A.; Dietrich, S.; Boldrini, B.; Kessler, R.W. Quantitative Determination of Fat and Total Protein in Milk Based on Visible Light Scatter. Food Chem. 2012, 134, 412–418. [Google Scholar] [CrossRef]
- Yang, B.; Guo, W.; Liang, W.; Zhou, Y.; Zhu, X. Design and Evaluation of a Miniature Milk Quality Detection System Based on UV/Vis Spectroscopy. J. Food Compos. Anal. 2022, 106, 104341. [Google Scholar] [CrossRef]
- Karoui, R.; Martin, B.; Dufour, É. Potentiality of Front-Face Fluorescence Spectroscopy to Determine the Geographic Origin of Milks from the Haute-Loire Department (France). Lait 2005, 85, 223–236. [Google Scholar] [CrossRef]
- Birlouez-Aragon, I.; Sabat, P.; Gouti, N. A New Method for Discriminating Milk Heat Treatment. Int. Dairy J. 2002, 12, 59–67. [Google Scholar] [CrossRef]
- Hougaard, A.B.; Lawaetz, A.J.; Ipsen, R.H. Front Face Fluorescence Spectroscopy and Multi-Way Data Analysis for Characterization of Milk Pasteurized Using Instant Infusion. LWT—Food Sci. Technol. 2013, 53, 331–337. [Google Scholar] [CrossRef]
- Domingo, E.; Tirelli, A.A.; Nunes, C.A.; Guerreiro, M.C.; Pinto, S.M. Melamine Detection in Milk Using Vibrational Spectroscopy and Chemometrics Analysis: A Review. Food Res. Int. 2014, 60, 131–139. [Google Scholar] [CrossRef]
- Vázquez-Diosdado, J.A.; Paul, V.; Ellis, K.A.; Coates, D.; Loomba, R.; Kaler, J. A Combined Offline and Online Algorithm for Real-Time and Long-Term Classification of Sheep Behaviour: Novel Approach for Precision Livestock Farming. Sensors 2019, 19, 3201. [Google Scholar] [CrossRef]
- Kaplan, A.; Haenlein, M. Siri, Siri, in My Hand: Who’s the Fairest in the Land? On the Interpretations, Illustrations, and Implications of Artificial Intelligence. Bus. Horiz. 2019, 62, 15–25. [Google Scholar] [CrossRef]
- Niloofar, P.; Francis, D.P.; Lazarova-Molnar, S.; Vulpe, A.; Vochin, M.-C.; Suciu, G.; Balanescu, M.; Anestis, V.; Bartzanas, T. Data-Driven Decision Support in Livestock Farming for Improved Animal Health, Welfare and Greenhouse Gas Emissions: Overview and Challenges. Comput. Electron. Agric. 2021, 190, 106406. [Google Scholar] [CrossRef]
- Norton, T.; Berckmans, D. Developing Precision Livestock Farming Tools for Precision Dairy Farming. Anim. Front. 2017, 7, 18–23. [Google Scholar] [CrossRef]
- VanderWaal, K.; Morrison, R.B.; Neuhauser, C.; Vilalta, C.; Perez, A.M. Translating Big Data into Smart Data for Veterinary Epidemiology. Front. Vet. Sci. 2017, 4, 110. [Google Scholar] [CrossRef]
- Benjamin, M.; Yik, S. Precision Livestock Farming in Swine Welfare: A Review for Swine Practitioners. Animals 2019, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Morota, G.; Ventura, R.V.; Silva, F.F.; Koyama, M.; Fernando, S.C. Big data analytics and precision animal agriculture symposium: Machine learning and data mining advance predictive big data analysis in precision animal agriculture. J. Anim. Sci. 2018, 96, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.; Yan, Z. Supervised Machine Learning Approaches: A Survey. Ictact. J. Soft Comput. 2015, 5, 946–952. [Google Scholar] [CrossRef]
- Soofi, A.A.; Awan, A. Classification techniques in machine learning: Applications and issues. J. Basic Appl. Sci. 2017, 13, 459–465.112. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer International Publishing: Cham, Switzerland, 2022; ISBN 978-3-030-89009-4. [Google Scholar]
- Nasir, N.; Kansal, A.; Alshaltone, O.; Barneih, F.; Sameer, M.; Shanableh, A.; Al-Shamma’a, A. Water Quality Classification Using Machine Learning Algorithms. J. Water Process Eng. 2022, 48, 102920. [Google Scholar] [CrossRef]
- Akalin, A. 5.13 Logistic Regression and Regularization | Computational Genomics with R. Available online: https://compgenomr.github.io/book/logistic-regression-and-regularization.html (accessed on 18 September 2024).
