Recent Advances in Rational Design and Engineering of Signal-Amplifying Substrates for Surface-Enhanced Raman Scattering-Based Bioassays
Abstract
:1. Introduction
2. Basic Theory of SERS
2.1. Electromagnetic Enhancement
2.2. Chemical Enhancement
3. Substrates in Colloidal Suspension
3.1. Dispersed Particles
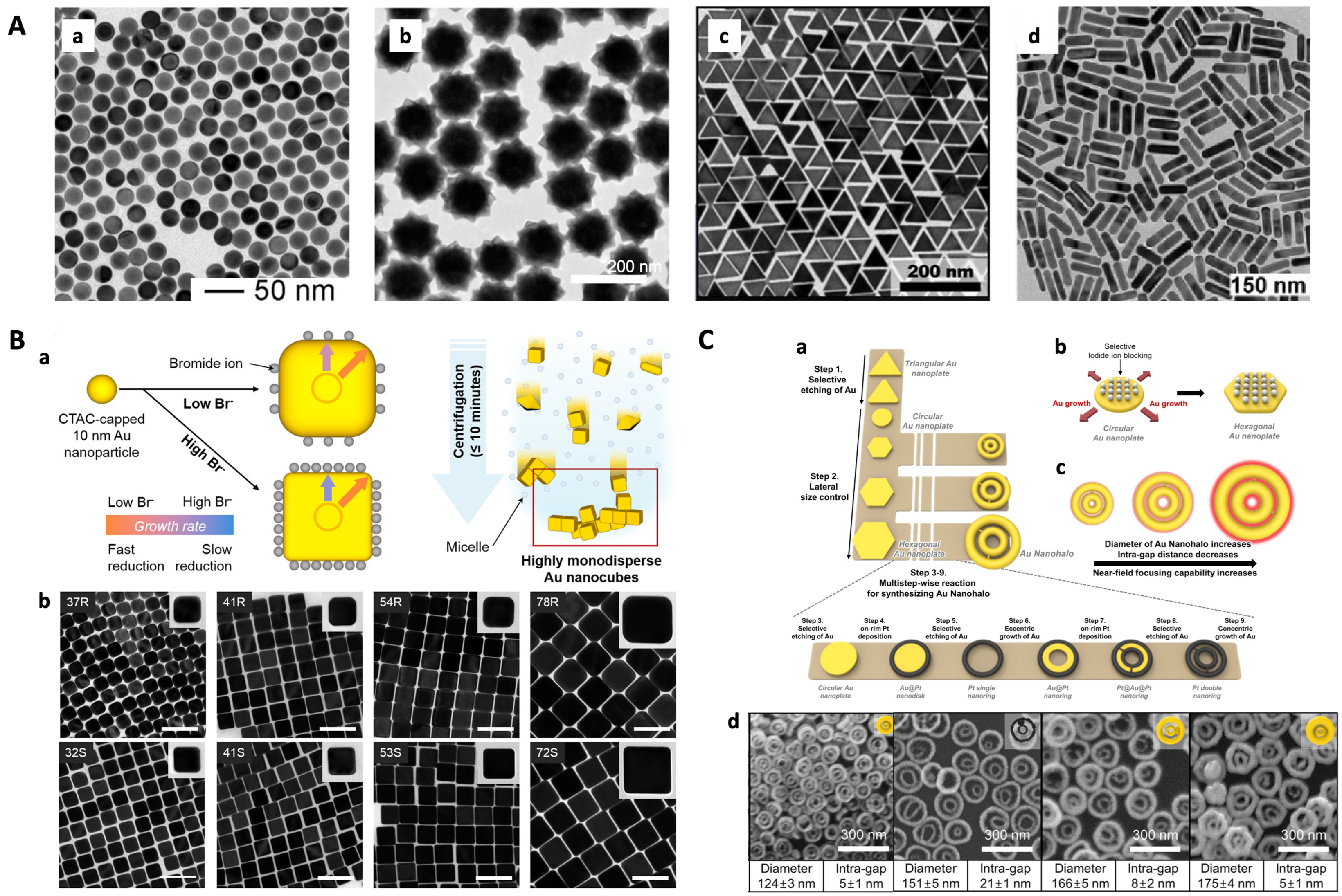
3.2. Coupling of Particles
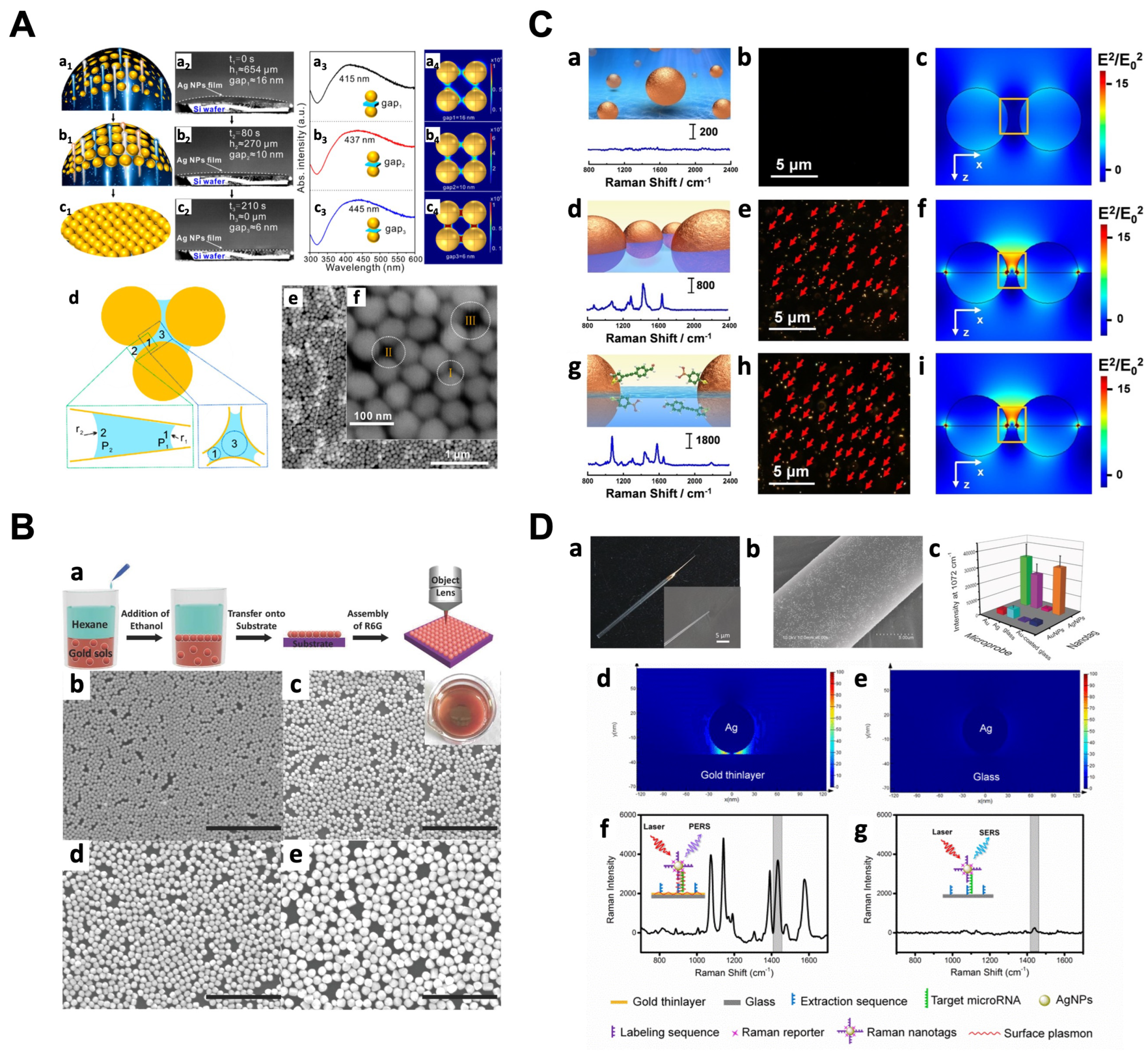
4. Substrates on a Solid Support
4.1. Bottom-Up Strategies
4.2. Top-Down Strategies
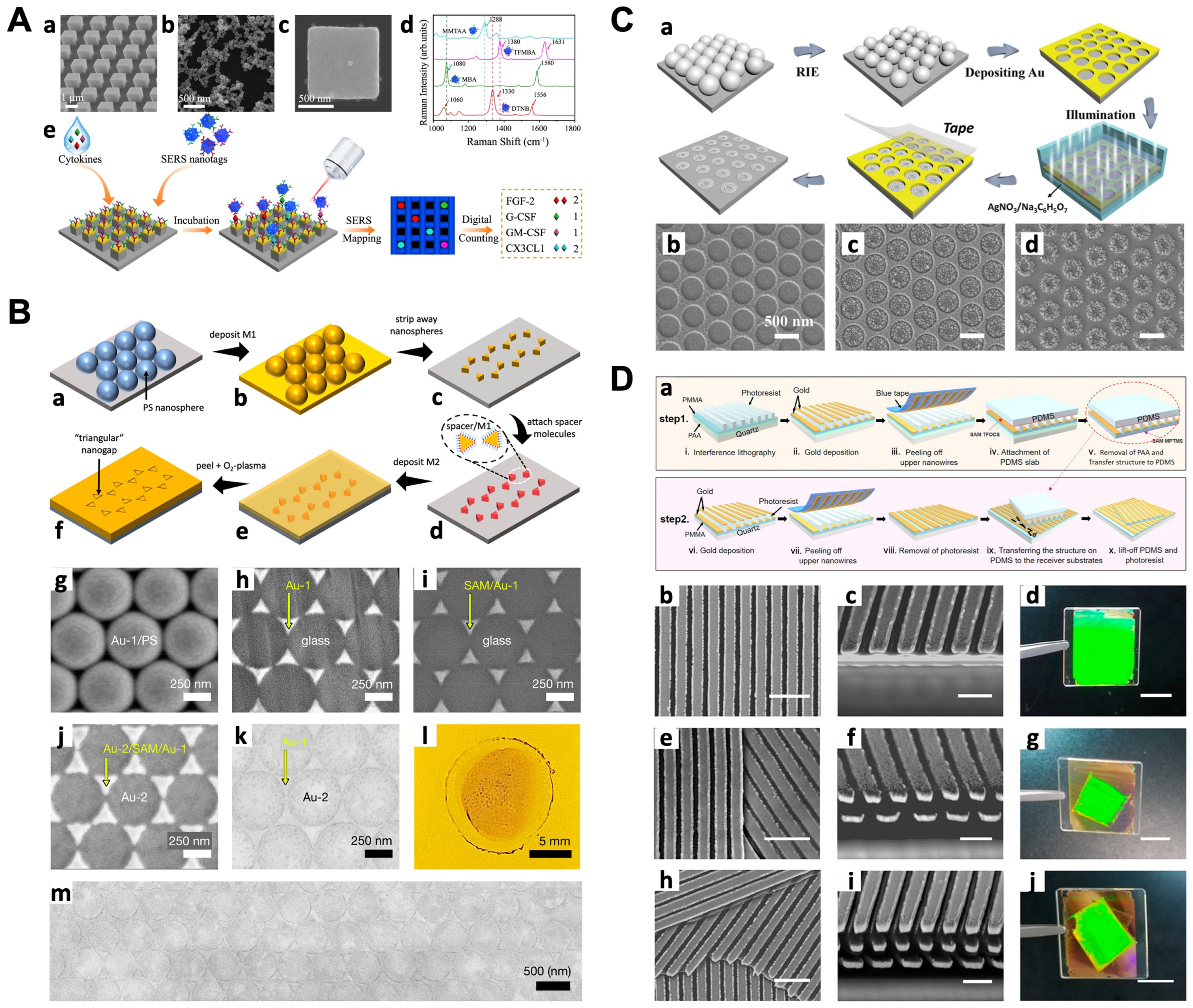
5. Applications
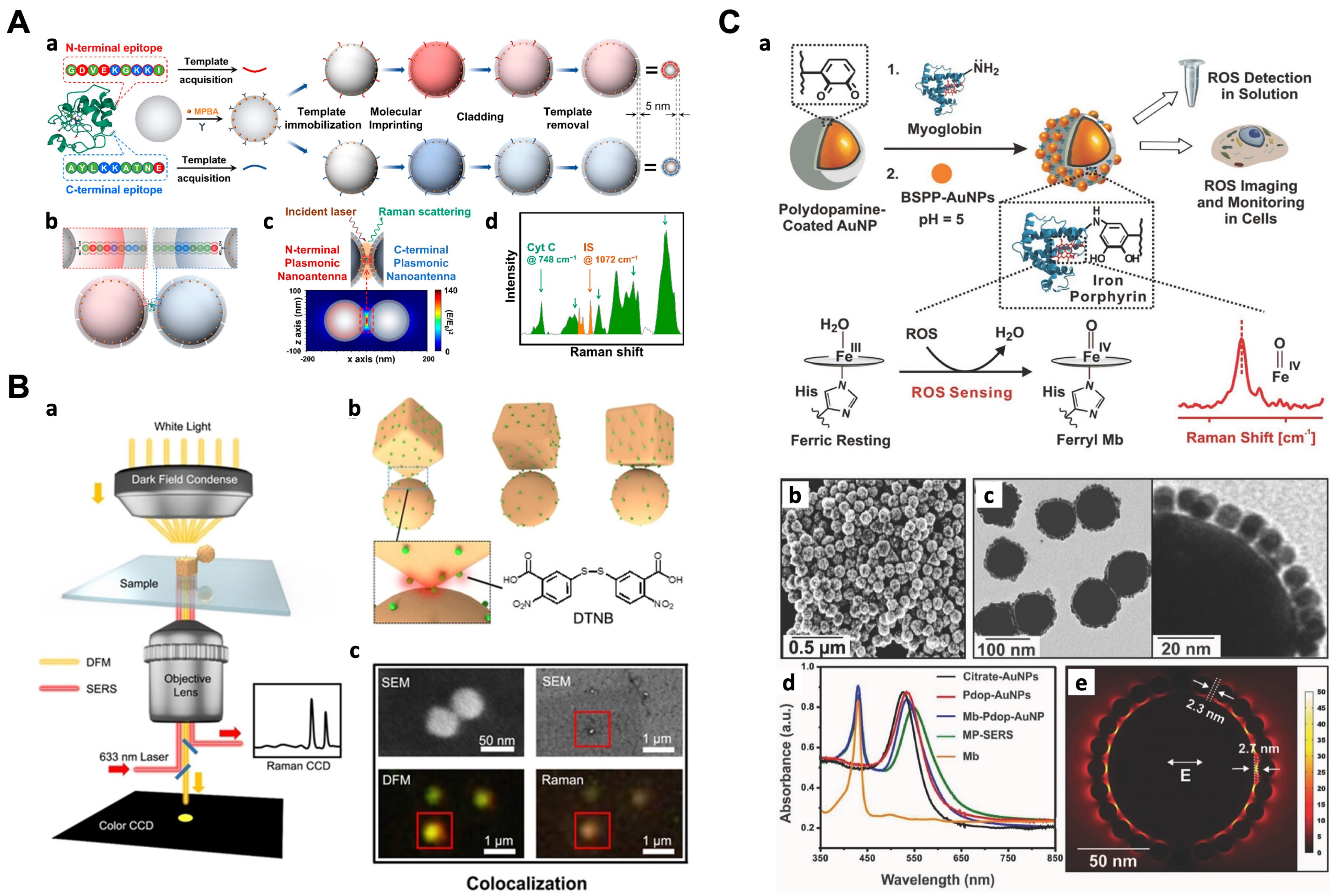
5.1. Detection of Proteins
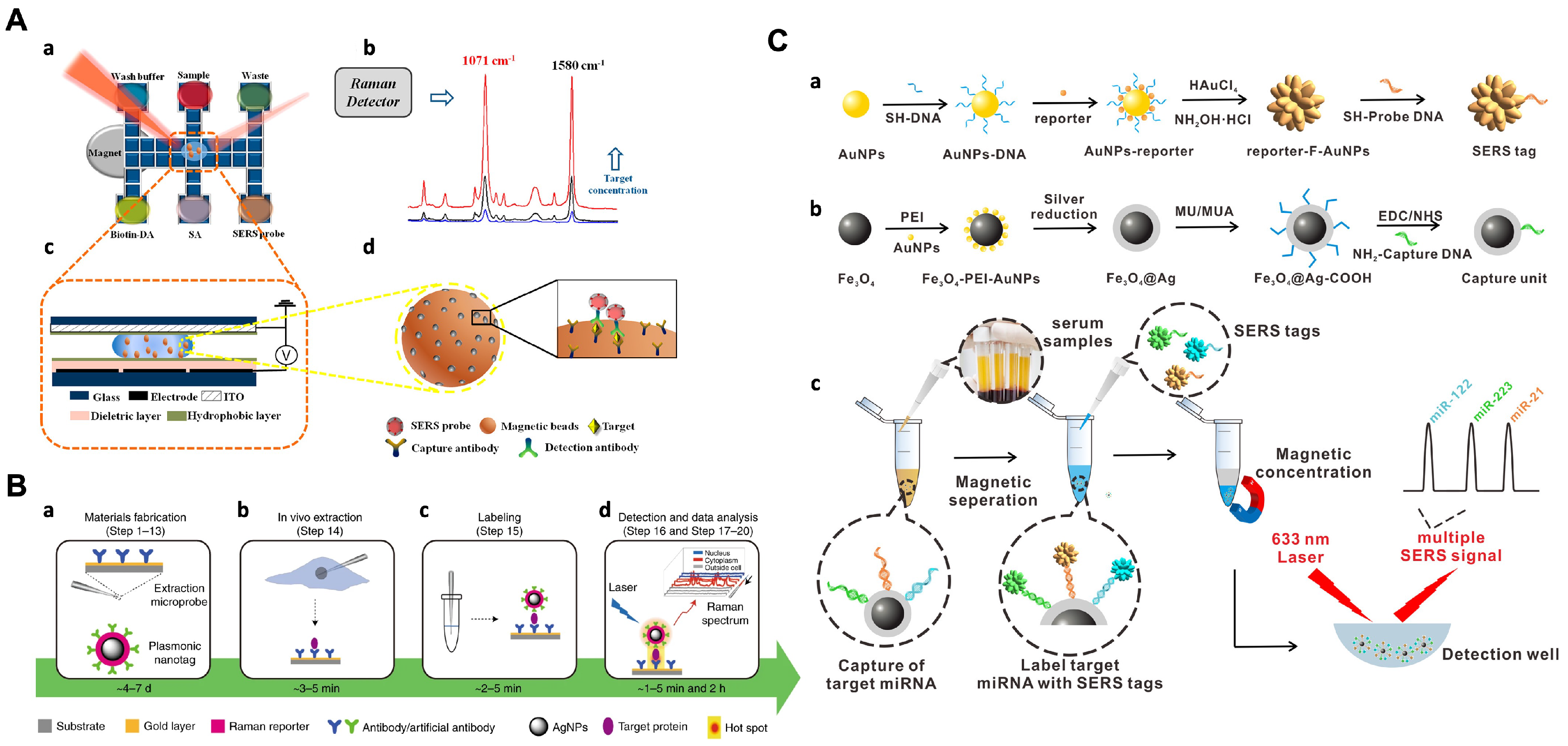
5.2. Detection of Nucleic Acids
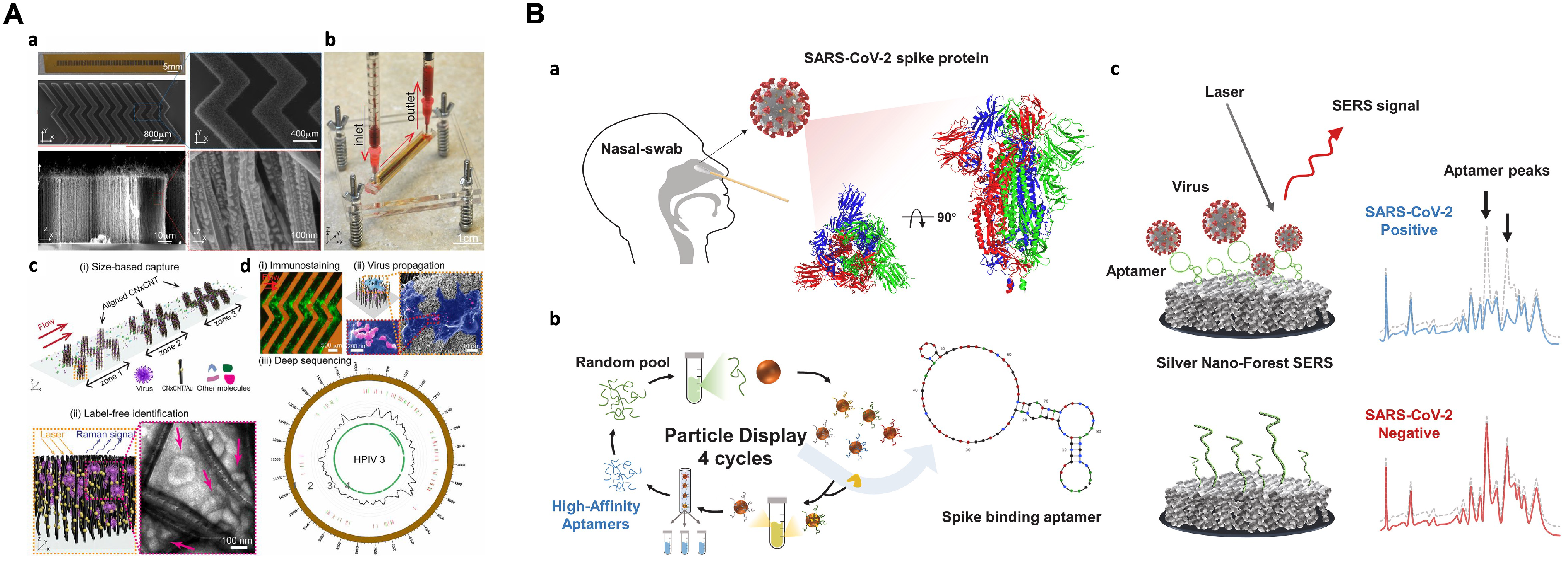
5.3. Detection of Viruses
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raman, C.V.; Krishnan, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346, 1247390. [Google Scholar] [CrossRef] [PubMed]
- Farka, Z.; Jurik, T.; Kovar, D.; Trnkova, L.; Skládal, P. Nanoparticle-based immunochemical biosensors and assays: Recent advances and challenges. Chem. Rev. 2017, 117, 9973–10042. [Google Scholar] [CrossRef]
- Lane, L.A.; Qian, X.; Nie, S. SERS nanoparticles in medicine: From label-free detection to spectroscopic tagging. Chem. Rev. 2015, 115, 10489–10529. [Google Scholar] [CrossRef]
- Kallaway, C.; Almond, L.M.; Barr, H.; Wood, J.; Hutchings, J.; Kendall, C.; Stone, N. Advances in the clinical application of Raman spectroscopy for cancer diagnostics. Photodiagn. Photodyn. Ther. 2013, 10, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Popp, J. The many facets of Raman spectroscopy for biomedical analysis. Anal. Bioanal. Chem. 2015, 407, 699–717. [Google Scholar] [CrossRef]
- Stone, N.; Matousek, P. Advanced transmission Raman spectroscopy: A promising tool for breast disease diagnosis. Cancer Res. 2008, 68, 4424–4430. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Mosca, S.; Conti, C.; Stone, N.; Matousek, P. Spatially offset Raman spectroscopy. Nat. Rev. Methods Primers 2021, 1, 21. [Google Scholar] [CrossRef]
- Ellis, D.I.; Cowcher, D.P.; Ashton, L.; O’Hagan, S.; Goodacre, R. Illuminating disease and enlightening biomedicine: Raman spectroscopy as a diagnostic tool. Analyst 2013, 138, 3871–3884. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface Raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Ye, J.; Chen, Y.; Liu, Z. A boronate affinity sandwich assay: An appealing alternative to immunoassays for the determination of glycoproteins. Angew. Chem. Int. Ed. 2014, 53, 10386–10389. [Google Scholar] [CrossRef]
- Ahn, K.C.; Zhao, B.; Chen, J.; Cherednichenko, G.; Sanmarti, E.; Denison, M.S.; Lasley, B.; Pessah, I.N.; Kültz, D.; Chang, D.P.; et al. In vitro biologic activities of the antimicrobials triclocarban, its analogs, and triclosan in bioassay screens: Receptor-based bioassay screens. Environ. Health Perspect. 2008, 116, 1203–1210. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, S.; Zhang, F. Optical Multiplexed Bioassays for Improved Biomedical Diagnostics. Angew. Chem. Int. Ed. 2019, 58, 13208–13219. [Google Scholar] [CrossRef] [PubMed]
- Kerns, E.H.; Di, L.; Carter, G.T. In vitro solubility assays in drug discovery. Curr. Drug Metab. 2008, 9, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.; Hutchings, M.; Benstead, R.; Thain, J.; Whitehouse, P. Bioassay selection, experimental design and quality control/assurance for use in effluent assessment and control. Ecotoxicology 2004, 13, 437–447. [Google Scholar] [CrossRef]
- Sprague, J. Measurement of pollutant toxicity to fish I. Bioassay methods for acute toxicity. Water Res. 1969, 3, 793–821. [Google Scholar] [CrossRef]
- Schuetzle, D.; Lewtas, J. Bioassay-directed chemical analysis in environmental research. Anal. Chem. 1986, 58, 1060A–1075A. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-enhanced Raman spectroscopy for in vivo biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar] [CrossRef]
- Francis, M.K.; Sahu, B.K.; Bhargav, P.B.; Balaji, C.; Ahmed, N.; Das, A.; Dhara, S. Ag nanowires based SERS substrates with very high enhancement factor. Physica E Low Dimens. Syst. Nanostruct. 2022, 137, 115080. [Google Scholar] [CrossRef]
- Xu, S.; Man, B.; Jiang, S.; Wang, J.; Wei, J.; Xu, S.; Liu, H.; Gao, S.; Liu, H.; Li, Z.; et al. Graphene/Cu nanoparticle hybrids fabricated by chemical vapor deposition as surface-enhanced Raman scattering substrate for label-free detection of adenosine. ACS Appl. Mater. Interfaces 2015, 7, 10977–10987. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, X.; Zhao, Y.; Chang, L.; Qi, J. Graphene oxide shell-isolated Ag nanoparticles for surface-enhanced Raman scattering. Carbon 2015, 81, 767–772. [Google Scholar] [CrossRef]
