Abstract
Molecularly imprinted polymers (MIPs) and aptamers (Apts) are widely used in substance detection due to their specific recognition abilities. However, both of them have limitations in terms of stability or sensitivity. Therefore, an increasingly employed strategy is to combine MIPs and aptamers to form mixed components for detecting various substances, such as viruses, bacteria, proteins, heavy-metal ions, and hormones. The aim of this review is to provide a comprehensive summary of the scientific research conducted on the construction and application of aptamer–MIP multiple-recognition components in the past five years. It also aims to analyze their research and development strategies, construction mechanisms, advantages, and potential applications, as well as limitations and current challenges that need to be addressed.
1. Introduction
The molecular recognition theory has been widely applied in various fields, including separation [1], sensing [2], material manufacturing [3], disease diagnosis [4], and drug delivery [5], due to its significant practical value. Nature provides a plethora of biometric pairs, such as glycans and glycan-binding proteins [6], enzymes and substrates [7], and antigens and antibodies [8]. They are widely used for detecting target molecules because of their outstanding specific recognition and binding ability. However, the identification of target molecules during application is often hindered by environmental interference. Additionally, their practical application is hampered by natural molecular instability, high costs, modification complexities, and limitations on mild conditions. Therefore, it is crucial to develop molecular recognition systems with enhanced properties.
In recent years, molecularly imprinted polymers have emerged as a cost-effective and highly stable alternative [9] to traditional molecular recognition systems, offering superior reusability and performance. Molecular imprinting is a biomimetic technology that utilizes template molecules and crosslinking agents to form three-dimensional cavity structures [10] with specific selection and high affinity through polymerization. Based on the “key-and-lock” principle [11], the obtained cavity can be used for the selective binding of targeted molecules, thus achieving high sensitivity. MIPs have been successfully applied in various scientific research fields, such as environmental pollutant adsorption, biomimetic sensing [12], and solid-phase extraction. However, the complexity of actual samples may result in the retention of large molecules like proteins on the surface of MIPs, leading to the blockage of specific recognition sites. Additionally, solutions still need to be sought for the limited analytical window [13] and limitations posed by non-polar environments.
Aptamers are short RNA/DNA oligonucleotides that specifically bind to targeted complementary molecules [14]. These molecules were first reported by Ellington [15] and Gold [16] in 1990. Aptamers possess several advantages, including high specificity, facile modification, small size, non-immunogenicity [17], and high affinity. As a potential substitute for natural antibodies, aptamers have been widely used in biosensors, clinical diagnosis and therapy, imaging technology, and cellular immunity. Particularly in the field of sensors, aptamer-based biosensors exhibit superior biocompatibility and reproducible functions [18] compared to other types of sensors, such as natural antibody sensors. However, the presence of nucleases everywhere can cause the rapid degradation of aptamers [19], leading to the loss of their three-dimensional structure and function. Meanwhile, there is a need to improve the thermal stability of aptamers and enhance the diversity of the functional groups they possess [20,21,22].
To address the functional limitations of MIPs and aptamers when used alone, researchers have proposed a hybrid detection system that combines both materials to achieve inherent affinity [23]. The aptamer–MIP hybrid creates a “best of both worlds” scenario. When the aptamer is made to be polymerizable, it can become the recognition part of a MIP by incorporating it into the polymer matrix via polymerizable groups on the aptamer. In this way, the high affinity and specificity of the aptamer are preserved whilst imparting the robustness and added shape specificity generated by the MIPs. The presence of the polymer should protect the aptamer from environmental degradation and potentially widen the scope of use of aptamers for recognition. An aptamer could also be used as an additional recognition monomer and further amplify the sensing signal of the system as a marker. By combining the specificity of MIPs and aptamers, a hybrid can have high sensitivity, a low limit of detection, high stability, and excellent detection results. The successful development of aptamer–MIP hybrid materials would pave the way for a new generation of MIPs and for their use to become more mainstream, offering significant commercial gain. In 2014, Bai and Spivak [24] successfully verified this idea by using the molecularly imprinted aptamer multiple-recognition strategy to construct an efficient virus detection sensor. Since then, more researchers have developed a plethora of hybrids [25,26] by combining the advantages of MIPs and aptamers, which have been effectively employed in detecting antibiotics, viruses, hormones, and other molecules. This review summarizes the preparation methods of the multiple-recognition imprinting layer and MIP–aptamer, as well as comprehensively reviews the application of MIP–aptamer recognition systems in various sensor applications (Figure 1). Furthermore, it discusses and analyzes current shortcomings and future prospects for this technology. This review is expected to provide researchers with a clearer understanding of the advantages of multiple-recognition systems based on molecular imprinting and aptamers while encouraging them to seek solutions for future challenges.
Figure 1.
Overview of different types of aptamer–MIP multiple-recognition systems.
2. Acquisition of Molecularly Imprinted Layer
In the aptamer–MIP multiple-recognition system, molecular-imprinting sites play a crucial role in recognition. These sites refer to three-dimensional imprinting cavities obtained through the elution of polymerized imprinting molecules, and the number of these sites depends on the effectiveness of the polymerization process. Currently, thermal polymerization, photopolymerization, the sol–gel method, and electrochemistry are commonly used for polymerizing molecularly imprinted films. Thermal polymerization is generally carried out by heating in a water bath with methacrylic acid (MAA) as a functional monomer, EDGMA (ethylene glycol di-methacrylate) as a crosslinking agent, and AIBN (azodiisobutyronitrile) as an initiator. The severe limitation of the method is the requirement for an organic solvent. This presents obvious drawbacks to their use in environmental and biological applications. Photopolymerization, on the other hand, is typically performed by triggering free radicals using either a high-pressure mercury lamp or a purple interconnector to complete the polymerization of the imprinted layer. The photopolymerization method is relatively easy to operate; however, it has the following disadvantages: long polymerization time, expensive photoinitiators, and poor surface regeneration ability. The sol–gel method possesses numerous significant advantages, such as being simple and low-cost, requiring mild conditions, and providing products with high thermal and mechanical stability [27]. The sol–gel imprinting process occurs by dissolving a metal oxide precursor or metal halides (M(OR)n or MXn, with M = Si, Al, Ti, etc.) in a low-molecular-weight solvent medium using a catalyst (acid, base, or ions such as F−), followed by a hydrolysis (water) and polycondensation step [28]. Silicon alkoxides have been regarded as the preferred molecular precursors for the sol–gel process because they have moderate reactivity but the chemical stability of the Si–C bond is high. During the molecular-imprinting process, alkoxysilane units (Si–OR) are transformed into silanol (Si–OH) with the release of alcohol (R–OH), and then the condensation reaction occurs with the formed silanol group. Both the hydrolysis and condensation reactions will continue simultaneously, leading to the formation of a three-dimensional network. However, the low capacity hinders its wide application. Electrically driven polymerization methods are mainly used for the fabrication of MIP films directly on a conducting substrate. Electropolymerization involves placing a pre-treated electrode into a solution containing a template and specific functional monomers, which undergo electrolysis to generate free radicals under the influence of an electric current. Polymerization is then completed in the presence of template molecules. The mechanism is simply explained as an alternation between chemical and electrode reaction steps. A radical cation is most likely formed during the electro-oxidation step, and then the radical reacts with the monomer and the protonated dimer of the radical formed. These steps follow one after another to form the polymer [29]. The electropolymerization method is low-cost and allows for adjusting the thickness and morphology of the imprinted film while also exhibiting excellent reproducibility. Although there are numerous advantages, the absence of a crosslinker during the electropolymerization process results in fewer recognition sites and poor recognition ability [30].
3. Multiple-Recognition Systems Based on Aptamer and MIPs
3.1. Sandwich Type
In a sandwich assay, the target is bound between a capture antibody and a detection antibody. The capture antibody is immobilized on a surface, while the detection antibody (conjugated to an enzyme or fluorophore label) is applied as the last step before quantitation. Because two antibodies against the same antigen are used, this method is flexible and sensitive. However, it is not always easy or possible to have pairs of antibodies that work well in this type of assay. As described in the introduction section, MIPs and aptamers can be obtained at a large scale using simple methods. They are good candidates for antibodies. The sandwich type consists of a MIP/target/aptamer [31], which combines the specific recognition capabilities of MIPs and aptamers to achieve signal amplification [32]. The MIP material acts as a pre-concentration part for the targets, while the aptamer is responsible for signaling the presence of the target on the MIP platform. The costs and time of MIP and aptamer production are substantially low. Meanwhile, incorporating nanoparticles into this approach serves to independently amplify the signal intensity following secondary recognition, thereby achieving stable and enhanced sensing of signal output. Based on this strategy, Li Tang et al. [33] developed a sensor for the ultrasensitive visual detection of EV71 (Enterovirus 71), which is a plus-stranded RNA virus that causes hand-foot-mouth disease and central nervous system infection [34]. The fabrication process of the sensor is shown in Figure 2. Firstly, Fe3O4-coated CD (carbon quantum dot) carriers were constructed, and MIPs were prepared using the sol–gel method with APTES (3-aminopropyltriethoxysilane) and TEOS (ethyl silicate) as functional monomers and crosslinkers. Then, the aptamers were introduced into the imprinting layer. The surface of the imprinting layer was modified with EV71 aptamers, and phenolphthalein was coated on the surface of ZIF-8 (Zeolitic Imidazolate Framework-8). When EV71 was captured by imprinted particles and combined with ZIF-8, the fluorescence signal was quenched, achieving the first signal amplification. Then, when the pH of the solution was adjusted to 12, ZIF-8 decomposed and released loaded phenolphthalein molecules, turning the solution system red and achieving the second signal amplification. Efficient visual detection can be achieved within 20 min of sensor application, with fluorescence and visual detection limits reaching 8.33 fM and 2.08 pM, respectively. The repeatability and stability of this sensor are commendable. After conducting five repeated determination experiments on the same sensor, the fluorescence signal maintains 82% of the initial value. Additionally, even after being stored for 6 months, the sensor still maintains a satisfactory color-rendering function, with an absorbance level at 91.9% of the initial value. Compared with traditional virus detection methods, which are time-consuming, expensive, and insensitive, this sensor allows for low-cost and highly accurate clinical diagnosis from the beginning. The research team’s work is expected to be used in future virus-screening efforts to reduce medical staff workload and clinical diagnosis costs.