- García, R.; Aguilar, J.; Toro, M.; Pinto, A.; Rodríguez, P. A Systematic Literature Review on the Use of Machine Learning in Precision Livestock Farming. Comput. Electron. Agric. 2020, 179, 105826. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; Taylor & Francis: New York, NY, USA, 2017; ISBN 978-0-412-04841-8. [Google Scholar]
- Mu, F.; Gu, Y.; Zhang, J.; Zhang, L. Milk Source Identification and Milk Quality Estimation Using an Electronic Nose and Machine Learning Techniques. Sensors 2020, 20, 4238. [Google Scholar] [CrossRef]
- Sun, S.; Huang, R. An Adaptive K-Nearest Neighbor Algorithm. In 2010 Seventh International Conference on Fuzzy Systems and Knowledge Discovery; IEEE: Yantai, China, 2010; Volume 1, pp. 91–94. [Google Scholar]
- Japkowicz, N. Learning from imbalanced data sets: A comparison of various strategies. In AAAI Workshop on Learning from Imbalanced Data Sets; AAAI Press: Menlo Park, CA, USA, 2000; Volume 68, pp. 10–15. [Google Scholar]
- Tan, S. Neighbor-Weighted K-Nearest Neighbor for Unbalanced Text Corpus. Expert Syst. Appl. 2005, 28, 667–671. [Google Scholar] [CrossRef]
- Zeng, Y.; Yang, Y.; Zhao, L. Pseudo Nearest Neighbor Rule for Pattern Classification. Expert Syst. Appl. 2009, 36, 3587–3595. [Google Scholar] [CrossRef]
- Shinde, T.A.; Prasad, J.R. IoT based animal health monitoring with naive Bayes classification. Int. J. Eng. Trends Technol. 2017, 1, 252–257. [Google Scholar]
- Rish, I. An empirical study of the naive Bayes classifier. In IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence; International Joint Conferences on Artificial Intelligence: Seattle, WA, USA, 2001; Volume 3, pp. 41–46. [Google Scholar]
- Rong, S.; Bao-wen, Z. The Research of Regression Model in Machine Learning Field. MATEC Web Conf. 2018, 176, 01033. [Google Scholar] [CrossRef]
- Sharma, A.; Paliwal, K.K. Linear Discriminant Analysis for the Small Sample Size Problem: An Overview. Int. J. Mach. Learn. Cyber. 2015, 6, 443–454. [Google Scholar] [CrossRef]
- Tharwat, A.; Gaber, T.; Ibrahim, A.; Hassanien, A.E. Linear Discriminant Analysis: A Detailed Tutorial. Agric. Ind. Confed. 2017, 30, 169–190. [Google Scholar] [CrossRef]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis, 6th ed.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007; ISBN 978-0-13-187715-3. [Google Scholar]
- Ferreira, A.J.; Figueiredo, M.A.T. Boosting Algorithms: A Review of Methods, Theory, and Applications. In Ensemble Machine Learning; Zhang, C., Ma, Y., Eds.; Springer New York: New York, NY, USA, 2012; pp. 35–85. ISBN 978-1-4419-9325-0. [Google Scholar]
- Schapire, R.E. The Boosting Approach to Machine Learning: An Overview. In Nonlinear Estimation and Classification; Denison, D.D., Hansen, M.H., Holmes, C.C., Mallick, B., Yu, B., Eds.; Lecture Notes in Statistics; Springer New York: New York, NY, USA, 2003; Volume 171, pp. 149–171. ISBN 978-0-387-95471-4. [Google Scholar]
- Ganaie, M.A.; Hu, M.; Malik, A.K.; Tanveer, M.; Suganthan, P.N. Ensemble Deep Learning: A Review. Eng. Appl. Artif. Intell. 2022, 115, 105151. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Pence, I.; Kumaş, K.; Cesmeli, M.S.; Akyüz, A. Future Prediction of Biogas Potential and CH4 Emission with Boosting Algorithms: The Case of Cattle, Small Ruminant, and Poultry Manure from Turkey. Environ. Sci. Pollut. Res. 2024, 31, 24461–24479. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Xue, H.; Jiang, X.; Zhou, Y. Recognition of Bovine Milk Somatic Cells Based on Multi-Feature Extraction and a GBDT-AdaBoost Fusion Model. Mol. Biol. Evol. 2022, 19, 5850–5866. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Z.; He, F.; Wang, R.; Yu, W.; Nie, F. Feature Learning Viewpoint of Adaboost and a New Algorithm. IEEE Access 2019, 7, 149890–149899. [Google Scholar] [CrossRef]
- Sun, Y.; Kamel, M.; Wang, Y. Boosting for Learning Multiple Classes with Imbalanced Class Distribution. In Proceedings of the Sixth International Conference on Data Mining (ICDM); IEEE: Hong Kong, China, 2006; pp. 592–602. [Google Scholar]
- Çelik, A. Using Machine Learning Algorithms to Detect Milk Quality. Eurasian J. Food Sci. Technol. 2022, 6, 76–87. [Google Scholar]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Otchere, D.A.; Ganat, T.O.A.; Ojero, J.O.; Tackie-Otoo, B.N.; Taki, M.Y. Application of Gradient Boosting Regression Model for the Evaluation of Feature Selection Techniques in Improving Reservoir Characterisation Predictions. J. Pet. Sci. Eng. 2022, 208, 109244. [Google Scholar] [CrossRef]
- Forecasting: Principles and Practice (2nd Ed) 11.3 Neural Network Models. Available online: https://otexts.com/fpp2/nnetar.html (accessed on 3 December 2024).