- Wang, X.; Huang, S.C.; Hu, S.; Yan, S.; Ren, B. Fundamental understanding and applications of plasmon-enhanced Raman spectroscopy. Nat. Rev. Phys. 2020, 2, 253–271. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.; Boisen, A.; Brolo, A.G.; et al. Present and future of surface-enhanced Raman scattering. ACS Nano 2019, 14, 28–117. [Google Scholar] [CrossRef]
- Karthick Kannan, P.; Shankar, P.; Blackman, C.; Chung, C.H. Recent advances in 2D inorganic nanomaterials for SERS sensing. Adv. Mater. 2019, 31, 1803432. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, L.; Tian, Y. Rational Design of Surface-Enhanced Raman Scattering Substrate for Highly Reproducible Analysis. Anal. Sens. 2023, 3, e202200064. [Google Scholar] [CrossRef]
- Lee, H.K.; Lee, Y.H.; Koh, C.S.L.; Phan-Quang, G.C.; Han, X.; Lay, C.L.; Sim, H.Y.F.; Kao, Y.C.; An, Q.; Ling, X.Y. Designing surface-enhanced Raman scattering (SERS) platforms beyond hotspot engineering: Emerging opportunities in analyte manipulations and hybrid materials. Chem. Soc. Rev. 2019, 48, 731–756. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef]
- Boujday, S.; Lamy de la Chapelle, M.; Srajer, J.; Knoll, W. Enhanced vibrational spectroscopies as tools for small molecule biosensing. Sensors 2015, 15, 21239–21264. [Google Scholar] [CrossRef]
- Ding, H.; Hu, D.J.J.; Yu, X.; Liu, X.; Zhu, Y.; Wang, G. Review on all-fiber online raman sensor with hollow core microstructured optical fiber. Photonics 2022, 9, 134. [Google Scholar] [CrossRef]
- Jorgenson, E.; Ianoul, A. Biofunctionalization of plasmonic nanoparticles with short peptides monitored by SERS. J. Phys. Chem. B 2017, 121, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xianyu, Y.; Jiang, X. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc. Chem. Res. 2017, 50, 310–319. [Google Scholar] [CrossRef]
- He, J.; Unser, S.; Bruzas, I.; Cary, R.; Shi, Z.; Mehra, R.; Aron, K.; Sagle, L. The facile removal of CTAB from the surface of gold nanorods. Colloids Surf. 2018, 163, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhong, X.; Li, Z.; Xia, Y. Successive, Seed-Mediated Growth for the Synthesis of Single-Crystal Gold Nanospheres with Uniform Diameters Controlled in the Range of 5–150 nm. Part. Part. Syst. Charact. 2014, 31, 266–273. [Google Scholar] [CrossRef]
- Niu, W.; Chua, Y.A.A.; Zhang, W.; Huang, H.; Lu, X. Highly symmetric gold nanostars: Crystallographic control and surface-enhanced Raman scattering property. J. Am. Chem. Soc. 2015, 137, 10460–10463. [Google Scholar] [CrossRef]
- Khoury, C.G.; Vo-Dinh, T. Gold nanostars for surface-enhanced Raman scattering: Synthesis, characterization and optimization. J. Phys. Chem. C 2008, 112, 18849–18859. [Google Scholar] [CrossRef] [PubMed]
- Childs, A.; Vinogradova, E.; Ruiz-Zepeda, F.; Velazquez-Salazar, J.J.; Jose-Yacaman, M. Biocompatible gold/silver nanostars for surface-enhanced Raman scattering. J. Raman Spectrosc. 2016, 47, 651–655. [Google Scholar] [CrossRef]
- Scarabelli, L.; Liz-Marzan, L.M. An extended protocol for the synthesis of monodisperse gold nanotriangles. ACS Nano 2021, 15, 18600–18607. [Google Scholar] [CrossRef]
- Khanal, B.P.; Zubarev, E.R. Chemical transformation of nanorods to nanowires: Reversible growth and dissolution of anisotropic gold nanostructures. ACS Nano 2019, 13, 2370–2378. [Google Scholar] [CrossRef] [PubMed]
- González-Rubio, G.; Díaz-Núñez, P.; Rivera, A.; Prada, A.; Tardajos, G.; González-Izquierdo, J.; Bañares, L.; Llombart, P.; Macdowell, L.G.; Alcolea Palafox, M.; et al. Femtosecond laser reshaping yields gold nanorods with ultranarrow surface plasmon resonances. Science 2017, 358, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Qu, H.; Liang, L.; Zhang, J.; Zhang, B.; Huang, D.; Xu, S.; Liang, C.; Xu, W. Tracing the therapeutic process of targeted aptamer/drug conjugate on cancer cells by surface-enhanced Raman scattering spectroscopy. Anal. Chem. 2017, 89, 2844–2851. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Lee, Y.; Nam, J.M. Precisely shaped, uniformly formed gold nanocubes with ultrahigh reproducibility in single-particle scattering and surface-enhanced Raman scattering. Nano Lett. 2018, 18, 6475–6482. [Google Scholar] [CrossRef]
- Matteini, P.; Cottat, M.; Tavanti, F.; Panfilova, E.; Scuderi, M.; Nicotra, G.; Menziani, M.C.; Khlebtsov, N.; de Angelis, M.; Pini, R. Site-selective surface-enhanced Raman detection of proteins. ACS Nano 2017, 11, 918–926. [Google Scholar] [CrossRef]