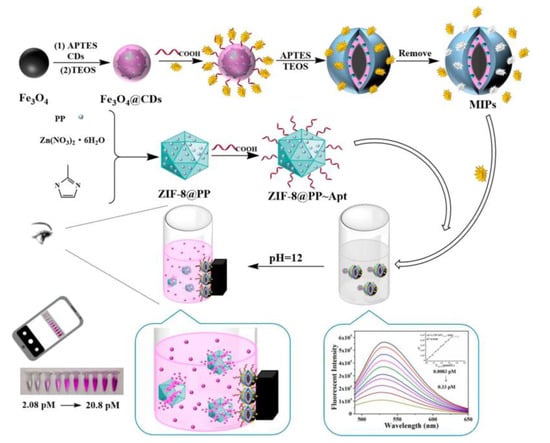
Figure 2.
Construction principle and flow diagram of the virus sensor. Reprinted with permission from ref. [33].
Exosomes, which are extracellular vesicles composed of nanoscale lipid bilayers, have emerged as promising biomarkers for early cancer diagnosis and monitoring due to their ability to provide rich molecular information similar to that of parental cells. However, the lack of efficient methods for separating and capturing intact exosomes has hindered the widespread application of exosome analysis platforms in clinical practice. To address this issue, Liao’s team [35] developed a more sensitive “on” fluorescent MIP sensor for detecting lysozyme, which is one of the innate immunity proteins carried by the intact exosome. The captured lysozyme was detected through selective fluorescence induced by aptamer-mediated aggregation. A linear relationship between the lysozyme content and fluorescence intensity was obtained. The specific working process is illustrated in Figure 3, where, in the first step, both lysozyme and characteristic exosomes from the sample are added to the surface polymerization system. After the reaction of the template and crosslinker, customized complementary cavities are formed. In the second step, AIMIP (“Artificial intelligence” imprinted polymers) particles selectively capture and enrich targeted analytes. In the third step, after magnetic collection is completed, the aptamers are incubated with a MIP. Utilizing a sandwich induction strategy, the sensor selectively captures AIIP target molecules and modulates the spin of water-soluble AIE (aggregation-induced emission) fluorescence agents through conformational changes in DNA aptamers. The fluorescence effects are enhanced, enabling the specific detection of lysozyme and exosomes within complex proteins through selective “on” fluorescent lighting. This technology has been preliminarily applied to the detection of clinical serum samples. The fluorescence intensity showed a strong linear correlation with the exosome concentration (0.9823), indicating a significant relationship between the two variables. Moreover, the detection limit was approximately 1.3 × 106 capsules/mL, demonstrating high sensitivity and accurate quantification capabilities. Additionally, the sensor exhibited good reproducibility, with coefficients of variation of 2.10 × 10−7 and 2.38 × 10−5, corresponding to percentages of 6% and 4.5%, respectively. Furthermore, the recovery rate in repeated tests reached as high as 107%, highlighting its potential application in future medical fields.
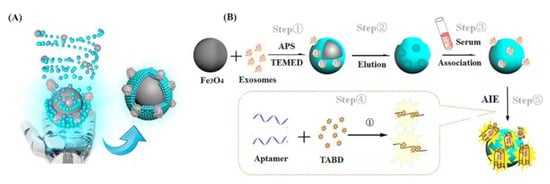
Figure 3.
(A) Conceptual graph of self-assembly of AIMIPs; (B) schematic of the preparation of AIMIPs and AIMIP separation integrated aptamer/AIE fluorescent sensors for exosomes. Reprinted with permission from ref. [35].
In conclusion, the use of a MIP and an aptamer in the sandwich system greatly improves the specific recognition ability of the target and realizes the secondary amplification of the signal, which would improve the sensitivity of the sensor. However, the susceptibility of the aptamer to thermal, chemical, and enzymatic degradation is still the main bottleneck limiting its wide application because of the lack of protective shells around aptamers. Further work should be conducted to improve the stability of aptamers.
3.2. Hybrid Type
Unlike the sandwich method, this hybrid strategy involves directly embedding the aptamer into the molecularly imprinted structure. After the hybrid structure is formed, the molecularly imprinted polymer acts as a protective scaffold for the aptamer molecule, confining it to a specific conformation and effectively separating it from potential interference from DNA enzymes present in the substrate. At the same time, the aptamer can bind to the target molecule specifically to make up for the deficiency of the sensitivity of the molecular imprinting itself. This enhances the stability and specificity of aptamer-mediated recognition. The interaction forces, such as a hydrogen bond, between the target molecule and MIP greatly improve the affinity between the recognition system and the target molecule.
This strategy has been successfully applied to the detection of antibiotics, such as amoxicillin (AMOX). In 2021, Lu’s group [36] developed a sensor that specifically detects amoxicillin using a co-deposition technique and a dopamine electropolymerization technique. The construction scheme of the sensor was as follows: (1) a large-surface-area conductive sensing platform was constructed by electrodepositing AuNPs (Au nanoparticles) on the electrode surface; (2) the aptamer–amoxicillin complex was fixed on the electrode surface with the Au-S bond; (3) the molecular-imprinting layer was prepared by dopamine electropolymerization; (4) the imprinted cavity was obtained after elution. CV (cyclic voltammetry) and DPV (differential pulse voltammetry) techniques were used for characterization and quantification by using Fe(CN)63−/Fe(CN)64− as probes. Compared to the curve depicted in Figure 4, Curve a presents a pair of standard REDOX peaks. Curve b shows a significant increase in the peak current, indicating that AuNPs/ZnO-rGO can accelerate the electron transfer rate on the electrode surface. In contrast, Curve c exhibits a decrease in peak value due to the formation of a self-assembled insulation layer on the electrode surface, which renders the molecular-imprinting layer non-conductive and impedes electron transfer. Curve d shows the peak current after electropolymerizing dopamine to obtain the blotting layer. Because the imprinting layer is non-conductive, the charge transfer of the probe is blocked, resulting in a significant decrease in the peak current. However, after the removal of template molecules, leaving behind an imprinting cavity, aptamer–AMOX probes can reach the electrode surface through pores and complete electron transfer, resulting in an increase in the peak value for Curve e. Finally, upon the completion of the recognition process and the occupation of imprinting sites, charge transfer is hindered again, leading to a reduction in the peak current for Curve f. During the process of sensor construction, the author utilized an aptamer as a functional monomer, combining its recognition ability with a molecular-imprinting layer to reduce non-specific recognition in complex systems. Additionally, it was discovered that the optimal mole ratio of the aptamer to the template molecule is 1:2. By using this ratio to construct the sensor system, the best detection effect can be achieved, with a detection limit of 3.3 × 10−15 M. Three electrodes were selected under identical conditions to construct a MIEAS for the determination of AMOX at equimolar concentrations, yielding an RSD of 2.99% upon repeated experimentation. Furthermore, in order to assess sensor stability, response signals from the same electrode were measured after 30 days and found to be 95.1% of their initial values. These results demonstrate that the constructed MIEAS exhibits excellent repeatability and stability. However, in order to optimize the utilization of this sensor system, further research on electrode material development is necessary due to its poor selectivity.
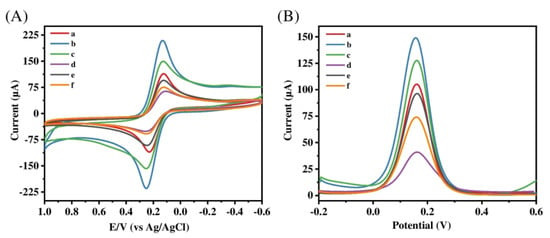
Figure 4.
(A) CV and (B) DPV for (a) bare GCE, (b) AuNPs/ZnO-rGO/GCE, (c) Apt-AMOX/AuNPs/ZnO-rGO/GCE, (d) MIP-Apt/AuNPs/ZnO-rGO before elution, (e) MIP-Apt/AuNPs/ZnO-rGO after elution, and (f) MIP-Apt/AuNPs/ZnO-rGO after adsorption in 5 mM [Fe(CN)6]3−/4− and 0.1 M KCl (CV scan rate: 100 mV·s−1, scan range: −0.2–+0.6 V; DPV scan range: −0.2–+0.6 V). Reprinted with permission from ref. [36].
Taking Krishnan’s work [37] as an example, hemophilia B, a genetic disease that can cause cardiovascular disease and paralysis, is often clinically diagnosed by the human coagulation factor IX protein (FIX). The authors designed the MIP embedded with the aptamer as a biomimetic biosensor, which can quickly and sensitively recognize FIX specifically. The author simultaneously utilized the resilience of the molecularly imprinted polymer layer to mitigate biological contamination on the electrode surface. The preliminary work of the research and development process is shown in Figure 5. (a) Human coagulation FIX-imprinted polymer was prepared by free radical initiation polymerization, and an aptamer–target complex was obtained by incubating the aptamer and imprinted template. (b) The working electrode was fabricated through the process of aluminum deposition. Then, the working electrode was modified with carbon nanomaterials and metal nanomaterials, and the aptamer–target complex was fixed on the modified electrode. After the modification, the performance of the electrode was greatly improved in terms of conductivity, sensitivity, and affinity. The amperometry method was used for the detection of FIX. Compared to a traditional Apt sensor without MIPs, it was discovered that the MIP-incorporated sensor has a 3-fold higher sensitivity and a 40 fM detection limit. However, experimental data show that the aptamer–MIP hybrid AIDE’s (Archimedean Interdigitated Sensor) detection capability is limited to the decreasing current response as surface mass loads are increased. Based on the author’s research, it can be inferred that potential advancements for this strategy primarily involve incorporating nanomaterials, eliminating impurities from the electrode surface [38], introducing additional affinity detection monomers to enhance sensitivity, and enabling the detection of various substances through the modification of a single component with the same synthesis approach.