- Pirouz, D.M. An Overview of Partial Least Squares. SSRN J. 2006. [Google Scholar] [CrossRef]
- Garthwaite, P.H. An Interpretation of Partial Least Squares. J. Am. Stat. Assoc. 1994, 89, 122–127. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Cheng, J.-H.; Sun, D.-W. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle. Food Eng. Rev. 2017, 9, 36–49. [Google Scholar] [CrossRef]
- Meisel, S.; Stöckel, S.; Elschner, M.; Melzer, F.; Rösch, P.; Popp, J. Raman Spectroscopy as a Potential Tool for Detection of Brucella Spp. in Milk. Appl. Environ. Microbiol. 2012, 78, 5575–5583. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Pegolo, S.; Baba, T.; Peñagaricano, F.; Morota, G.; Bittante, G.; Cecchinato, A. Evaluating the Performance of Machine Learning Methods and Variable Selection Methods for Predicting Difficult-to-Measure Traits in Holstein Dairy Cattle Using Milk Infrared Spectral Data. J. Dairy Sci. 2021, 104, 8107–8121. [Google Scholar] [CrossRef]
- Frizzarin, M.; O’Callaghan, T.F.; Murphy, T.B.; Hennessy, D.; Casa, A. Application of Machine-Learning Methods to Milk Mid-Infrared Spectra for Discrimination of Cow Milk from Pasture or Total Mixed Ration Diets. J. Dairy Sci. 2021, 104, 12394–12402. [Google Scholar] [CrossRef]
- Giannuzzi, D.; Mota, L.F.M.; Pegolo, S.; Gallo, L.; Schiavon, S.; Tagliapietra, F.; Katz, G.; Fainboym, D.; Minuti, A.; Trevisi, E.; et al. In-Line near-Infrared Analysis of Milk Coupled with Machine Learning Methods for the Daily Prediction of Blood Metabolic Profile in Dairy Cattle. Sci. Rep. 2022, 12, 8058. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, D.; Mota, L.F.M.; Pegolo, S.; Tagliapietra, F.; Schiavon, S.; Gallo, L.; Marsan, P.A.; Trevisi, E.; Cecchinato, A. Prediction of Detailed Blood Metabolic Profile Using Milk Infrared Spectra and Machine Learning Methods in Dairy Cattle. J. Dairy Sci. 2023, 106, 3321–3344. [Google Scholar] [CrossRef] [PubMed]
- Soyeurt, H.; Grelet, C.; McParland, S.; Calmels, M.; Coffey, M.; Tedde, A.; Delhez, P.; Dehareng, F.; Gengler, N. A Comparison of 4 Different Machine Learning Algorithms to Predict Lactoferrin Content in Bovine Milk from Mid-Infrared Spectra. J. Dairy Sci. 2020, 103, 11585–11596. [Google Scholar] [CrossRef]
- Teixeira, J.L.D.P.; Caramês, E.T.D.S.; Baptista, D.P.; Gigante, M.L.; Pallone, J.A.L. Vibrational Spectroscopy and Chemometrics Tools for Authenticity and Improvement the Safety Control in Goat Milk. Food Control 2020, 112, 107105. [Google Scholar] [CrossRef]
- Ullah, R.; Khan, S.; Ali, H.; Bilal, M. Potentiality of Using Front Face Fluorescence Spectroscopy for Quantitative Analysis of Cow Milk Adulteration in Buffalo Milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 225, 117518. [Google Scholar] [CrossRef]