- Guo, P.; Sikdar, D.; Huang, X.; Si, K.J.; Xiong, W.; Gong, S.; Yap, L.W.; Premaratne, M.; Cheng, W. Plasmonic core–shell nanoparticles for SERS detection of the pesticide thiram: Size-and shape-dependent Raman enhancement. Nanoscale 2015, 7, 2862–2868. [Google Scholar] [CrossRef]
- Jana, D.; Gorunmez, Z.; He, J.; Bruzas, I.; Beck, T.; Sagle, L. Surface enhanced Raman spectroscopy of a Au@Au core–shell structure containing a spiky Shell. J. Phys. Chem. C 2016, 120, 20814–20821. [Google Scholar] [CrossRef]
- Li, J.F.; Zhang, Y.J.; Ding, S.Y.; Panneerselvam, R.; Tian, Z.Q. Core–shell nanoparticle-enhanced Raman spectroscopy. Chem. Rev. 2017, 117, 5002–5069. [Google Scholar] [CrossRef]
- Zhang, P.; Li, Y.; Wang, D.; Xia, H. High–Yield Production of Uniform Gold Nanoparticles with Sizes from 31 to 577 nm via One-Pot Seeded Growth and Size-Dependent SERS Property. Part. Part. Syst. Charact. 2016, 33, 924–932. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, L.; Chen, J.; Tang, Z.; Liang, P.; Huang, Y.; Cao, M.; Zou, M.; Ni, D.; Chen, J.; et al. Controllable self-assembly of SERS hotspots in liquid environment. Langmuir 2021, 37, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Lee, S.; Yoo, S.; Jung, I.; Lee, S.; Kim, J.; Son, J.; Kim, J.E.; Kim, J.M.; Nam, J.M.; et al. Enormous enhancement in single-particle surface-enhanced Raman scattering with size-controllable Au double nanorings. Chem. Mater. 2022, 34, 2197–2205. [Google Scholar] [CrossRef]
- Cho, N.H.; Byun, G.H.; Lim, Y.C.; Im, S.W.; Kim, H.; Lee, H.E.; Ahn, H.Y.; Nam, K.T. Uniform chiral gap synthesis for high dissymmetry factor in single plasmonic gold nanoparticle. ACS Nano 2020, 14, 3595–3602. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhou, L.; Guo, Z.; Li, P.; Gao, S.; Liu, Z. Dual Biomimetic Recognition-Driven Plasmonic Nanogap-Enhanced Raman Scattering for Ultrasensitive Protein Fingerprinting and Quantitation. Nano Lett. 2022, 22, 9664–9671. [Google Scholar] [CrossRef] [PubMed]
- Niu, R.; Song, C.; Gao, F.; Fang, W.; Jiang, X.; Ren, S.; Zhu, D.; Su, S.; Chao, J.; Chen, S.; et al. DNA Origami-Based Nanoprinting for the Assembly of Plasmonic Nanostructures with Single-Molecule Surface-Enhanced Raman Scattering. Angew. Chem. Int. Ed. 2021, 60, 11695–11701. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, A.; Kim, G.H.; Rhim, W.K.; Hartman, K.L.; Nam, J.M. Myoglobin and polydopamine-engineered Raman nanoprobes for detecting, imaging, and monitoring reactive oxygen species in biological samples and living cells. Small 2017, 13, 1701584. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef]
- Liu, N.; Liedl, T. DNA-assembled advanced plasmonic architectures. Chem. Rev. 2018, 118, 3032–3053. [Google Scholar] [CrossRef]
- Liu, W.; Halverson, J.; Tian, Y.; Tkachenko, A.V.; Gang, O. Self-organized architectures from assorted DNA-framed nanoparticles. Nat. Chem. 2016, 8, 867–873. [Google Scholar] [CrossRef]
- Liu, Z.; Pei, H.; Zhang, L.; Tian, Y. Mitochondria-targeted DNA nanoprobe for real-time imaging and simultaneous quantification of Ca2+ and pH in neurons. ACS Nano 2018, 12, 12357–12368. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, W.; Chen, X.; Li, J.; Ye, H.; Li, C.; Yin, X.; Zhou, X.; Qiao, X.; Xue, Z.; et al. Gold nanoparticle-bridge array to improve DNA hybridization efficiency of SERS sensors. J. Am. Chem. Soc. 2022, 144, 17533–17539. [Google Scholar] [CrossRef] [PubMed]
- Niu, R.; Gao, F.; Wang, D.; Zhu, D.; Su, S.; Chen, S.; YuWen, L.; Fan, C.; Wang, L.; Chao, J. Pattern recognition directed assembly of Plasmonic gap nanostructures for single-molecule SERS. ACS Nano 2022, 16, 14622–14631. [Google Scholar] [CrossRef]
- Fang, W.; Jia, S.; Chao, J.; Wang, L.; Duan, X.; Liu, H.; Li, Q.; Zuo, X.; Wang, L.; Wang, L.; et al. Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Sci. Adv. 2019, 5, eaau4506. [Google Scholar] [CrossRef] [PubMed]
- Simoncelli, S.; Roller, E.M.; Urban, P.; Schreiber, R.; Turberfield, A.J.; Liedl, T.; Lohmuller, T. Quantitative single-molecule surface-enhanced Raman scattering by optothermal tuning of DNA origami-assembled plasmonic nanoantennas. ACS Nano 2016, 10, 9809–9815. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Song, C.; Zhu, D.; Wang, X.; Zhao, M.; Yang, Y.; Zhang, Y.; Su, S.; Shi, J.; Chao, J.; et al. DNA-Origami-based assembly of anisotropic plasmonic gold nanostructures. Small 2017, 13, 1603991. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, S.; Haldar, K.K.; Sen, T. DNA origami directed Au nanostar dimers for single-molecule surface-enhanced Raman scattering. J. Am. Chem. Soc. 2017, 139, 17639–17648. [Google Scholar] [CrossRef]