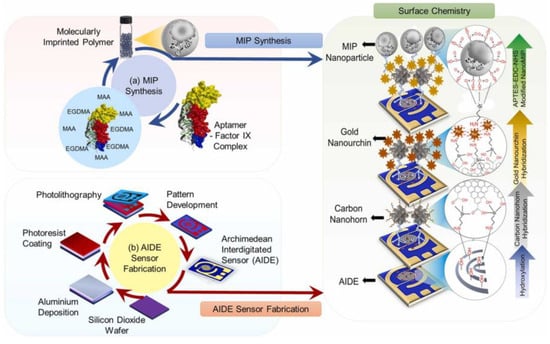
Figure 5.
Schematic illustration of the overall research. It comprised of MIP synthesis (a), AIDE sensor fabrication by photolithography process (b). Reprinted with permission from ref. [37].
For the hybrid assay, the affinity and specificity of the prepared aptamer–MIP hybrid toward the target were improved dramatically as compared with the aptamer or MIP alone. However, the hybrid process is still based on a trial-and-error process. The monomer in the imprinting process may not be ideal, leading to a low imprinting factor. Rational computer modeling would help to improve the aptamer–MIP hybrid preparation and make the hybrid more efficient.
3.3. Recapture–Detection–Separate Sensing Strategy
Unlike the sandwich and hybrid modes, the recapture–detection–separate sensing strategy incorporates a microfluidic control system with miniature valves on the chip. By regulating the opening and closing of the valves, it detects signals produced by target molecules as they flow through narrow channels. In this system, the molecularly imprinted polymer and aptamer function as two distinct recognition elements. Once captured and eluted by the MIP site, analytes are transferred to the aptamer for secondary capture. A furan sensor is taken as an example, as furan is an insecticide widely used in agriculture, but its residue poses a serious threat to human health. Although the commonly used methods for detecting furan include chromatography [39,40] and electrochemical methods [41,42], due to the operational complexity and poor targeting selectivity of these two technologies, there is a general trend to develop a new type of sensor. Under such requirements, Li et al. [43] developed an interesting design idea by etching a microfluidic channel, and two functional regions were etched on polydimethylsiloxane (PDMS). These two functional areas serve as recognition areas for the MIP and aptamer. In this sensing process, the target furan molecule is initially transported through the microchannel and subsequently captured by the MIP recognition site. Following elution, the furan is conveyed to the aptamer’s functional region, where it undergoes a secondary capture event facilitated by DNA aptamer binding. The Pt-Au-Ag/AgCl three-electrode system generates a current signal at the second capture, while GO (graphene oxide)-AuNPs amplify the detected signal. This research group realized the high-sensitivity detection of furan in complex samples, and the detection limit was up to 67 pM. After optimizing the chip preparation process, this strategy is expected to expand to the detection of other harmful substances and promote the development process in the field of sensors. In this method, molecular imprinting and the aptamer work independently without interference. However, the production process of the working chip for microfluidics is often costly, and the construction of microchannels and recognition units remains a technical problem that needs improvement. The unoccupied imprinting sites on the chip are prone to binding with aptamers and generating interference signals. Therefore, future research should focus on improving controllability.
4. Applications of MIP–Aptamer Multiple-Recognition Systems
4.1. Virus
Viruses are microorganisms with the smallest and simplest structures, yet their types are complex. Most viruses exhibit strong infectivity and can cause life-threatening diseases in severe cases. Although vaccines are available for most viruses, an outbreak can still result in significant damage to the economy and public health, as seen during the three-year pandemic caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) worldwide [44]. Enhanced virus detection is a crucial element in managing and resolving epidemics. Currently, the enzyme-linked immunosorbent assay [2] and the isolation culture and detection of virus strains [45] are often used for virus-related detection, but these methods can be expensive and susceptible to complex substrates, resulting in low sensitivity and poor selectivity. To overcome these shortcomings, Chen S.Y. [46] designed a molecularly imprinted polymer–aptamer sensor for the non-autofluorescence detection of the H5N1 virus (Influenza A virus). In this work, the author fixed three functional monomers, including acrylamide, to the modified magnetic Fe3O4 and used AIBN to initiate polymerization. The H5N1 virus template was embedded into the polymer, and the subsequent elution of viral molecules yielded an imprinted cavity with selective recognition for the H5N1 virus, serving as the identification probe. Additionally, the aptamer was immobilized onto amino-modified ZGO (Zn2GeO4:Mn2+) via amide bonding to serve as a secondary recognition probe. When the target H5N1 virus particle is introduced into the recognition system, it is captured by the imprinting chamber to form MIP-H5N1. Furthermore, the cavity selectively captures ZGO-H5N1 Apt from the solution, resulting in the formation of a magnetic sandwich structure; however, nanoparticles without a sandwich structure remain in the solution system. As the concentration of virus molecules increases, there is a continuous decrease in the residual aptamer amount, leading to changes in the persistent photoluminescence (PL) signal. The correlation curve between the PL signal and template molecule concentration was obtained. Through these processes, a sandwich sensor with a detection limit of 1.16 fM was constructed.
Hepatitis B virus is a common blood-borne pathogen that can lead to death from cirrhosis and liver cancer in carriers [47]. To detect this virus, Chen S.Y. [48] introduced a resonance light-scattering sensor in 2021 based on molecular imprinting and an aptamer for the early diagnosis of hepatitis B virus (HBV) using the same sandwich strategy, as shown in Figure 6. The imprinting factor of the sensor reached 7.56, surpassing other molecularly imprinted sensors in virus detection. The sensing system employs a molecular-imprinting layer supported by carbon spheres as probe 1 and utilizes a specific aptamer as probe 2. Simultaneously, glucose is introduced during the construction of the imprinting vector to generate hydroxyl and carboxyl groups on the carrier’s surface, thereby enhancing hydrogen bonding and improving the binding capacity of the imprinting layer toward the target virus molecules. The successful binding of the imprinted probe to the target molecule generates a resonant light-scattering response. Upon the double binding of the aptamer to the target molecule, this light signal is further enhanced, as depicted in Figure 7, demonstrating excellent detection efficacy. The authors’ study indicates that an imprinting layer of 25 nm thickness made with 2.5 μL mL−1 TEOS yields the optimal signal strength and imprinting effect. When the imprinting layer is too thin, it results in a loose network structure for the constructed imprinting cavity, which lacks specificity. Meanwhile, the thickness of the imprinting layer should be carefully considered during the construction of the dual-recognition system to prevent excessive crosslinking, as an excessively thick layer hinders the removal of target molecules from the imprinting cavity, thereby impeding secondary signal generation by aptamers. Only an appropriate thickness of the imprinting layer can ensure sufficient recognition sites for aptamer binding. The ratio of MIPs and SiO2@Apt was also optimized. A dosage ratio of MIPs and SiO2@Apt of 1:2 and a dosage of MIPs of 80 ng/mL−1 were used. In general, this sensing approach holds significant implications for the timely detection of a wide range of viruses.
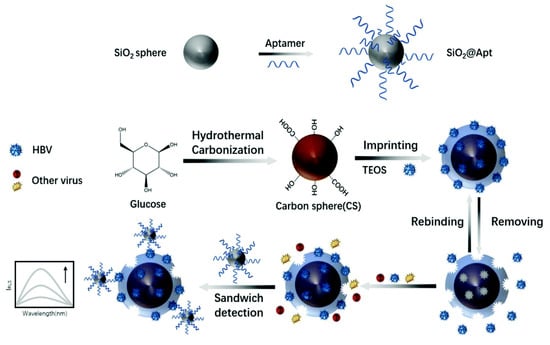
Figure 6.
Principle of the preparation of virus–MIPs. Reprinted with permission from ref. [48].
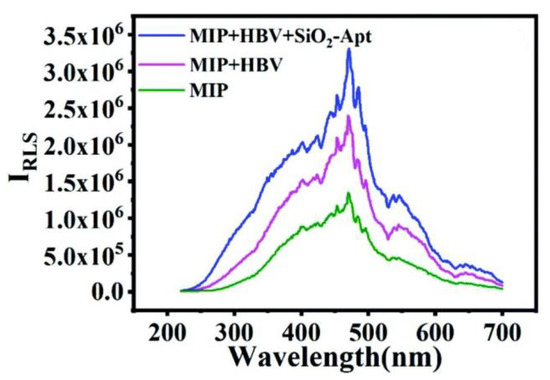
Figure 7.
Detection possibility verification: MIP, MIP + HBV, and MIP + HBV + SiO2-Apt. Reprinted with permission from ref. [48].