- Sowmya, N.; Ponnusamy, V. Development of Spectroscopic Sensor System for an IoT Application of Adulteration Identification on Milk Using Machine Learning. IEEE Access 2021, 9, 53979–53995. [Google Scholar] [CrossRef]
- Lima, J.S.; Ribeiro, D.C.S.Z.; Neto, H.A.; Campos, S.V.A.; Leite, M.O.; Fortini, M.E.D.R.; De Carvalho, B.P.M.; Almeida, M.V.O.; Fonseca, L.M. A Machine Learning Proposal Method to Detect Milk Tainted with Cheese Whey. J. Dairy Sci. 2022, 105, 9496–9508. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-T.; Ren, H.-B.; Liang, W.-Y.; Jin, X.; Yuan, Q.; Liu, Z.-R.; Chen, D.-M.; Zhang, Y.-H. A Novel Approach to Temperature-Dependent Thermal Processing Authentication for Milk by Infrared Spectroscopy Coupled with Machine Learning. J. Food Eng. 2021, 311, 110740. [Google Scholar] [CrossRef]
- Behkami, S.; Zain, S.M.; Gholami, M.; Khir, M.F.A. Classification of Cow Milk Using Artificial Neural Network Developed from the Spectral Data of Single- and Three-Detector Spectrophotometers. Food Chem. 2019, 294, 309–315. [Google Scholar] [CrossRef]
- Nychas, G.-J.E.; Panagou, E.Z.; Mohareb, F. Novel Approaches for Food Safety Management and Communication. Curr. Opin. Food Sci. 2016, 12, 13–20. [Google Scholar] [CrossRef]
- Nychas, G.-J.; Sims, E.; Tsakanikas, P.; Mohareb, F. Data Science in the Food Industry. Annu. Rev. Biomed. Data Sci. 2021, 4, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.H. Review: Sensor Techniques in Ruminants: More than Fitness Trackers. Animal 2020, 14, s187–s195. [Google Scholar] [CrossRef] [PubMed]


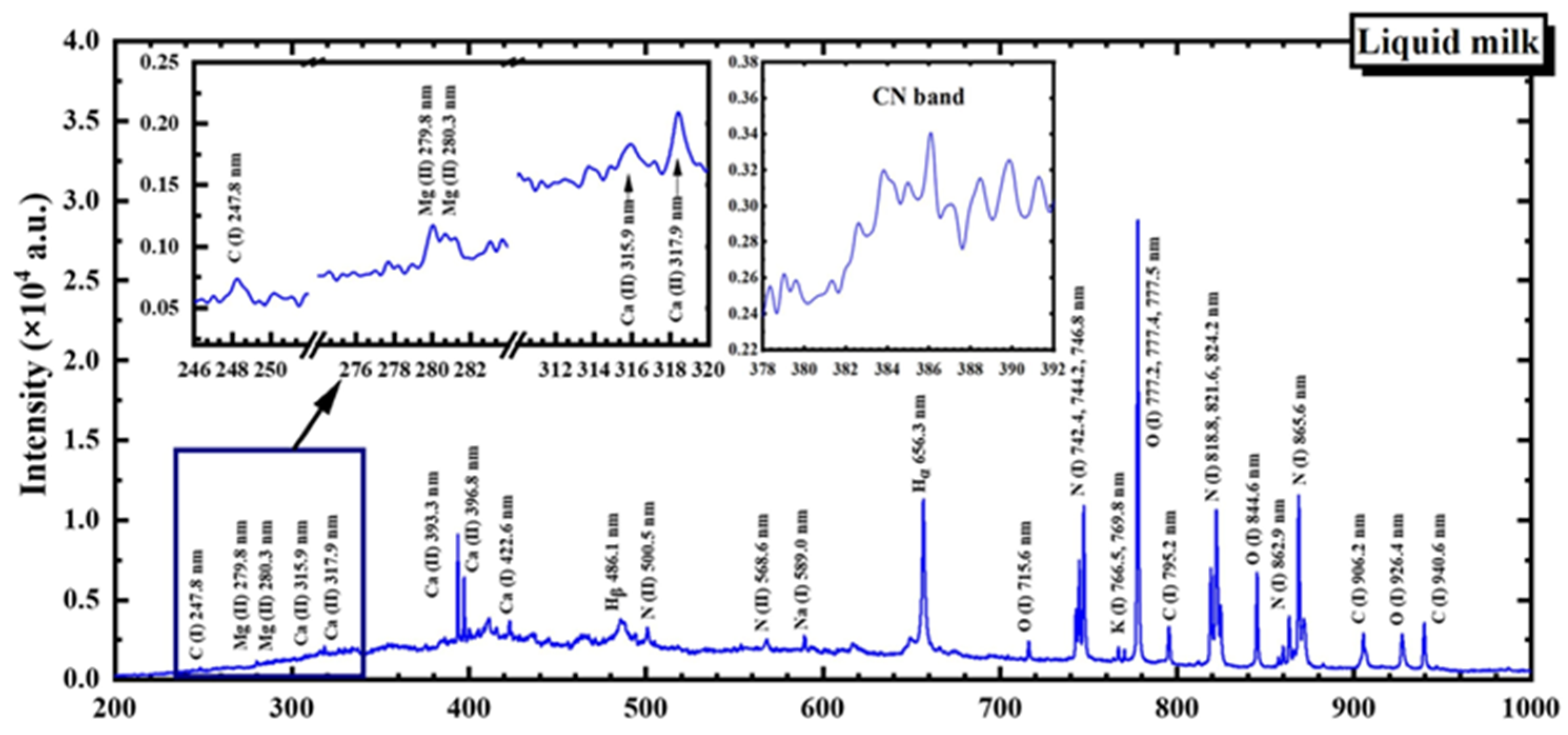
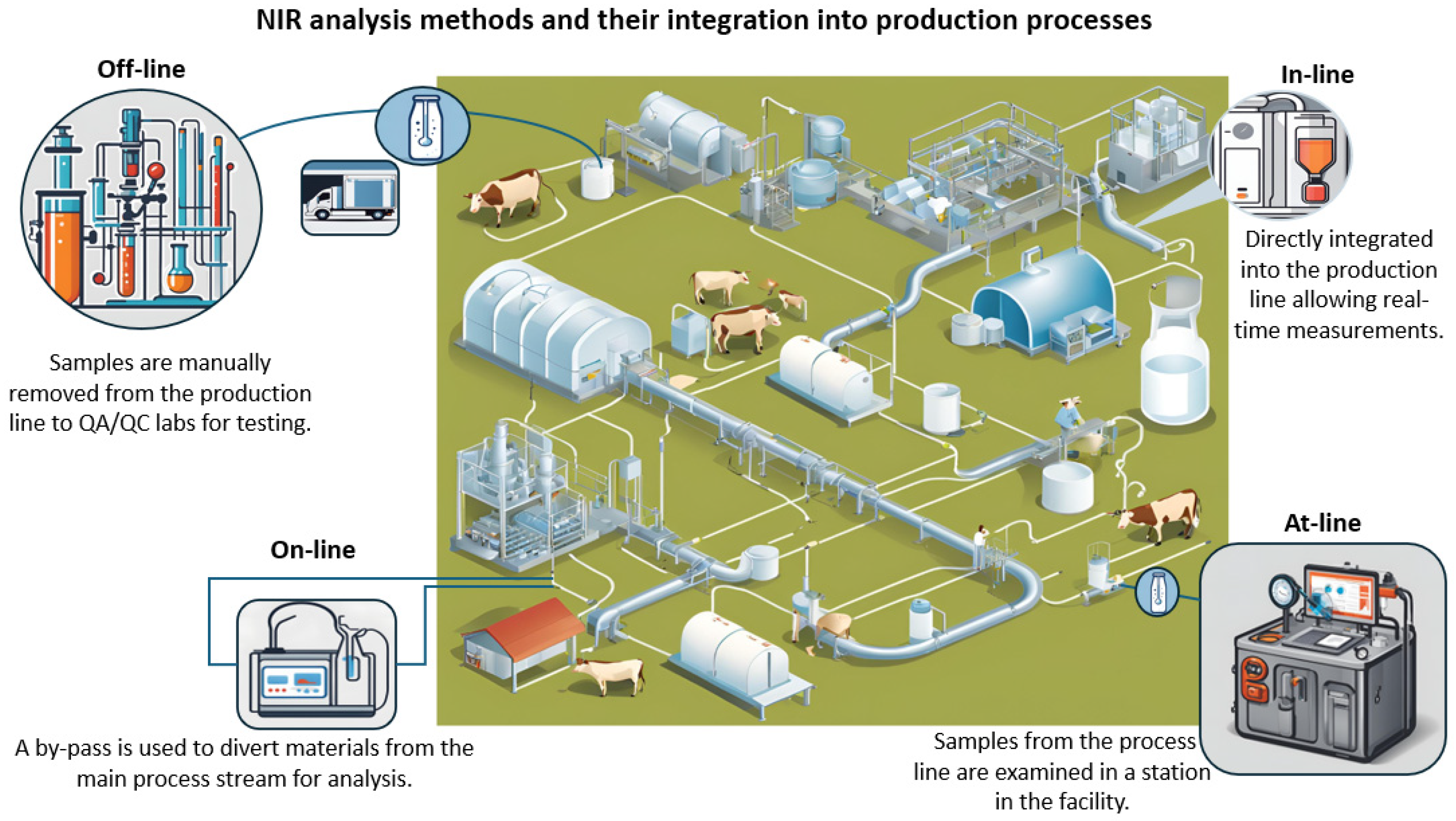

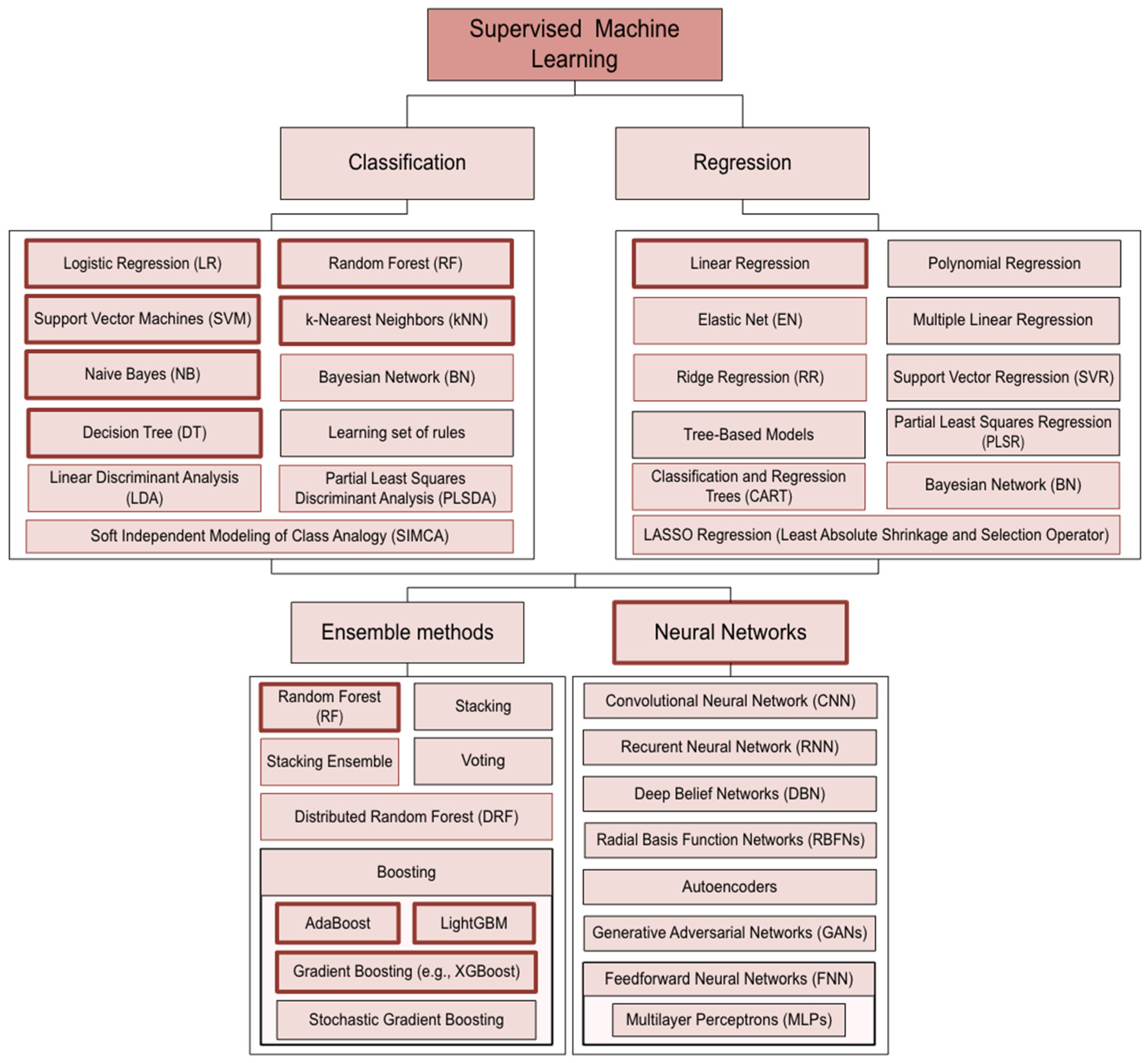

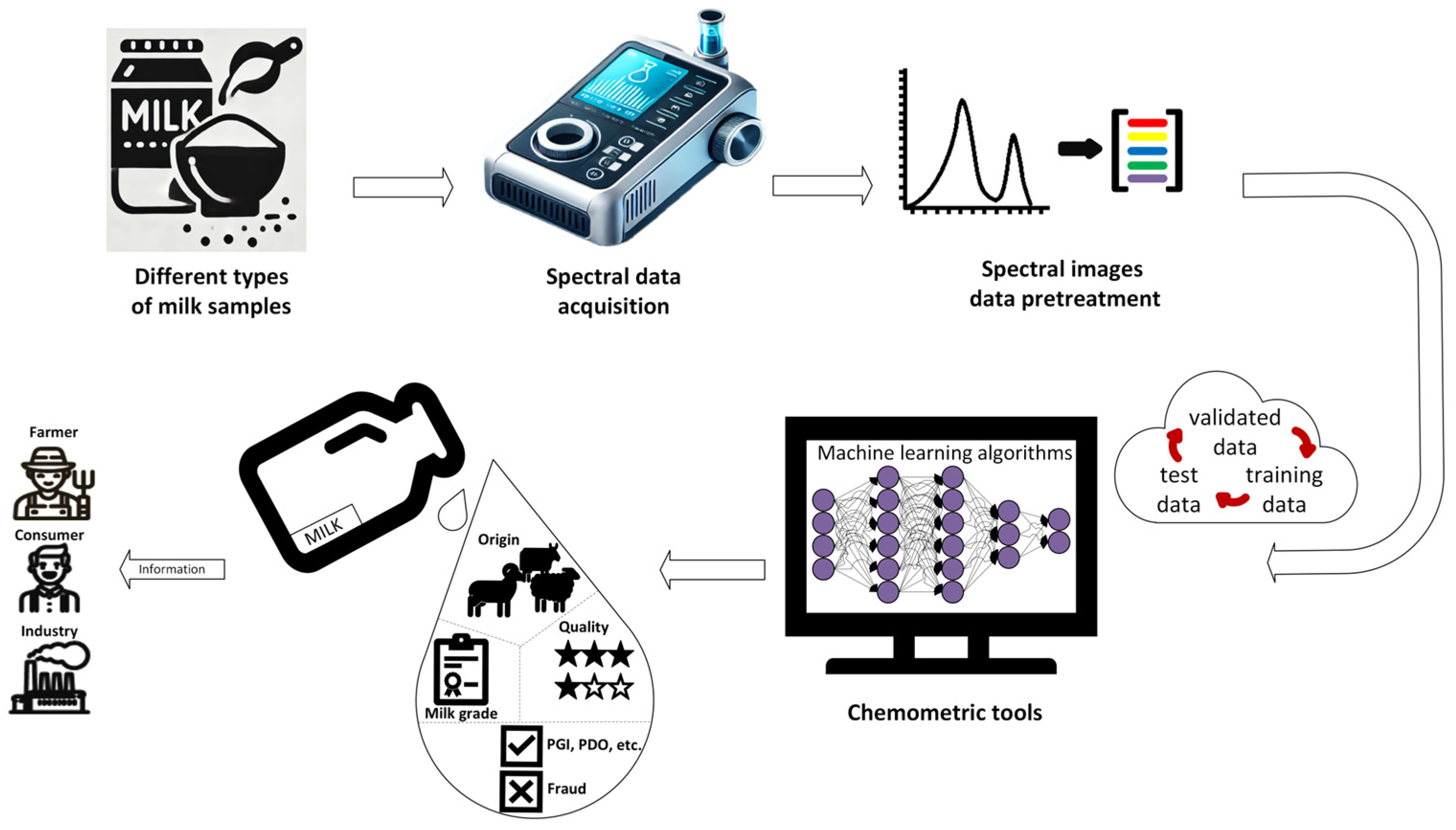
| Bovine | Sheep | Goat | |
|---|---|---|---|
| Fat (%) | 3.6 | 7.9 | 3.8 |
| Lactose (%) | 4.7 | 4.9 | 4.1 |
| Protein (%) | 3.2 | 6.2 | 3.4 |
| Calcium (mg/100 g) | 122 | 193 | 134 |
| Phosphorus (mg/100 g) | 119 | 158 | 121 |
| Vitamin A (IU) | 126 | 146 | 185 |
| Vitamin D (IU) | 2.0 | 0.18 | 2.3 |
| Wavelength (cm−1) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE | Diagnostic Performance | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 300–1700 | powder | ND | retail | spiked samples | lactose | 0.91 | - | - | [23] |
| 250–3500 | powder | 136 | retail | untreated samples | fat protein | - | 0.21–0.31% w/w p 0.14–0.35% w/w p | - | [42] |
| 800–3050 | liquid * liquid ** powder * | 13 | retail | untreated samples | fat | 0.97 v 0.97 v 0.97 v | 0.16% v 0.06% v 0.18% v | - | [39] |
| 8, 16, 32 | liquid | 75 | retail | mixed/diluted samples | fat protein carbohydrates dry matter | - | 5.3–5.8% sp 5.6–6.1% sp 3.5–4.8% sp 3.4–4.8% sp | - | [38] |