- Zhan, P.; Wen, T.; Wang, Z.g.; He, Y.; Shi, J.; Wang, T.; Liu, X.; Lu, G.; Ding, B. DNA origami directed assembly of gold bowtie nanoantennas for single-molecule surface-enhanced Raman scattering. Angew. Chem. Int. Ed. 2018, 57, 2846–2850. [Google Scholar] [CrossRef]
- Lim, D.K.; Jeon, K.S.; Kim, H.M.; Nam, J.M.; Suh, Y.D. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat. Mater. 2010, 9, 60–67. [Google Scholar] [CrossRef]
- Gandra, N.; Abbas, A.; Tian, L.; Singamaneni, S. Plasmonic planet–satellite analogues: Hierarchical self-assembly of gold nanostructures. Nano Lett. 2012, 12, 2645–2651. [Google Scholar] [CrossRef]
- Li, A.; Tang, L.; Song, D.; Song, S.; Ma, W.; Xu, L.; Kuang, H.; Wu, X.; Liu, L.; Chen, X.; et al. A SERS-active sensor based on heterogeneous gold nanostar core–silver nanoparticle satellite assemblies for ultrasensitive detection of aflatoxinB1. Nanoscale 2016, 8, 1873–1878. [Google Scholar] [CrossRef]
- Li, Q.; Ge, X.; Ye, J.; Li, Z.; Su, L.; Wu, Y.; Yang, H.; Song, J. Dual ratiometric SERS and photoacoustic core–satellite nanoprobe for quantitatively visualizing hydrogen peroxide in inflammation and cancer. Angew. Chem. Int. Ed. 2021, 60, 7323–7332. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hu, Y.; Fan, C.; Yang, L.; Zhang, Y.; Li, H.; Xie, W. Surface-enhanced Raman spectroscopic evidence of key intermediate species and role of NiFe dual-catalytic center in water oxidation. Angew. Chem. Int. Ed. 2021, 60, 19774–19778. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lorenzo, L.; Alvarez-Puebla, R.A.; Pastoriza-Santos, I.; Mazzucco, S.; Stéphan, O.; Kociak, M.; Liz-Marzán, L.M.; García de Abajo, F.J. Zeptomol detection through controlled ultrasensitive surface-enhanced Raman scattering. J. Am. Chem. Soc. 2009, 131, 4616–4618. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Ma, X.; Dong, J.; Jiang, C.; Qian, W. A reproducible SERS substrate based on electrostatically assisted APTES-functionalized surface-assembly of gold nanostars. ACS Appl. Mater. Interfaces 2011, 3, 1873–1879. [Google Scholar] [CrossRef] [PubMed]
- Fortuni, B.; Fujita, Y.; Ricci, M.; Inose, T.; Aubert, R.; Lu, G.; Hutchison, J.A.; Hofkens, J.; Latterini, L.; Uji-i, H. A novel method for in situ synthesis of SERS-active gold nanostars on polydimethylsiloxane film. Chem. Commun. 2017, 53, 5121–5124. [Google Scholar] [CrossRef]
- Lee, J.; Hua, B.; Park, S.; Ha, M.; Lee, Y.; Fan, Z.; Ko, H. Tailoring surface plasmons of high-density gold nanostar assemblies on metal films for surface-enhanced Raman spectroscopy. Nanoscale 2014, 6, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Zrimsek, A.B.; Henry, A.I.; Van Duyne, R.P. Single molecule surface-enhanced Raman spectroscopy without nanogaps. J. Phys. Chem. Lett. 2013, 4, 3206–3210. [Google Scholar] [CrossRef]
- Das, G.; Chirumamilla, M.; Toma, A.; Gopalakrishnan, A.; Zaccaria, R.P.; Alabastri, A.; Leoncini, M.; Di Fabrizio, E. Plasmon based biosensor for distinguishing different peptides mutation states. Sci. Rep. 2013, 3, 1792. [Google Scholar] [CrossRef]
- Chirumamilla, M.; Toma, A.; Gopalakrishnan, A.; Das, G.; Zaccaria, R.P.; Krahne, R.; Rondanina, E.; Leoncini, M.; Liberale, C.; De Angelis, F.; et al. 3D nanostar dimers with a sub-10-nm gap for single-/few-molecule surface-enhanced Raman scattering. Adv. Mater. 2014, 26, 2353–2358. [Google Scholar] [CrossRef]
- Gopalakrishnan, A.; Chirumamilla, M.; De Angelis, F.; Toma, A.; Zaccaria, R.P.; Krahne, R. Bimetallic 3D nanostar dimers in ring cavities: Recyclable and robust surface-enhanced Raman scattering substrates for signal detection from few molecules. ACS Nano 2014, 8, 7986–7994. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, H.; Ma, L.; Zou, S.; Ling, Y.; Zhang, Z. Highly stable and active SERS substrates with Ag–Ti alloy nanorods. Nanoscale 2018, 10, 19863–19870. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Stepula, E.; Philippi, M.; Schlücker, S.; Steinhart, M. Evaluation of 3D gold nanodendrite layers obtained by templated galvanic displacement reactions for SERS sensing and heterogeneous catalysis. Nanoscale 2018, 10, 20671–20680. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Li, P.; Zhou, G.; Chen, S.; Han, W.; Qin, F.; Nie, Y.; Wang, Y.; Qin, M.; Huang, G.; et al. General surface-enhanced Raman spectroscopy method for actively capturing target molecules in small gaps. J. Am. Chem. Soc. 2021, 143, 7769–7776. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Han, H.S.; Choi, I.; Lee, C.; Hong, S.; Suh, S.H.; Lee, L.P.; Kang, T. Interfacial liquid-state surface-enhanced Raman spectroscopy. Nat. Commun. 2013, 4, 2182. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Liang, W.; Sun, Y.; Huang, J.; Ma, W.; Liang, Z.; Bao, Q.; Jiang, L. Facile fabrication of high-density sub-1-nm gaps from Au nanoparticle monolayers as reproducible SERS substrates. Adv. Funct. Mater. 2016, 26, 8137–8145. [Google Scholar] [CrossRef]