Hepatitis C virus (HCV) is also a causative agent of liver disease. In Ghanbari’s work [49], the aptamer was fixed on a multi-wall carbon-nanotube–chitosan nanocomposite (MWCNT-CHIT), and after the imprinting layer was obtained by dopamine electropolymerization, the HCV core antigen sensor was prepared. When the aptamer binds to the target molecule, the peak current is gradually increased by the DPV method, and then the antigen changes conformation to overcome the electrostatic effect during the process of separation from the electrode surface, so the current density changes with the antigen concentration. The method combines the high stability of the nanocomposite platform, the specific recognition capability provided by the MIP–aptamer, and the excellent sensitivity of the hybrid receptor to successfully detect HCV core antigens in human serum, maintaining an initial response of 95% and a relative standard deviation of 5.4% after 50 cycles, with good repeatability. These findings demonstrate the effectiveness of this method in detecting HCV and its potential for widespread application in analyzing complex samples.
4.2. Carcinogens
As one of the most lethal diseases, cancer is caused by various carcinogenic factors, among which aflatoxin is included. Aflatoxin B1 (AFB1) [50] is the representative, which has the highest yield, the highest toxicity, and the strongest carcinogenicity. Once AFB1 enters the food chain, it can result in significant economic losses and pose a grave threat to human health through biological accumulation. In 2022 and 2023, Roushani [51] and Chi Hai [52] put forward a summary of the work on the multiple-recognition strategy for AFB1 detection. In Roushani’s study, a novel electrochemical biological device with a detection limit of 12.0 pg/L was designed by hybridizing MIPs and aptamers on a glassy carbon electrode modified with Cu2O NCs (nanocubes) with a large surface area. The experimental data were measured by the impedance method. In Chi’s work, the recognition probe MIP/PC (porous carbon) and signal probe CdTe/ZnS-Apt were used, which have good matching and stable fluorescence signals. In this detection process, the intensity of the fluorescence signal is determined by the synthesis ratio of CdTe/ZnS and Apt, and the concentration is insufficient to bind to all modified aptamers in the sensor, resulting in fluorescence interference. When the optimal concentration of CdTe/ZnS was 0.8 mg/mL and the Apt content was 10 nmol/L, the fluorescence intensity reached its maximum efficacy. The results show that there is satisfactory compatibility and synergy between the aptamer and the molecular-imprinting layer, so the detection of AFB1 is realized with a very low detection limit of 4.0 pg/mL−1. The simultaneous determination of 10 ng/mL samples using five identical sensors has a relative standard deviation of 4.1%. In addition, after three times of repeated use, the sensor system showed a fluorescence signal reduction of only 9.7%, which indicates that the sensor has great commercial potential in the food industry.
Ochratoxin (OTA) is another mycotoxin that has attracted worldwide attention. The toxicity, carcinogenicity, and teratogenicity of OTA seriously harm human health [53]. In view of the fact that the aptamer–MIP materials developed in the past have not been applied to the detection of OTA, Lyu [54] first proposed a novel aptamer–MIP material for OTA-specific recognition in 2019. In this study, APT-OTA complexes were obtained by mixing an aptamer with a concentration of 60 μmolL−1 with template molecules, and then the raw materials, such as the monomer, crosslinker, initiator, and complex, were placed in the purple linkage instrument to complete photopolymerization. The hydrogen bonding between the monomer AMPS (2-acrylamide-2-methylpropanesulfonic acid) and template OTA and the specific affinity of the aptamer greatly increased the adsorption effect. Based on these advantages, the material was successfully applied to actual beer samples, and the OTA-sensitive LOD and LOQ were measured to be 0.07 ng/mL−1 and 0.14 ng/mL−1, respectively, realizing the specific enrichment and high-sensitivity detection of OTA. To evaluate the stability of the aptamer–MIP monolithic column, changes in flow velocity were measured in five mobile phases with different polarities. The linear correlation coefficients R2 of these five groups of data are in the range of 0.9908~0.9985, indicating that the measurement precision and accuracy are high. Furthermore, after one month of use, the recovery rate of aptamer–MIP was found to be 87.3%, demonstrating excellent mechanical stability and reproducibility. However, as the components have an impact on the overall pore structure and permeability of the aptamer–MIP, the next step is to optimize the composition of the polymeric mixture in order to comprehensively enhance the performance of this multiple-recognition material.
There are heavy-metal ions [55] with potent carcinogenicity, among which the bivalent chromium ion is a representative. Upon entering the human body, cadmium will affect the formation of human bone cells, leading to deformation and pain and inducing cancer and even death in patients in severe cases [56]. To achieve the effective detection of Cd2+, Li [57] devised a fluorescence-quenching sensor. The team fixed SN-CQD/Au (carbon quantum dots co-doped with sulfur and nitrogen atoms) fluorescent particles on the surface of the indium tin oxide glass electrode, and then they combined the self-assembled aptamer–Cd2+ complex with fluorescent particles and obtained the molecular-imprinting sites by electropolymerization. Two signal units, molecular imprinting and the aptamer, are used to show two signals in the detection. For electrical signals, when Cd2+ is captured by the imprinting cavity formed, the diffusion channel will be blocked due to the density of the MIP structure, and the charge transfer will be blocked, showing a high impedance value. The fluorescence response signal is caused by the quenching of Cd2+. When the aptamer binds to the gold nanoparticles, it will show a strong fluorescence signal, but because the polymerized MIP layer on the electrode surface will cover the original structure after binding Cd2+, resulting in the partial absorption and refraction of the fluorescence, it will show weakened signal intensity. Based on the specific affinity of MIP and the aptamer in the sensor for Cd2+, interference and non-specific recognition are greatly reduced. The actual measurement results show that the method has a good resolution, and the detection limit is 1.2 × 10−12 mol/L−1. To ensure the reproducibility and stability of the sensors, a relative standard deviation of 2.65% was achieved for 5.0 × 10−9 mol/L−1 Cd2+ using nine sensors under identical conditions, while a relative standard deviation of 3.14% was obtained for fluorescence signals when utilizing the same sensors. Furthermore, after storing the sensor in an environment at 4 °C for twenty days, there was a decrease in the fluorescence signal of 7.48% compared to its initial value; these results demonstrate that this study’s sensor system exhibits exceptional performance. However, due to the unsatisfactory number of aptamers and target metal ions, the sensor must overcome this limitation in order to improve its adsorption capacity.
4.3. Antibiotics
Antibiotics are either metabolites or synthetic analogs produced by microorganisms. The most common classes of antibiotics include beta-lactams, quinolones, and aminoglycosides. Antibiotics are utilized to treat infections caused by pathogenic bacteria and have widespread use in clinical medicine. However, due to widespread use and even abuse, bacteria gradually adapt to the environment where antibiotics exist and develop resistance, which has become a potential threat to global health [58,59]. Lincomycin is a kind of amide drug with an antibacterial effect against bacteria, especially Gram-positive bacteria, and also protozoans bioactive [60]. Li [61] proposed a new method for detecting lincomycin based on the energy resonance transfer between the Au-GO nanomaterial and C-dots. In the absence of target molecules, C-dots exhibit an enhanced light signal after energy absorption. After the introduction of the target molecule, the aptamer and MIP compete to bind with lincomycin within the sensor system. Due to the structural change in the aptamer after binding with lincomycin, the electron supply and energy transfer between GO-Au and C-dots are hindered, resulting in a decrease in the electrochemical luminescence signal strength in the monitoring system. Through this signal change, the trace detection of lincomycin can be directly detected. At the same time, this experiment shows that when the adsorption time is 8 min and the solution pH is maintained at 8.0, the current intensity is the best. Based on this signal change, the method can achieve multiple identifications with high resolution, and the detection limit is 1.6 × 10−13 mol/L.
Kanamycin (KAN) is an aminoglycoside antibiotic commonly employed for the treatment of Gram-positive bacterial infections. However, its neurotoxicity and nephrotoxicity necessitate the development of an efficient sensor for monitoring kanamycin levels. In response to this societal need, Bi [62] proposed an electrochemical detection strategy in 2019. In this study, Fe3O4 material was loaded with gold nanoparticles and modified with a KAN aptamer. The imprinted cavity formed by pre-polymerization on the electrode surface selectively captures and recognizes kanamycin molecules in the substrate, followed by the specific binding of the aptamer to kanamycin during sensor application. The sensor system goes through two processes of capture and binding to generate electrical signals, which are quickly detected by cyclic voltammetry. The presence of sufficient binding sites on the electrode surface enables the sensor to have a wide linear range. Furthermore, when the molar ratio of the template molecule to the functional monomer is 1:2, the physical properties and performance of the material are the best, the current response is the largest, and the recovery rate is 96.5% to 101.5% when applied to the detection of groundwater samples. For the stability and repeatability of the sensor system, the relative standard deviation of the corresponding current obtained by the determination of 100 nM KAN by the DPV method is 3.0%. At the same time, the detached electrode was stored at a low temperature for 2 days and measured again. After 7 repeated cycles, the relative deviation was only 1.8% compared with the initial peak value. These data confirm that the stability and repeatability of the research work are excellent.
Later in 2022, there was also a new testing strategy for tetracycline (TC), another threat in the antibiotic family. Tetracycline is a natural antibiotic produced by actinomycetes [63] and has important applications in aquaculture and other fields due to its antibacterial activity. However, the accumulation of tetracycline in the environment inevitably leads to the emergence of drug-resistant strains. Ma [64] reported a magnetic material for separating and enriching tetracycline. As shown in Figure 8, the authors utilized aptamers and β-CD (β-cyclodextrin) as replacements for traditional functional monomers and employed free radical polymerization to construct molecular-imprinting recognition sites on the outer layer of the material. In practical applications, the functional monomer modified on the material works cooperatively with the imprinting chamber to selectively adsorb tetracycline. The magnetic properties of the material enable the adsorbed magnet to remove tetracycline from the substrate, thereby achieving TC enrichment. Compared with other molecularly imprinted materials, the material has a core–shell structure, which ensures ecological friendliness and non-toxicity. This effectively isolates the aptamer from nucleases in the environment and prevents inactivation due to enzymatic hydrolysis. Due to these advantages, the material has a blot factor of 7.6 and a detection limit of 1.0 μg/L−1 by UV spectroscopy, which shows the special potential for the trace detection of TC in complex samples. In the selection of a material for testing, reusability and stability are crucial parameters for evaluating its practicality. The adsorption capacity of the material was assessed after six cycles of use, yielding a result of 84.6% compared to the initial treatment. This demonstrates the excellent reusability of the material. Furthermore, the sustained adsorption effectiveness, even after repeated usage, indicates that the constructed imprinted cavity remains intact and exhibits remarkable stability. Additionally, TC was detected in five samples with varying concentrations under identical experimental conditions, resulting in an RSD value of 0.81% (n = 5), confirming the method’s exceptional reproducibility and stability. In summary, this study presents a novel approach for utilizing molecularly imprinting technology in environmental protection and has promising applications for detecting and enriching other hazardous substances in the environment.