| 400–3500 | powder | 45 | retail | spiked samples | lactose high/low classification maltodextrin adulteration | - | - | Se: 98.6% Sp: 100.0% Se: 88.6% Sp: 100.0% | [40] |
| 750–1800 | liquid | 10 batches | retail | spiked samples | urea adulteration | >0.95 | - | Acc + 100 mg/dL: >97% 50–100 mg/dL: 90–95% <50 mg/dL: ≈60% | [41] |
| 600–1800 | liquid | 602 | cow human buffalo goat | untreated samples | milk origin | - | - | Se: 93.0% Sp: 97.0% Acc: 93.7% | [43] |
| Element | Wavelength (nm) |
|---|---|
| H | ) |
| N (I) | 742.4, 744.2, 746.8, 818.8, 821.6, 824.2, 862.9, 865.6 |
| N (II) | 500.5, 568.6 |
| O (I) | 715.6, 777.2, 777.4, 777.5, 844.6, 926.4 |
| C (I) | 247.8, 795.2, 906.2, 940.6 |
| Mg (II) | 279.8, 280.3 |
| Ca (I) | 422.6, 428.3 *, 428.9 *, 430.2 *, 431.9 *, 442.5 *, 443.6 *, 445.5 *, 559.4 *, 612.2 *, 616.2 *, 643.9 *, 646.3 *, 649.4 * |
| Ca (II) | 315.9, 317.9, 393.3, 396.8 |
| Na (I) | 589.0 |
| K (I) | 766.5, 769.8 |
| Wavelength (nm) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE/SEP | Accuracy (%) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 534.9 766.5 285.2 | powder | 23 | retail | digested samples | Ca K Mg | 0.92 0.80 0.91 | 2614 mg kg−1 SEP 1549 mg kg−1 SEP 91 mg kg−1 SEP | - | [47] |
| Laser excitation: 1064 and 532 | liquid, ashed L/ph powder | ND | cow R, goat R, sheep R | untreated samples | major minerals † minor minerals †† | - | - | - | [36] |
| 181–904 | powder | 5 | infant formula | spiked samples | Ca | 0.85 pr | 0.68 mg/g p | - | [54] |
| 200–700 | dried | 60 ND | maternal infant formula | untreated samples | composition quality (Mg, Ca, Fe, Na) | - | - | - | [52] |
| 200–900 | liquid | 300 | cow | untreated samples | fat, protein, lactose, SNF, density, SCC | - | - | - | [53] |
| 200–1000 | liquid L/ph powder | 1296 683 | cow, goat, sheep | untreated samples | milk origin | - | - | 92.8 95.5 | [49] |
| Mg, Ca, Na, K spectral lines | liquid L/ph powder | 1296 683 | cow, goat, sheep | untreated samples | milk origin | - | - | 87.6 92.9 | [49] |
| ≈185–1048 | powder | 50 | vetch root | pelleted samples | milk origin | - | - | 73.1 | [55] |
| 190–450 | blended powder | 12 | cow R, goat R, sheep R | pelleted samples | melamine A, p/b clss. | 0.99 (melamine) | - | 98 (clss. rate) | [50] |
| 540–900 | powder | 36 | cow | lyophilized, pasteurized, spiked, centrifuged | sweet whey A acid whey A | 0.981 0.985 | - | - | [51] |
| 186–900 | gel | 13 13 14 | cow goat sheep | homogenized, gel formed | caprine adult. with bovine ovine adult. with bovine | 0.993 0.995 | 4.53 μg mL−1 p 3.56 μg mL−1 p | - | [56] |
| 196–874 | powder | 25 | infant formula | spiked samples | exogenous protein | - | - | 93.9 (SVM) 97.8 (CNN) | [57] |
| Compound Assignment | Wavelength (nm) |
|---|---|
| N-H, protein | 904, 1014, 1031, 1720, 1758, 2196, 2296, 2334 [71,72] |