- Tian, L.; Su, M.; Yu, F.; Xu, Y.; Li, X.; Li, L.; Liu, H.; Tan, W. Liquid-state quantitative SERS analyzer on self-ordered metal liquid-like plasmonic arrays. Nat. Commun. 2018, 9, 3642. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, M.; Zhang, L.; Tian, Y. A liquid interfacial SERS platform on a nanoparticle array stabilized by rigid probes for the quantification of norepinephrine in rat brain microdialysates. Angew. Chem. Int. Ed. 2022, 61, e202117125. [Google Scholar]
- Liu, J.; Yin, D.; Wang, S.; Chen, H.Y.; Liu, Z. Probing Low-Copy-Number Proteins in a Single Living Cell. Angew. Chem. Int. Ed. 2016, 55, 13215–13218. [Google Scholar] [CrossRef]
- Liu, J.; Wen, Y.; He, H.; Chen, H.Y.; Liu, Z. Probing cytoplasmic and nuclear microRNAs in single living cells via plasmonic affinity sandwich assay. Chem. Sci. 2018, 9, 7241–7246. [Google Scholar] [CrossRef]
- Tian, T.; Yi, J.; Liu, Y.; Li, B.; Liu, Y.; Qiao, L.; Zhang, K.; Liu, B. Self-assembled plasmonic nanoarrays for enhanced bacterial identification and discrimination. Biosens. Bioelectron. 2022, 197, 113778. [Google Scholar] [CrossRef]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; Wu, D.Y.; et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.D.; Liu, B.J.; Li, J.F.; Yang, Z.L.; Ren, B.; Tian, Z.Q. SHINERS and plasmonic properties of Au Core SiO2 shell nanoparticles with optimal core size and shell thickness. J. Raman Spectrosc. 2013, 44, 994–998. [Google Scholar] [CrossRef]
- Duan, H.; Hu, H.; Kumar, K.; Shen, Z.; Yang, J.K. Direct and reliable patterning of plasmonic nanostructures with sub-10-nm gaps. ACS Nano 2011, 5, 7593–7600. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Xia, B.; Kravchenko, D.E.; Tietze, M.L.; Cruz, A.J.; Stassen, I.; Hauffman, T.; Teyssandier, J.; De Feyter, S.; Wang, Z.; et al. Direct X-ray and electron-beam lithography of halogenated zeolitic imidazolate frameworks. Nat. Mater. 2021, 20, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Scherrer, D.; Vogel, D.; Drechsler, U.; Olziersky, A.; Sparr, C.; Mayor, M.; Lörtscher, E. Monitoring Solid-Phase Reactions in Self-Assembled Monolayers by Surface-Enhanced Raman Spectroscopy. Angew. Chem. Int. Ed. 2021, 60, 17981–17988. [Google Scholar] [CrossRef]
- Schröder, T.; Trusheim, M.E.; Walsh, M.; Li, L.; Zheng, J.; Schukraft, M.; Sipahigil, A.; Evans, R.E.; Sukachev, D.D.; Nguyen, C.T.; et al. Scalable focused ion beam creation of nearly lifetime-limited single quantum emitters in diamond nanostructures. Nat. Commun. 2017, 8, 15376. [Google Scholar] [CrossRef] [PubMed]
- Park, S.G.; Xiao, X.; Min, J.; Mun, C.; Jung, H.S.; Giannini, V.; Weissleder, R.; Maier, S.A.; Im, H.; Kim, D.H. Self-assembly of nanoparticle-spiked pillar arrays for plasmonic biosensing. Adv. Funct. Mater. 2019, 29, 1904257. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Son, J.; Kim, J.M.; Lee, J.; Yoo, S.; Haddadnezhad, M.; Shin, J.; Kim, J.; Nam, J.M.; et al. Web-above-a-Ring (WAR) and Web-above-a-Lens (WAL): Nanostructures for Highly Engineered Plasmonic-Field Tuning and SERS Enhancement. Small 2021, 17, 2101262. [Google Scholar] [CrossRef]
- Flauraud, V.; Mastrangeli, M.; Bernasconi, G.D.; Butet, J.; Alexander, D.T.; Shahrabi, E.; Martin, O.J.; Brugger, J. Nanoscale topographical control of capillary assembly of nanoparticles. Nat. Nanotechnol. 2017, 12, 73–80. [Google Scholar] [CrossRef]
- Li, J.; Wuethrich, A.; Sina, A.A.; Cheng, H.H.; Wang, Y.; Behren, A.; Mainwaring, P.N.; Trau, M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat. Commun. 2021, 12, 1087. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, M.; Zhou, F.; Liu, D.; Liu, G.; Duan, G.; Cai, W.; Li, Y. Physical deposition improved SERS stability of morphology controlled periodic micro/nanostructured arrays based on colloidal templates. Small 2015, 11, 844–853. [Google Scholar] [CrossRef]
- Luo, S.; Mancini, A.; Wang, F.; Liu, J.; Maier, S.A.; de Mello, J.C. High-throughput fabrication of triangular nanogap arrays for surface-enhanced Raman spectroscopy. ACS Nano 2022, 16, 7438–7447. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ai, B.; Wang, Z.; Chen, C.; Zhang, W.; Wang, Y.; Zhang, G. In situ chemical patterning technique. Adv. Funct. Mater. 2022, 32, 2107945. [Google Scholar] [CrossRef]