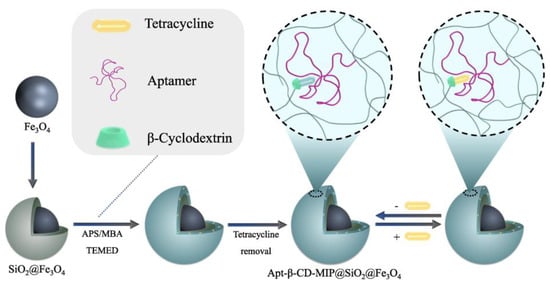
Figure 8.
Schematic illustration of the Apt-β-CD-MIP@SiO2@Fe3O4 preparation and its application to detect TC. Reprinted with permission from ref. [64].
4.4. Bacteria
As the most abundant organisms in nature, bacteria are widely distributed in soil and water. On the one hand, bacteria are closely related to human production and life, such as food production, digestion and absorption, battery manufacturing, and antibiotic extraction. On the other hand, these prokaryotes pose a serious threat to human health due to their diversity of infectivity and transmission routes. Pseudomonas aeruginosa is an opportunistic bacterium that is one of the main sources of infection in hospitals. It is more common in postoperative patients, and infected individuals may experience a range of uncomfortable symptoms, including death in severe cases [65]. Therefore, it is imperative to develop a rapid method for detecting Pseudomonas aeruginosa. In Sarabaegi’s work [66], they designed an electrochemical sensor for multiple molecular recognition. The preparation process is shown in Figure 9: (1) gold nanoparticles are deposited on the surface of the glassy carbon electrode; (2) the aptamer is modified on the electrode surface, and the molecularly imprinted polymerization layer is obtained by the electrochemical polymerization of dopamine; (3) the eluting layer obtains the cavity and adds the target for detection. In order to observe the cavity obtained after elution, FE-SEM (Field-Emission Scanning Electron Microscope) was used to analyze the morphology and structure of the electrode. According to the scanning images, the removal of template molecules made the surface of the molecular-imprinting layer rougher, and a large number of imprinting cavities were observed, indicating that the elution process was successful, and identification sites with a satisfactory number and specific surface area were obtained. During the construction of the recognition system, when gold nanoparticles are deposited on the exposed glassy carbon electrode, these nanoparticles increase the electrode area, and the current peak value is increased. Then, when the aptamer is modified on the electrode surface, the carboxylate group in the aptamer generates a repulsive force to reduce the current peak value. Finally, when the molecular-imprinting site captures the target molecule and undergoes desorption, the peak current is enhanced again, and by observing the changes in this series of currents, the direct detection of Pseudomonas aeruginosa can be realized. The detection limit of the sensor is 1 CFU/mL−1.
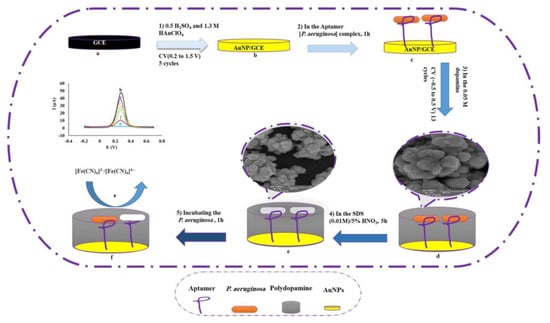
Figure 9.
Schematic diagram of aptamer–MIP preparation for detection of P. aeruginosa. Reprinted with permission from ref. [66].
Staphylococcus aureus is a prevalent pathogenic bacterium that exhibits robust antibacterial resistance and frequently induces pneumonia and sepsis in humans [67,68]. The simple sensor constructed by El-Wekil using electrical analysis technology shows outstanding application potential [69]. In this study, El-Wekil immobilized aptamers onto composite materials via Au-S bonds and electropolymerized imprinting layers on the material surface using o-phenylenediamine as a functional monomer in the presence of Staphylococcus aureus. As the recognition sites formed are complementary to the target molecules, the binding of S. aureus to the imprinted cavity and aptamer can result in a decrease in the peak current on the sensing electrode, thereby achieving the effective separation of S. aureus from complex matrices. In this study, the MIP–aptamer dual-identification system was successfully implemented for the first time to identify Staphylococcus aureus. The linear range of the sensor is 101 to 107 CFU mL−1, and the detection limit LOD is 1 CFU mL−1. Given its robust detection capabilities and high recovery rates, we anticipate widespread adoption of this technology in the future.
4.5. Protein
Proteins are substances that possess specific spatial conformations, participate in the formation of vital components within the human body, and serve as primary mediators of organismal life processes. Given the functional diversity exhibited by human proteins, characteristic protein detection can effectively facilitate disease diagnosis and screening. For example, the medical diagnosis of Alzheimer’s disease is often based on amyloid-beta oligomers (AβOs) [70]. In 2020, You Min [31] proposed a diagnostic strategy that could be used as an alternative to these methods. As shown in Figure 10, the author introduced silver and silicon dioxide nanoparticles, followed by the self-assembly of the aptamer on silica particles, thereby accomplishing the construction of a sandwich structure on the MIP membrane. Once the imprinted cavity captures AβOs, the captured object is transferred to the aptamer for secondary binding, resulting in electron transfer and electrical signal transmission within the three-dimensional imprinted cavity. This method combines site-specific binding with aptamer affinity toward targeted molecules, enabling highly sensitive electrochemical signals to be obtained even when a small number of AβOs enter the detection system. During the construction process of this detection system, the number of imprinted cavities is directly determined by the concentration of the template, which, in turn, determines the number of captured template molecules. Only when an appropriate concentration of template molecules is used can a stable current response signal be generated. Additionally, it is important to consider the concentration of the AβO-specific aptamer added to the system. When its concentration exceeds 5 μM, excessive aptamers will inhibit the transmission of electrons and weaken the current signal. It was determined that the combination of 1 mg/mL−1 Aβ1-42 oligomer and 5μM aptamer is the best concentration choice and shows the best electrochemical signal. The sensor based on this strategy has successfully achieved the microdetection of AβOs in human serum. It exhibits good linearity within the concentration range of 5 pg/mL−1 to 10 ng/mL−1 and a detection limit of 1.22 pg/mL−1.
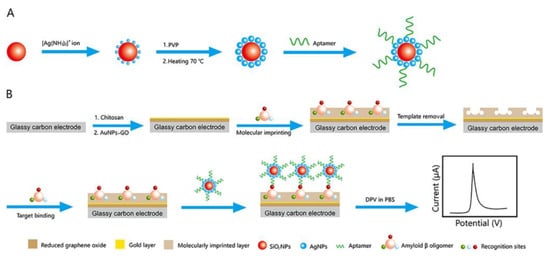
Figure 10.
Schematic illustration of (A) the preparation of the SiO2@Ag–aptamer composite and (B) the fabrication of the MIP-based antibody-free biosensor and the electrochemical detection of AβO via a sandwich-type assay. Reprinted with permission from ref. [31].
Similar strategies have also been applied in the detection of troponin (cTnI), which is a marker of myocardial injury and necrosis and is often used as a diagnostic basis for myocardial infarction [71]. In Mokhtari’s study [72], a non-immune dual-recognition approach was employed to fabricate a sensing platform for cTnI detection. After immobilizing the cTnl aptamer on the electrode surface, the imprinted polymer was obtained by the electropolymerization of methylene blue monomers around the aptamer. During the detection process, cTnl was selectively adsorbed by the hybrid receptor of MIPs and the aptamer due to their selective affinity and cavity effects, respectively. The determination of the adsorbed cTnl was achieved through differential pulse voltammetry. The detection limit of the sensor platform was 1.04 pM (2.61 × 10−5 μg/mL), and the RSD of the sensor system designed in this study was 5.37% for cTnl in a protein mixture containing cardiac troponin, and several electrochemical values obtained in this study demonstrate that the novel hybrid probe developed by the authors possesses superior performance.
In addition, Wang [73] successfully detected alpha-fetoprotein (AFP) and insulin using a nanoprobe. The team utilized static electricity between gold nanoparticles and the abundant amino groups, as well as Au-S bonds, to increase the fixed number of aptamers on the substrate. Additionally, a synergistic interaction between the imprinted cavity and aptamer lowered the detection limit. When insulin is used as the template molecule, the detection limit reaches approximately 0.5 ng/mL−1, whereas when AFP serves as the template molecule, this magnetic nanoprobe can detect AFP in samples ranging from 1000 ng/mL−1 to 20 ng/mL−1. It is worth mentioning that this study demonstrates the controllability of preparing an imprinting layer through the sol–gel method, as evidenced by the direct relationship between the consumption time and thickness change in the SiO2 coating. The optimal coating time of 90 min results in the highest specific recognition ability. These findings highlight the significant potential of our magnetic nanoprobe for the efficient detection of disease serum protein markers.