| O-H, C-H lipids | 2076, 2376 [71] |
| Carotenoids | 400–700 [71] |
| O-H, water | 1454, 1984, 1953 [73] |
| O-H, N-H | 1953, 2048 [73] |
| Attributed to high somatic cell count | 782, 788, 908, 980, 1068 [74] |
| Wavelength (nm) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE/SEP | Accuracy (%) | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1000–1700 refl 1000–2500 tranms | liquid | 300 | cow | untreated samples | fat crude protein lactose urea | refl 0.997 0.959 0.300 - | tranms 0.997 0.927 0.768 - | refl 0.047% p 0.099% p 0.282% p - | tranms 0.043% p 0.133% p 0.162% p - | - | [61] |
| 1445–2348 | liquid HM liquid UM | 166 | goat | mixed samples | fat protein casein total solid SCC | 0.98 HM, R 0.96 HM, R 0.91 HM, R 0.94 HM, R 0.79 HM, R | 0.98 UM, R 0.95 UM, R 0.92 UM, R 0.95 UM, R 0.74 UM, R | - | - | [68] | |
| 851–1649 | liquid | 785 | cow | untreated samples | fat protein lactose urea SCClog | 0.998 0.98 0.92 0.82 0.85 | 0.09% SEP 0.05% SEP 0.06% SEP 19.3 mg/L SEP 0.18 SEP | - | [28] | ||
| 1500–2500 | powder | 409 | retail | spiked, tableted samples | protein | 0.966 p | 0.547% p | - | [77] | ||
| 700–1100 | liquid | 384 | cow | heated samples | SCC | 0.76 | - | - | [74] | ||
| 400–2500 | oven-dried | 242 | cow | homogenized samples | carotenoids vitamins FAs | 0.09–0.63 0.01–0.69 0.07–0.96 | 0.01–0.15 μg/mL SEP 0.15 μg/mL–611.82 pg/mL SEP 0.12–4.13 g/100 g SEP | - | [78] | ||
| 400–2498 refl | oven-dried | 805 | goat | untreated samples | FAs | 0.80–0.47 | 0.06–2.99 g/100 g SEP | - | [76] | ||
| 400–2498 trans | liquid oven dried | 220 220 | goat | untreated samples | FAs | 0.11–0.79 0.23–0.78 | 0.05–2.81 g/100 g SEP 0.05–3.35 g/100 g SEP | - | [76] | ||
| 400–2498 | liquid oven-dried | 468 | cow, bulk | lyophilized or untreated samples | FAs | 0.00–0.91 v 0.20–0.95 v | 0.11–3.93 g/100 g SEP 0.03–3.25 g/100 g SEP | - | [75] | ||
| 400–2498 | liquid oven-dried | 215 | cow | untreated samples | FAs | 0.29–0.92 v 0.46–0.97 v | 0.08–2.34 g/100 g SEP 0.05–1.00 g/100 g SEP | - | [79] | ||
| 600–1100 | liquid | ND | retail | diluted samples | pH | - | 0.031 pH unit | 88.0–93.0 | [72] | ||
| ≈1100–2500 | powder | 50 | vetch root | pelleted samples | milk origin | - | - | 91.5 | [55] | ||
| 1100–2500 | liquid powder infant formula | 690 660 660 | retail | homogenized, spiked samples | melamine A | - | - | - | [80] | ||
| 1000–2500 | powder | 110 | infant formula | mixed, spiked, homogenized samples | melamine A | - | 0.28–0.31% p | - | [81] | ||
| 1000–2500 | liquid | 150 | cow | centrifuged samples | scattering in NIR absorption | - | - | - | [73] | ||
| 1100–2498 | liquid dried | 219 | sheep | thawed, heated, homogenized samples | summer milk winter milk | - | - | liquid: 79.0 dried: 89.0 liquid: 78.0 dried: 93.0 | [69] | ||