- Zheng, C.; Shen, Y.; Liu, M.; Liu, W.; Wu, S.; Jin, C. Layer-by-layer assembly of three-dimensional optical functional nanostructures. ACS Nano 2019, 13, 5583–5590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Feng, L.; Dong, B.; Xu, T.; Li, D.; Jiang, L.; Chi, L. Tape-imprinted hierarchical lotus seedpod-like arrays for extraordinary surface-enhanced Raman spectroscopy. Small 2019, 15, 1804527. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wu, Z.; Li, S.; Zhao, W.; Ma, C.; Wang, J.; Jiang, Z.; Zhong, Z.; Zheng, Y.; Yang, X. Large-area Au-nanoparticle-functionalized Si nanorod arrays for spatially uniform surface-enhanced Raman spectroscopy. ACS Nano 2017, 11, 1478–1487. [Google Scholar] [CrossRef]
- Kamil Reza, K.; Wang, J.; Vaidyanathan, R.; Dey, S.; Wang, Y.; Trau, M. Electrohydrodynamic-induced SERS immunoassay for extensive multiplexed biomarker sensing. Small 2017, 13, 1602902. [Google Scholar] [CrossRef]
- Lai, Y.; Schlücker, S.; Wang, Y. Rapid and sensitive SERS detection of the cytokine tumor necrosis factor alpha (tnf-α) in a magnetic bead pull-down assay with purified and highly Raman-active gold nanoparticle clusters. Anal. Bioanal. Chem. 2018, 410, 5993–6000. [Google Scholar] [CrossRef]
- Sánchez-Purrà, M.; Carré-Camps, M.; de Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Surface-enhanced Raman spectroscopy-based sandwich immunoassays for multiplexed detection of Zika and Dengue viral biomarkers. ACS Infect. Dis. 2017, 3, 767–776. [Google Scholar] [CrossRef]
- Hu, S.W.; Qiao, S.; Pan, J.B.; Kang, B.; Xu, J.J.; Chen, H.Y. A paper-based SERS test strip for quantitative detection of Mucin-1 in whole blood. Talanta 2018, 179, 9–14. [Google Scholar] [CrossRef]
- Tran, V.; Walkenfort, B.; König, M.; Salehi, M.; Schlücker, S. Rapid, quantitative, and ultrasensitive point-of-care testing: A portable SERS reader for lateral flow assays in clinical chemistry. Angew. Chem. Int. Ed. 2019, 58, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ruan, Q.; Lei, Z.C.; Lin, S.C.; Zhu, Z.; Zhou, L.; Yang, C. Highly sensitive and automated surface enhanced Raman scattering-based immunoassay for H5N1 detection with digital microfluidics. Anal. Chem. 2018, 90, 5224–5231. [Google Scholar] [CrossRef]
- Liu, J.; He, H.; Xie, D.; Wen, Y.; Liu, Z. Probing low-copy-number proteins in single living cells using single-cell plasmonic immunosandwich assays. Nat. Protoc. 2021, 16, 3522–3546. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Wen, Y.; Chen, J.; Guo, Z.; Li, P.; Liu, Z. Probing Protein 4′-Phosphopantetheinylation in Single Living Cells. Anal. Chem. 2023, 95, 7229–7236. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Zhao, J.; He, H.; Zhao, Q.; Liu, Z. Multiplexed single-cell plasmonic immunoassay of intracellular signaling proteins enables non-destructive monitoring of cell fate. Anal. Chem. 2021, 93, 14204–14213. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, P.; Tu, X.; Liu, J.; Wang, Y.; Liu, Z. Molecularly imprinted plasmonic substrates for specific and ultrasensitive immunoassay of trace glycoproteins in biological samples. ACS Appl. Mater. Interfaces 2017, 9, 12082–12091. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Xing, R.; Chen, J.; Liu, J.; Li, W.; Liu, Z. Orthogonal dual molecularly imprinted polymer-based plasmonic immunosandwich assay: A double characteristic recognition strategy for specific detection of glycoproteins. Biosens. Bioelectron. 2019, 145, 111729. [Google Scholar] [CrossRef]
- Xing, R.; Wen, Y.; Dong, Y.; Wang, Y.; Zhang, Q.; Liu, Z. Dual molecularly imprinted polymer-based plasmonic immunosandwich assay for the specific and sensitive detection of protein biomarkers. Anal. Chem. 2019, 91, 9993–10000. [Google Scholar] [CrossRef]
- Pang, J.; Li, P.; He, H.; Xu, S.; Liu, Z. Molecularly imprinted polymers outperform lectin counterparts and enable more precise cancer diagnosis. Chem. Sci. 2022, 13, 4589–4597. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Q.; Xing, R.; Liu, Z. Molecularly imprinted and cladded polymers for constructing a portable plasmonic immunoassay for peptides in biofluids. Chem. Commun. 2023, 59, 3075–3078. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, X.; Li, P.; Lin, X.; Wang, J.; Hu, Z.; Zhang, P.; Chen, D.; Cai, H.; Niessner, R.; et al. Ultrasensitive and simultaneous SERS detection of multiplex microRNA using fractal gold nanotags for early diagnosis and prognosis of hepatocellular carcinoma. Anal. Chem. 2021, 93, 8799–8809. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Tian, Y.F.; Yin, B.C.; Ye, B.C. Simultaneous surface-enhanced Raman spectroscopy detection of multiplexed microRNA biomarkers. Anal. Chem. 2017, 89, 6120–6128. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Kim, W.H.; Lee, H.S.; Park, K.H.; Sim, S.J. Quantitative and specific detection of exosomal miRNAs for accurate diagnosis of breast cancer using a surface-enhanced Raman scattering sensor based on plasmonic head-flocked gold nanopillars. Small 2019, 15, 1804968. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wang, C.; Lu, L.; Wang, C.; Sun, Z.; Xiao, R. Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer. Biosens. Bioelectron. 2019, 130, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, Q.; Wang, C.; Pang, Y.; Sun, Z.; Xiao, R. In situ exosomal MicroRNA determination by target-triggered SERS and Fe3O4@ TiO2-based exosome accumulation. ACS Sens. 2021, 6, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, J.; Li, B.; Liu, J.; Xu, J.J.; Chen, H.Y. Dual-mode SERS and electrochemical detection of miRNA based on popcorn-like gold nanofilms and toehold-mediated strand displacement amplification reaction. Anal. Chem. 2021, 93, 6120–6127. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, J.; Dong, Y.; Li, W.; Xing, R.; Ma, Y.; Liu, Z. Gold nanoparticle-decorated Ag@SiO2 nanocomposite-based plasmonic affinity sandwich assay of circulating microRNAs in human serum. ACS Appl. Nano Mater. 2019, 2, 3960–3970. [Google Scholar] [CrossRef]
- Xie, D.; Wen, Y.; Chen, J.; Lu, H.; He, H.; Liu, Z. Probing Queuosine Modifications of Transfer RNA in Single Living Cells via Plasmonic Affinity Sandwich Assay. Anal. Chem. 2022, 94, 12828–12835. [Google Scholar] [CrossRef]
- Zhou, Q.; Zheng, J.; Qing, Z.; Zheng, M.; Yang, J.; Yang, S.; Ying, L.; Yang, R. Detection of circulating tumor DNA in human blood via DNA-mediated surface-enhanced Raman spectroscopy of single-walled carbon nanotubes. Anal. Chem. 2016, 88, 4759–4765. [Google Scholar] [CrossRef]
- Fu, X.; Cheng, Z.; Yu, J.; Choo, P.; Chen, L.; Choo, J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016, 78, 530–537. [Google Scholar] [CrossRef]
- Wu, L.; Xiao, X.; Chen, K.; Yin, W.; Li, Q.; Wang, P.; Lu, Z.; Ma, J.; Han, H. Ultrasensitive SERS detection of Bacillus thuringiensis special gene based on Au@ Ag NRs and magnetic beads. Biosens. Bioelectron. 2017, 92, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Freedman, E.; Odion, R.A.; Strobbia, P.; De Silva Indrasekara, A.S.; Vohra, P.; Taylor, S.M.; Vo-Dinh, T. Direct detection of unamplified pathogen RNA in blood lysate using an integrated lab-in-a-stick device and ultrabright SERS nanorattles. Sci. Rep. 2018, 8, 4075. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Wu, L.; Zong, S.; Yun, B.; Cui, Y. Combining multiplex SERS nanovectors and multivariate analysis for in situ profiling of circulating tumor cell phenotype using a microfluidic chip. Small 2018, 14, 1704433. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Teixeira, A.; Garrido-Maestu, A.; Muinelo-Romay, L.; Lima, L.; Santos, L.L.; Prado, M.; Diéguez, L. Profiling DNA mutation patterns by SERS fingerprinting for supervised cancer classification. Biosens. Bioelectron. 2020, 165, 112392. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lyu, N.; Rajendran, V.K.; Piper, J.; Rodger, A.; Wang, Y. Sensitive and direct DNA mutation detection by surface-enhanced Raman spectroscopy using rational designed and tunable plasmonic nanostructures. Anal. Chem. 2020, 92, 5708–5716. [Google Scholar] [CrossRef] [PubMed]
- Moitra, P.; Chaichi, A.; Hasan, S.M.A.; Dighe, K.; Alafeef, M.; Prasad, A.; Gartia, M.R.; Pan, D. Probing the mutation independent interaction of DNA probes with SARS-CoV-2 variants through a combination of surface-enhanced Raman scattering and machine learning. Biosens. Bioelectron. 2022, 208, 114200. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Shin, M.; Yang, L.; Conley, B.; Yoon, J.; Lee, S.N.; Lee, K.B.; Choi, J.W. Clustered regularly interspaced short palindromic repeats-mediated amplification-free detection of viral DNAs using surface-enhanced Raman spectroscopy-active nanoarray. ACS Nano 2021, 15, 13475–13485. [Google Scholar] [CrossRef]
- Yeh, Y.T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef]
- Park, K.S.; Choi, A.; Kim, H.J.; Park, I.; Eom, M.S.; Yeo, S.G.; Son, R.G.; Park, T.I.; Lee, G.; Soh, H.T.; et al. Ultra-sensitive label-free SERS biosensor with high-throughput screened DNA aptamer for universal detection of SARS-CoV-2 variants from clinical samples. Biosens. Bioelectron. 2023, 228, 115202. [Google Scholar] [CrossRef]