To expand the detection range, Yang [74] developed a novel electrochemiluminescence (ECL) “signal on” sensor platform for the efficient detection of thrombin, a bioactive protein that plays a crucial role in blood clotting and has significant clinical implications in the treatment of various diseases. In this detection system, the author employed molecularly imprinted nanocavities to bind target molecules to thrombin aptamers. Simultaneously, the synergistic effect between polyetherimide (PEI) and Ru(bpy)32+ (RuNP) is utilized to expedite electron transfer, thereby enhancing the signal strength of the sensing platform by sevenfold compared with conventional methods. The sensor platform exhibits excellent stability and detection efficacy, with the ECL sensor signal strength remaining robust even after ten cycles of testing at concentrations of 10−10 M and 10−12 M, yielding corresponding RSDs of 2.0972% and 2.1509%, respectively. Following thirty days of low-temperature storage, the ECL signal strength remains at approximately 90% of its initial value, while the detection limit is as low as 1.73 × 10−15 M, indicating significant potential for application in food safety monitoring. It is worth mentioning that previous molecularly imprinted aptamer sensors are the signal closure type, but the signal flow type was used in this study, which provides a new research direction for the construction of a new small-molecule detection platform in the future.
4.6. Others
In view of the remarkable benefits and immense potential of the multiple-recognition strategy based on aptamer–MIPs, researchers have increasingly adopted this approach. In addition to well-known molecular categories, even relatively obscure substances such as cortisol, bisphenol A [75], and histamine can be detected using this method.
Histamine (HIS) is a hydrophilic biogenic amine that is produced by bacterial fermentation and is commonly found in food. Studies have shown that high levels of histamine in food can indirectly lead to Gram-negative bacterial infections. In order to realize highly sensitive detection of HIS in canned tuna and human serum, Mahmoud [76] developed a nanosensor. Based on molecular-imprinting polymerization technology and DNA aptamer affinity characteristics, the MIP-apt/AuNPs/cCNTs/GCE architecture was fabricated by the surface modification of gold nanoparticles and carbonylated carbon nanotubes on the glassy carbon electrode, followed by the covalent immobilization of the thiolate aptamer and the electropolymerization of phenylenediamine. The system was applied to the detection of plasma and canned tuna. The detection limit was 0.11 nmol, the recovery rate was 96.2~105.2%, and the RSD% was 95.3~104.4%. These data confirmed that this detection system has higher accuracy and precision for the analysis of HIS in complex components.
Cortisol is a steroid hormone involved in the regulation of human metabolism. In the medical field, cortisol detection is frequently utilized for the diagnosis and treatment of depression and post-traumatic stress disorder. Due to the similarity in detecting cortisol levels between saliva and blood samples, Cheng [77] proposed a sensing method with a detection limit as low as 3.3 × 10−13 M. In this study, a bare glassy carbon electrode was initially coated with a dispersion of graphene and carbon quantum dots. Subsequently, the aptamers were immobilized on the electrode surface through electrostatic interaction. Finally, chitosan was utilized as a raw material to form an imprinting layer, and cortisol molecules were eluted to obtain the imprinting cavity. In this experiment, the authors’ success can be attributed to two factors. Firstly, the combination of graphene and carbon quantum dots increases the number of active sites, resulting in a more sensitive detection method. Secondly, optimization of the concentration of the template molecule (1μM) and the molar ratio of the template molecule to aptamer (1:2) further enhances sensitivity. The developed sensor is reported to outperform other methods for detecting cortisol.
As a steroid hormone, progesterone is often used for oral contraception. However, excessive progesterone released into the environment will enter the human body along with water circulation. In order to achieve the trace analysis of this substance, an ultrasensitive sensor platform was established based on SnO2-Gr and AuNPs by Huang Yan [78]. The construction process of the sensor is as follows. Firstly, SnO2-Gr is modified on the exposed glassy carbon electrode to increase the conductivity and specific surface area, then gold nanoparticles are electrodeposited, and aptamers are fixed by Au-S bonds. Finally, the imprinted cavity is obtained by electropolymerization and the elution process. The sensor platform exhibits excellent performance with a low detection limit (1.73 × 10−15 M) while achieving a relative standard deviation (RSD) of 2.79% at a concentration of 10−10 M using this hybrid system. Furthermore, the DPV current was monitored every 10 days after storage at low temperatures, and even after four cycles, the DPV value remained at an impressive 93.19% of its initial value, demonstrating the exceptional repeatability and stability of the developed sensor. Thus, it possesses immense potential for effective detection in the field of food safety.
Table 1 lists the MIP layer formation methods, recognition systems, and detection methods of aptamer–MIP hybrid systems in recent years. The results show that aptamer–MIP hybrids can be used as superior recognition elements for the detection of various targets.

Table 1.
Summary of polymerization methods, recognition systems, and detection methods of aptamer–MIP hybrid systems.
5. Conclusions
In summary, we review the work conducted in recent years on multiple-recognition systems utilizing MIPs and aptamers. Aptamers and molecular imprinting have made remarkable contributions to sensing as highly favorable detection elements. Numerous researchers have demonstrated that combining the specific recognition ability of molecular-imprinting technology with the affinity of aptamers can significantly enhance detection effectiveness. This recognition strategy finds applications in various fields, including medical treatment, environmental monitoring, and agricultural production. In comparison to a single-recognition-element approach, a multi-recognition strategy offers improved selectivity, sensitivity, stability, and reusability. Nevertheless, it still encounters numerous challenges.
5.1. Limitations
In the process of constructing a multiple-recognition system, the principle of the recognition mechanism is not perfect, the polymerization process is uncontrollable, the repeatability is unstable, and conformational changes will occur in the process of imprinting sites and the aptamer-specific recognition and capture of target molecules. In addition, the identification of large water-soluble biomacromolecules remains problematic because conformational integrity cannot be guaranteed during synthesis. These challenges hinder the wide application of multiple-recognition strategies.
5.2. Challenges
The method requires further refinement, including finding the best molecular-imprinting film thickness, optimizing the molar concentration ratio of the template molecule to the aptamer, enhancing the electron transfer effect on the electrode, finding a suitable electrode modifier to multiply the signal, finding a method to freely switch monomers to detect different or similar substances, and improving the commercial application range of the technology. This strategy has shown great promise in environmental monitoring, disease prevention, and medical diagnosis and treatment, and we believe that this approach, as researchers continue to optimize it, will open up a broad avenue for the analysis of multi-component complex components.
Author Contributions
Conceptualization, K.N. and Q.X.; writing—original draft preparation, writing—review and editing, K.N., Y.S., Y.Y., W.X., C.M. and Q.X.; visualization, K.N. and Q.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NSFC (22076161, 21675140 and 21705141), the Talent Support Program of Yangzhou University, Yangzhou University Interdisciplinary Research Foundation for Chemistry Discipline of Targeted Support (yzuxk202009), the project funded by the PAPD and TAPP.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Templates | |
| EV71 | Enterovirus 71 |
| AMOX | Amoxicillin |
| FIX | Factor IX protein |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| H5N1 virus | Influenza A virus |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| AFB1 | Aflatoxin B1 |
| OTA | Ochratoxin |
| KAN | Kanamycin |
| TC | Tetracycline |
| AβOs | Amyloid-beta oligomers |
| cTnI | Troponin |
| AFP | Alpha-fetoprotein |
| HIS | Histamine |
| Functional monomers | |