| 400–2498 | oven-dried | 486 | cow | untreated samples | cow feeding-type classification | - | - | 91.5–95.5 | [71] | ||
| Wavelength (nm) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE/SEP | Diagnostic Performance | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1600–2400 | liquid | 108 | cow | untreated samples | FAs | 0.01–0.92 | 0.01–1.57 g/100 g SEP | - | [82] | ||
| 908–1676 | liquid | 87 | retail | untreated samples | O/NO classification | - | - | Se: 59.0% Sp: 81.0% Acc: 73.0% | [83] | ||
| 1600–2400 | liquid | 542 | cow | untreated samples | fat protein SNF | 0.971 0.758 0.612 | 0.126% SEP 0.124% SEP 0.221% SEP | - | [84] | ||
| ≈1600–2400 | powder | 110 | infant formula | mixed, spiked, homogenized samples | melamine A | - | 0.33–0.35% p | - | [81] | ||
| ≈1100–2200 | powder | 110 | infant formula | mixed, spiked, homogenized samples | melamine A | - | 0.27–0.30% p | - | [81] | ||
| 960–1690 | liquid | 1270 | cow | untreated samples | fat protein lactose | 0.989 p_rl 0.894 p_rl 0.644 p_rl | 0.989 p_ph 0.947 p_ph 0.689 p_ph | 0.083 p_rl * 0.110 p_rl * 0.092 p_rl * | 0.078 p_ph * 0.080 p_ph * 0.077 p_ph * | - | [85] |
| 800–1060 | liquid | 81 | cow | mixed, diluted, homogenized samples | fat casein whey | 0.88 0.89 0.91 | 0.08% wt p 0.13% wt p 0.07% wt p | - | [86] | ||
| Wavelength (cm−1) | Type of Milk Sample | No of Samples | Origin of Milk | Samples Preparation | Application | R2 | RMSE/SEP | Accuracy (%) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1000–4000 | liquid | 235 | cow | heated, spiked, mixed or untreated samples | protein | - | PLS: 0.22% NN: 0.08% | - | [88] |
| 1470–1730 | L/ph powder | ND | cow | spiked, diluted samples | protein | 0.974 c | 0.765 mg mL−1 cv | - | [89] |
| 400–4000 | powder | 409 | retail | spiked, tableted samples | protein | 0.990 pr | 0.294% p | - | [77] |
| All MIR excluding: 1600–1710 2990–3690 >3822 | liquid | 730 | cow | untreated samples | CMS pH protein traits RCT | 0.08 0.65 0.19–0.47 0.50 | 25.286 mm cv 0.061 pH unit cv 0.255–1.759 g/L cv 6.397 min cv | 0.62 0.80 0.41–0.48 0.75 | [90] |
| 525–4000 | liquid | 242 | cow | heated, homogenized samples | carotenoids vitamins FAs | 0.01–0.50 0.02–0.40 0.01–0.34 | 0.01–0.19 μg/mL SEP 0.15 μg/mL–907.3 pg/mL SEP 0.13–12.63 g/100 g SEP | - | [78] |
| 1000–5000 | liquid | 215 | cow | untreated samples | FAs | 0.33–0.94 v | 0.06–1.14 g/100 g SEP | - | [79] |
| 900–4000 | liquid | 1064 | cow | spiked, diluted samples | RCT titratable acidity pH | 0.62 0.66 0.59 | 2.36 min cv 0.26 SHo/50 mL cv 0.08 Ph unit cv | - | [91] |
| 500–4000 | liquid powder infant formula | 690 660 660 | retail | homogenized, spiked samples | melamine A | - | - | - | [80] |