- Kamińska, A.; Witkowska, E.; Winkler, K.; Dzięcielewski, I.; Weyher, J.L.; Waluk, J. Detection of Hepatitis B virus antigen from human blood: SERS immunoassay in a microfluidic system. Biosens. Bioelectron. 2015, 66, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, L.; Zhang, F.; Song, Z.; Hu, Y.; Ji, Y.; Shen, J.; Li, B.; Lu, H.; Yang, H. A promising magnetic SERS immunosensor for sensitive detection of avian influenza virus. Biosens. Bioelectron. 2017, 89, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Camacho, S.A.; Sobral-Filho, R.G.; Aoki, P.H.B.; Constantino, C.J.L.; Brolo, A.G. Zika immunoassay based on surface-enhanced Raman scattering nanoprobes. ACS Sens. 2018, 3, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Park, S.G.; Choi, N.; Moon, J.I.; Dang, H.; Das, A.; Lee, S.; Kim, D.G.; Chen, L.; Choo, J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020, 167, 112496. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Zhao, Z.; Lian, K.; Yin, L.; Wang, J.; Man, S.; Liu, G.; Ma, L. SERS-based CRISPR/Cas assay on microfluidic paper analytical devices for supersensitive detection of pathogenic bacteria in foods. Biosens. Bioelectron. 2022, 207, 114167. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS based lateral flow immunoassay for point-of-care detection of SARS-CoV-2 in clinical samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Sitjar, J.; Liao, J.D.; Lee, H.; Tsai, H.P.; Wang, J.R.; Liu, P.Y. Challenges of SERS technology as a non-nucleic acid or-antigen detection method for SARS-CoV-2 virus and its variants. Biosens. Bioelectron. 2021, 181, 113153. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B 2021, 329, 129196. [Google Scholar] [CrossRef]
- Zhang, M.; Li, X.; Pan, J.; Zhang, Y.; Zhang, L.; Wang, C.; Yan, X.; Liu, X.; Lu, G. Ultrasensitive detection of SARS-CoV-2 spike protein in untreated saliva using SERS-based biosensor. Biosens. Bioelectron. 2021, 190, 113421. [Google Scholar] [CrossRef]
- Chen, H.; Park, S.G.; Choi, N.; Kwon, H.J.; Kang, T.; Lee, M.K.; Choo, J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sens. 2021, 6, 2378–2385. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.X.; Leong, Y.X.; Tan, E.X.; Sim, H.Y.F.; Koh, C.S.L.; Lee, Y.H.; Chong, C.; Ng, L.S.; Chen, J.R.T.; Pang, D.W.C.; et al. Noninvasive and point-of-care surface-enhanced Raman scattering (SERS)-based breathalyzer for mass screening of coronavirus disease 2019 (COVID-19) under 5 min. ACS Nano 2022, 16, 2629–2639. [Google Scholar] [CrossRef]
- Peng, Y.; Lin, C.; Li, Y.; Gao, Y.; Wang, J.; He, J.; Huang, Z.; Liu, J.; Luo, X.; Yang, Y. Identifying infectiousness of SARS-CoV-2 by ultra-sensitive SnS2 SERS biosensors with capillary effect. Matter 2022, 5, 694–709. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.g.; Choi, B.H.; Kang, K.W.; Jeong, H.; Park, Y.; et al. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Siddhanta, S.; Zheng, G.; Kickler, T.; Barman, I. Rapid, Label-free Optical Spectroscopy Platform for Diagnosis of Heparin-Induced Thrombocytopenia. Angew. Chem. Int. Ed. 2020, 59, 5972–5978. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Su, X.; Wen, Y.; Zheng, C.; Li, M. Artificial intelligent label-free SERS profiling of serum exosomes for breast cancer diagnosis and postoperative assessment. Nano Lett. 2022, 22, 7910–7918. [Google Scholar] [CrossRef]
- Shu, W.; Zhang, M.; Zhang, C.; Li, R.; Pei, C.; Zeng, Y.; Zhao, L.; Zhao, J.; Wan, J. An Alloy Platform of Dual-Fingerprints for High-Performance Stroke Screening. Adv. Funct. Mater. 2023, 33, 2210267. [Google Scholar] [CrossRef]
- Huang, L.; Sun, H.; Sun, L.; Shi, K.; Chen, Y.; Ren, X.; Ge, Y.; Jiang, D.; Liu, X.; Knoll, W.; et al. Rapid, label-free histopathological diagnosis of liver cancer based on Raman spectroscopy and deep learning. Nat. Commun. 2023, 14, 48. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Luo, R.; Popp, J.; Bocklitz, T. Deep learning for Raman spectroscopy: A review. Analytica 2022, 3, 287–301. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, S.; Guo, Z.; Liu, Z. Recent Advances in Rational Design and Engineering of Signal-Amplifying Substrates for Surface-Enhanced Raman Scattering-Based Bioassays. Chemosensors 2023, 11, 461. https://doi.org/10.3390/chemosensors11080461
Gao S, Guo Z, Liu Z. Recent Advances in Rational Design and Engineering of Signal-Amplifying Substrates for Surface-Enhanced Raman Scattering-Based Bioassays. Chemosensors. 2023; 11(8):461. https://doi.org/10.3390/chemosensors11080461
Chicago/Turabian StyleGao, Song, Zhanchen Guo, and Zhen Liu. 2023. "Recent Advances in Rational Design and Engineering of Signal-Amplifying Substrates for Surface-Enhanced Raman Scattering-Based Bioassays" Chemosensors 11, no. 8: 461. https://doi.org/10.3390/chemosensors11080461
APA StyleGao, S., Guo, Z., & Liu, Z. (2023). Recent Advances in Rational Design and Engineering of Signal-Amplifying Substrates for Surface-Enhanced Raman Scattering-Based Bioassays. Chemosensors, 11(8), 461. https://doi.org/10.3390/chemosensors11080461