| Apt | Aptamer |
| AA | Acrylic acid |
| MAA | Methacrylic acid |
| MBA | Methylene diacrylamide |
| MBAA | 5,5′-Methylenedianthranilic acid |
| APTES | 3-Aminopropyltriethoxysilane |
| AMPS | 2-Acrylamide-2-methylpropanesulfonic acid |
| β-CD | β-Cyclodextrin |
| Crosslinking agent | |
| EDGMA | Ethylene glycol dimethacrylate |
| APTES | 3-Aminopropyltriethoxysilane |
| TEOS | Ethyl silicate |
| Patterns | |
| AIMIP | “Artificial intelligence” imprinted polymers |
| AIE | Aggregation-induced emission |
| AIDE | Archimedean Interdigitated Sensor |
| Base | |
| ZIF-8 | Zeolitic Imidazolate Framework-8 |
| PDMS | Polydimethylsiloxane |
| PC | Porous carbon |
| MWCNT-CHIT | Multi-wall carbon-nanotube–chitosan nanocomposite |
| Surface modification | |
| MIPs | Molecularly imprinted polymers |
| CDs | Carbon quantum dots |
| AuNPs | Au nanoparticles |
| GO | Graphene oxide |
| ZGO | Zn2GeO4:Mn2+ |
| NCs | Nanocubes |
| SN-CQD/Au | Carbon quantum dots co-doped with sulfur and nitrogen atoms |
| Others | |
| AIBN | Azodiisobutyronitrile |
| CV | Cyclic voltammetry |
| DPV | Differential pulse voltammetry |
| PL | Persistent luminescence |
| FE-SEM | Field-Emission Scanning Electron Microscope |
| ECL | Electrochemiluminescence |
| PEI | Polyetherimide |
| RuNP | Ru(bpy)32+ |
| EIS | Electrochemical impedance spectroscopy |
References
- Cheng, Y.; Zhao, X.; Zhang, Q.; Li, X.; Wei, Z. Constructing imprinted reticular structure in molecularly imprinted hybrid membranes for highly selective separation of acteoside. Sep. Purif. Technol. 2022, 298, 121572. [Google Scholar] [CrossRef]
- Zhou, J.; Ni, Y.; Wang, D.; Fan, B.; Zhu, X.; Zhou, J.; Hu, Y.; Li, L.; Li, B. Development of a Competitive Enzyme-Linked Immunosorbent Assay Targeting the-p30 Protein for Detection of Antibodies against African Swine Fever Virus. Viruses 2023, 15, 154. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Gao, R.; Tian, X.; Heinlein, J.; Hussain, S.; Pfefferle, L.D.; Chen, X.; Zhang, X.; Hao, Y. Strategic design and fabrication of lightweight sesame ball-like hollow double-layer hybrid magnetic molecularly imprinted nanomaterials for the highly specific separation and recovery of tetracycline from milk. Green Chem. 2022, 24, 8036–8045. [Google Scholar] [CrossRef]
- Feng, D.; Ren, M.; Miao, Y.; Liao, Z.; Zhang, T.; Chen, S.; Ye, K.; Zhang, P.; Ma, X.; Ni, J.; et al. Dual selective sensor for exosomes in serum using magnetic imprinted polymer isolation sandwiched with aptamer/graphene oxide based FRET fluorescent ignition. Biosens. Bioelectron. 2022, 207, 114112. [Google Scholar] [CrossRef] [PubMed]
- Kakkar, V.; Narula, P. Role of molecularly imprinted hydrogels in drug delivery—A current perspective. Int. J. Pharm. 2022, 625, 121883. [Google Scholar] [CrossRef]
- Gao, C.; Wei, M.; McKitrick, T.R.; McQuillan, A.M.; Heimburg-Molinaro, J.; Cummings, R.D. Glycan Microarrays as Chemical Tools for Identifying Glycan Recognition by Immune Proteins. Front. Chem. 2019, 7, 833. [Google Scholar] [CrossRef]
- Fang, Y.Z.; Jiang, L.; He, Q.; Cao, J.; Yang, B. Deubiquitination complex platform: A plausible mechanism for regulating the substrate specificity of deubiquitinating enzymes. Acta Pharm. Sin. B 2023, 13, 2295–2816. [Google Scholar] [CrossRef]
- Kachhawa, P.; Mishra, S.; Jain, A.K.; Tripura, C.; Joseph, J.; Radha, V.; Chaturvedi, N. Antigen-Antibody Interaction-Based GaN HEMT Biosensor for C3G Detection. IEEE Sens. J. 2022, 22, 6256–6262. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Cheng, N.; Luo, Y.; Lin, Y.; Xu, W.; Du, D. Recent advances in nanomaterials-based electrochemical (bio)sensors for pesticides detection. TrAC Trends Anal. Chem. 2020, 132, 116041. [Google Scholar] [CrossRef]
- Cui, M.; Che, Z.; Gong, Y.; Li, T.; Hu, W.; Wang, S. A graphdiyne-based protein molecularly imprinted biosensor for highly sensitive human C-reactive protein detection in human serum. Chem. Eng. J. 2022, 431, 133455. [Google Scholar] [CrossRef]
- Carballido, L.; Karbowiak, T.; Cayot, P.; Gerometta, M.; Sok, N.; Bou-Maroun, E. Applications of molecularly imprinted polymers and perspectives for their use as food quality trackers. Chem 2022, 8, 2330–2341. [Google Scholar] [CrossRef]
- Yarman, A.; Scheller, F.W. Coupling Biocatalysis with Molecular Imprinting in a Biomimetic Sensor. Angew. Chem.-Int. Ed. 2013, 52, 11521–11525. [Google Scholar] [CrossRef] [PubMed]
- Rampey, A.M.; Umpleby, R.J.; Rushton, G.T.; Iseman, J.C.; Shah, R.N.; Shimizu, K.D. Characterization of the imprint effect and the influence of imprinting conditions on affinity, capacity, and heterogeneity in molecularly imprinted polymers using the Freundlich isotherm-affinity distribution analysis. Anal. Chem. 2004, 76, 1123–1133. [Google Scholar] [CrossRef]
- Aljohani, M.M.; Cialla-May, D.; Popp, J.; Chinnappan, R.; Al-Kattan, K.; Zourob, M. Aptamers: Potential Diagnostic and Therapeutic Agents for Blood Diseases. Molecules 2022, 27, 383. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef] [PubMed]
- Nxele, S.R.; Nkhahle, R.; Nyokong, T. The composites of asymmetric Co phthalocyanines-graphitic carbon nitride quantum dots-aptamer as specific electrochemical sensors for the detection of prostate specific antigen. J. Am. Chem. Soc. 2021, 900, 115730. [Google Scholar] [CrossRef]
- Poma, A.; Brahmbhatt, H.; Pendergraff, H.M.; Watts, J.K.; Turner, N.W. Generation of Novel Hybrid Aptamer-Molecularly Imprinted Polymeric Nanoparticles. Adv. Mater. 2015, 27, 750–758. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Rosenthal, M.; Siegl, J.; Ewers, J.; Mayer, G. Customised nucleic acid libraries selection for enhanced aptamer and performance. Curr. Opin. Biotechnol. 2017, 48, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Rothlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Gariano, N.A.; Spivak, D.A. Macromolecular Amplification of Binding Response in Superaptamer Hydrogels. J. Am. Chem. Soc. 2013, 135, 6977–6984. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Spivak, D.A. A Double-Imprinted Diffraction-Grating Sensor Based on a Virus-Responsive Super-Aptamer Hydrogel Derived from an Impure Extract. Angew. Chem.-Int. Ed. 2014, 53, 2095–2209. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ren, J.; Su, L.; Gao, X.; Tang, Y.; Ma, T.; Zhu, L.; Li, J. Novel hybrid probe based on double recognition of aptamer-molecularly imprinted polymer grafted on upconversion nanoparticles for enrofloxacin sensing. Biosens. Bioelectron. 2017, 87, 203–208. [Google Scholar] [CrossRef]
- Zhou, Q.Q.; Xu, Z.G.; Liu, Z.M. Molecularly Imprinting-Aptamer Techniques and Their Applications in Molecular Recognition. Biosensors 2022, 12, 576. [Google Scholar] [CrossRef]
- Moein, M.M.; Abdel-Rehim, A.; Abdel-Rehim, M. Recent Applications of Molecularly Imprinted Sol-Gel Methodology in Sample Preparation. Molecules 2019, 24, 2889. [Google Scholar] [CrossRef]
- Gutiérrez-Climente, R.; Clavié, M.; Dumy, P.; Mehdi, A.; Subra, G. Sol-gel process: The inorganic approach in protein imprinting. J. Mater. Chem. B 2021, 9, 2155–2178. [Google Scholar] [CrossRef]
- Malitesta, C.; Mazzotta, E.; Picca, R.A.; Poma, A.; Chianella, I.; Piletsky, S.A. MIP sensors—The electrochemical approach. Anal. Bioanal. Chem. 2012, 402, 1827–1846. [Google Scholar] [CrossRef]
- Palladino, P.; Bettazzi, F.; Scarano, S. Polydopamine: Surface coating, molecular imprinting, and electrochemistry-successful applications and future perspectives in (bio)analysis. Anal. Bioanal. Chem. 2019, 411, 4327–4338. [Google Scholar] [CrossRef]
- You, M.; Yang, S.; An, Y.; Zhang, F.; He, P. A novel electrochemical biosensor with molecularly imprinted polymers and aptamer-based sandwich assay for determining amyloid-beta oligomer. J. Electroanal. Chem. 2020, 862, 114017. [Google Scholar] [CrossRef]
- Ocana, C.; del Valle, M. Three different signal amplification strategies for the impedimetric sandwich detection of thrombin. Anal. Chim. Acta 2016, 912, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Liang, K.; Wang, L.; Chen, C.; Cai, C.; Gong, H. Construction of an Ultrasensitive Molecularly Imprinted Virus Sensor Based on an “Explosive” Secondary Amplification Strategy for the Visual Detection of Viruses. Anal. Chem. 2022, 94, 13879–13888. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.Y.; Lu, C.Y.; Tsao, K.C.; Shih, H.M.; Cheng, A.L.; Huang, L.M.; Chang, L.Y. Association of EV71 3C polymorphisms with clinical severity. J. Microbiol. Immunol. Infect. 2018, 51, 608–613. [Google Scholar] [CrossRef]
- Liao, Z.; Peng, J.; Chen, S.; Zhang, P.; Chen, H.; Feng, D.; Zhang, T.; Ye, K.; Deng, Y.; Dong, Y.; et al. Sensitive fluorescent sensor for the fuzzy exosomes in serum based on the exosome imprinted polymer sandwiched with aggregation induced emission. Sens. Actuators B-Chem. 2022, 358, 131182. [Google Scholar] [CrossRef]