| 1450–1600 | liquid | 310 | retail | centrifuged, spiked samples | (w, sm, su, u, hp) A | 0.96, 0.94, 0.98, 0.98, 0.90 | (2.33, 0.06, 0.41, 0.30, 0.01) g/L SEP | - | [87] |
| Spectral Technique | Cost | Adaptability | Convenience | Accuracy | Speed | Portability | Authority | Promotion |
|---|---|---|---|---|---|---|---|---|
| Raman | +++ | +++ | ++ | +++ | ++ | +++ | ++ | +++ |
| LIBS | +++ | ++ | ++ | +++ | +++ | +++ | ++ | ++ |
| NIRS (Benchtop) | ++ | +++ | ++ | +++ | +++ | + | +++ | +++ |
| NIRS (Portable) | ++ | ++++ | ++++ | +++ | ++++ | ++++ | +++ | +++ |
| MIRS | +++ | ++ | ++ | ++++ | ++ | + | +++ | ++ |
| FT-IR | +++ | + | + | ++++ | + | + | +++ | ++ |
| UV | + | ++ | +++ | ++ | +++ | +++ | + | ++ |
| Fluorescence | ++ | ++ | +++ | +++ | +++ | ++ | + | ++ |
| UV/Vis | + | ++ | +++ | ++ | +++ | ++ | ++ | ++ |
| ML | Tools | No and Type of Milk Samples | Application | Accuracy (%) | Ref. |
|---|---|---|---|---|---|
| NN | LIBS | 683 lyophilized 1296 liquid b, c, o | animal origin: liquid milk powdered milk Mg, Ca, Na, K | 97.2 (train), 86.3 (test) 97.5 (train), 94.5 (test), 98.6 (train), 92.7 (test) | [49] |
| ANN | UV-Vis/NIR, FT-NIR | 63 b | geographic origin of cow milk | 100 classification 95 train 92 validation | [158] |
| SVM | LIBS | 683 lyophilized 1296 liquid b, c, o | animal origin: liquid milk powdered milk | 96.6 (train), 91.3 (test) 96.2 (train), 93.1 (test) | [49] |
| GBM | LIBS | 683 lyophilized 1296 liquid b, c, o | animal origin: liquid milk powdered milk | 96.7 (train), 83.0 (test) 97.4 (train), 91.4 (test) | [49] |
| RF | Raman | 602 b, c, o, h | classify milk (cow, human, buffalo, goat) | 93.63 | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agiomavriti, A.-A.; Nikolopoulou, M.P.; Bartzanas, T.; Chorianopoulos, N.; Demestichas, K.; Gelasakis, A.I. Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants. Chemosensors 2024, 12, 263. https://doi.org/10.3390/chemosensors12120263
Agiomavriti A-A, Nikolopoulou MP, Bartzanas T, Chorianopoulos N, Demestichas K, Gelasakis AI. Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants. Chemosensors. 2024; 12(12):263. https://doi.org/10.3390/chemosensors12120263
Chicago/Turabian StyleAgiomavriti, Aikaterini-Artemis, Maria P. Nikolopoulou, Thomas Bartzanas, Nikos Chorianopoulos, Konstantinos Demestichas, and Athanasios I. Gelasakis. 2024. "Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants" Chemosensors 12, no. 12: 263. https://doi.org/10.3390/chemosensors12120263
APA StyleAgiomavriti, A.-A., Nikolopoulou, M. P., Bartzanas, T., Chorianopoulos, N., Demestichas, K., & Gelasakis, A. I. (2024). Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants. Chemosensors, 12(12), 263. https://doi.org/10.3390/chemosensors12120263