- Lu, H.; Huang, Y.; Cui, H.; Li, L.; Ding, Y. A molecularly imprinted electrochemical aptasensor based on zinc oxide and co-deposited gold nanoparticles/reduced graphene oxide composite for detection of amoxicillin. Microchim. Acta 2022, 189, 421. [Google Scholar] [CrossRef]
- Krishnan, H.; Gopinath, S.C.; Arshad, M.M.; Zulhaimi, H.I.; Anbu, P.; Subramaniam, S. Molecularly imprinted polymer enhances affinity and stability over conventional aptasensor for blood clotting biomarker detection on regimented carbon nanohorn and gold nanourchin hybrid layers. Sens. Actuators B-Chem. 2022, 363, 131842. [Google Scholar] [CrossRef]
- Shahdost-fard, F.; Roushani, M. Impedimetric detection of trinitrotoluene by using a glassy carbon electrode modified with a gold nanoparticle@fullerene composite and an aptamer-imprinted polydopamine. Microchim. Acta 2017, 184, 3997–4006. [Google Scholar] [CrossRef]
- Ho, I.P.; Yoo, S.J.; Tefera, S. Determination of furan levels in coffee using automated solid-phase microextraction and gas chromatography/mass spectrometry. J. AOAC Int. 2005, 88, 574–576. [Google Scholar]
- Hashemi-Moghaddam, H.; Ahmadifard, M. Novel molecularly-imprinted solid-phase microextraction fiber coupled with gas chromatography for analysis of furan. Talanta 2016, 150, 148–154. [Google Scholar] [CrossRef]
- Dounin, V.; Veloso, A.J.; Schulze, H.; Bachmann, T.T.; Kerman, K. Disposable electrochemical printed gold chips for the analysis of acetylcholinesterase inhibition. Anal. Chim. Acta 2010, 669, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Mariyappan, V.; Keerthi, M.; Chen, S.M. Highly Selective Electrochemical Sensor Based on Gadolinium Sulfide Rod-Embedded RGO for the Sensing of Carbofuran. J. Agric. Food Chem. 2021, 69, 2679–2688. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Luo, J.; Xu, Z.; Ma, X. A microfluidic chip containing a molecularly imprinted polymer and a DNA aptamer for voltammetric determination of carbofuran. Microchim. Acta 2018, 185, 295. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.V.; Allabush, F.; Flynn, H.; Balansethupathy, B.; Reed, J.A.; Barnes, E.T.; Robson, C.; O’Hara, P.; Milburn, L.J.; Bunka, D.; et al. Highly Selective Aptamer-Molecularly Imprinted Polymer Hybrids for Recognition of SARS-CoV-2 Spike Protein Variants. Glob. Chall. 2021, 12, 4394–4405. [Google Scholar] [CrossRef] [PubMed]
- Abe-Chayama, H.; Hayes, C.N.; Chayama, K. Pan-genotypic cell culture system for propagation of hepatitis C virus clinical isolates. Hepatology 2016, 64, 1356–1358. [Google Scholar] [CrossRef]
- Chen, S.; Cai, G.; Gong, X.; Wang, L.; Cai, C.; Gong, H. Non-autofluorescence Detection of H5N1 Virus Using Photochemical Aptamer Sensors Based on Persistent Luminescent Nanoparticles. ACS Appl. Mater. Interfaces 2022, 14, 46964–46971. [Google Scholar] [CrossRef]
- Shih, C.; Yang, C.C.; Choijilsuren, G.; Chang, C.H.; Liou, A.T. Hepatitis B Virus. Trends Microbiol. 2018, 26, 386–387. [Google Scholar] [CrossRef]
- Chen, S.; Luo, L.; Wang, L.; Chen, C.; Gong, H.; Cai, C. A sandwich sensor based on imprinted polymers and aptamers for highly specific double recognition of viruses. Analyst 2021, 146, 3924–3932. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M. A nanohybrid probe based on double recognition of an aptamer MIP grafted onto a MWCNTs-Chit nanocomposite for sensing hepatitis C virus core antigen. Sens. Actuators B Chem. 2018, 258, 1066–1071. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Q.L.; Wu, W. Progress on Structured Biosensors for Monitoring Aflatoxin B1 From Biofilms: A Review. Front. Microbiol. 2020, 11, 408. [Google Scholar] [CrossRef]
- Roushani, M.; Farokhi, S.; Rahmati, Z. Development of a dual-recognition strategy for the aflatoxin B1 detection based on a hybrid of aptamer-MIP using a Cu2O NCs/GCE. Microchem. J. 2022, 178, 107328. [Google Scholar] [CrossRef]
- Chi, H.; Liu, G. A fluorometric sandwich biosensor based on molecular imprinted polymer and aptamer modified CdTe/ZnS for detection of aflatoxin B1 in edible oil. Food Sci. Technol. 2023, 180, 114726. [Google Scholar] [CrossRef]
- Khalil, O.A.A.; Hammad, A.A.; Sebaei, A.S. Aspergillus flavus and Aspergillus ochraceus inhibition and reduction of aflatoxins and ochratoxin A in maize by irradiation. Toxicon 2021, 198, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.; Sun, H.; Zhu, Y.; Wang, J.; Xie, Z.; Li, J. A double-recognized aptamer-molecularly imprinted monolithic column for high-specificity recognition of ochratoxin A. Anal. Chim. Acta 2020, 1103, 97–105. [Google Scholar] [CrossRef]
- Aralekallu, S.; Palanna, M.; Hadimani, S.; Prabhu C.P., K.; Sajjan, V.A.; Thotiyl, M.O.; Sannegowda, L.K. Biologically inspired catalyst for electrochemical reduction of hazardous hexavalent chromium. Dalton Trans. 2020, 49, 15061–15071. [Google Scholar] [CrossRef]
- Ma, Y.; Ran, D.; Shi, X.; Zhao, H.; Liu, Z. Cadmium toxicity: A role in bone cell function and teeth development. Sci. Total Environ. 2021, 769, 144646. [Google Scholar] [CrossRef]
- Li, S.; Ma, X.; Pang, C.; Tian, H.; Xu, Z.; Yang, Y.; Lv, D.; Ge, H. Fluorometric aptasensor for cadmium(II) by using an aptamer-imprinted polymer as the recognition element. Microchim. Acta 2019, 186, 823. [Google Scholar] [CrossRef]
- Hauser, A.R.; Mecsas, J.; Moir, D.T. Beyond antibiotics: New therapeutic approaches for bacterial infections. Int. J. Antimicrob. Agents 2017, 50, S19–S20. [Google Scholar] [CrossRef]
- Hoeksema, M.; Brul, S.; Kuile, B.H.T. Influence of Reactive Oxygen Species on De Novo Acquisition of Resistance to Bactericidal Antibiotics. Antimicrob. Agents Chemother. 2018, 62, 17. [Google Scholar] [CrossRef]
- Spizek, J.; Rezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 2017, 133, 20–28. [Google Scholar] [CrossRef]
- Li, S.; Liu, C.; Yin, G.; Zhang, Q.; Luo, J.; Wu, N. Aptamer-molecularly imprinted sensor base on electrogenerated chemiluminescence energy transfer for detection of lincomycin. Biosens. Bioelectron. 2017, 91, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Wu, Y.; Wang, Y.; Liu, G.; Ning, G.; Xu, Z. A molecularly imprinted polymer combined with dual functional Au@Fe3O4 nanocomposites for sensitive detection of kanamycin. J. Electroanal. Chem. 2020, 870, 114216. [Google Scholar] [CrossRef]
- Sanchez, A.R.; Rogers, R.S.; Sheridan, P.J. Tetracycline and other tetracycline-derivative staining of the teeth and oral cavity. Int. J. Dermatol. 2004, 43, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liao, X.; Zhao, Y.; Qiu, L.; Yao, Y.; Wang, S.; Yang, X.; Hu, X. Fabrication of magnetic molecularly imprinted polymers based on aptamers and ss-cyclodextrin for synergistic recognition and separation of tetracycline. Anal. Chim. Acta 2022, 1236, 340572. [Google Scholar] [CrossRef] [PubMed]
- Paprocka, P.; Durnaś, B.; Mańkowska, A.; Król, G.; Wollny, T.; Bucki, R. Pseudomonas aeruginosa Infections in Cancer Patients. Pathogens 2022, 11, 679. [Google Scholar] [CrossRef]
- Sarabaegi, M.; Roushani, M. Rapid and sensitive determination of Pseudomonas aeruginosa by using a glassy carbon electrode modified with gold nanoparticles and aptamer-imprinted polydopamine. Microchem. J. 2021, 168, 106388. [Google Scholar] [CrossRef]
- Jarneborn, A.; Mohammad, M.; Engdahl, C.; Hu, Z.; Na, M.; Ali, A.; Jin, T. Tofacitinib treatment aggravates Staphylococcus aureus septic arthritis, but attenuates sepsis and enterotoxin induced shock in mice. Sci. Rep. 2020, 10, 10891. [Google Scholar] [CrossRef]
- Cai, R.; Yin, F.; Zhang, Z.; Tian, Y.; Zhou, N. Functional chimera aptamer and molecular beacon based fluorescent detection of Staphylococcus aureus with strand displacement-target recycling amplification. Anal. Chim. Acta 2019, 1075, 128–136. [Google Scholar] [CrossRef]
- El-Wekil, M.M.; Halby, H.M.; Darweesh, M.; Ali, M.E.; Ali, R. An innovative dual recognition aptasensor for specific detection of Staphylococcus aureus based on Au/Fe3O4 binary hybrid. Sci. Rep. 2022, 12, 12502. [Google Scholar] [CrossRef]
- De Oliveira, J.; Kucharska, E.; Garcez, M.L.; Rodrigues, M.S.; Quevedo, J.; Moreno-Gonzalez, I.; Budni, J. Inflammatory Cascade in Alzheimer’s Disease Pathogenesis: A Review of Experimental Findings. Cells 2021, 10, 2581. [Google Scholar] [CrossRef]
- Roos, A.; Edgren, G. Using historical cardiac troponins to identify patients at a high risk of myocardial infarction. Heart 2023, 109, 127–133. [Google Scholar] [CrossRef]
- Mokhtari, Z.; Khajehsharifi, H.; Hashemnia, S.; Solati, Z.; Azimpanah, R.; Shahrokhian, S. Evaluation of molecular imprinted polymerized methylene blue/aptamer as a novel hybrid receptor for Cardiac Troponin I (cTnI) detection at glassy carbon electrodes modified with new biosynthesized ZnONPs. Sens. Actuators B-Chem. 2020, 320, 108316. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, X.; Sun, N.; Deng, C. A rational route to hybrid aptamer-molecularly imprinted magnetic nanoprobe for recognition of protein biomarkers in human serum. Anal. Chim. Acta 2020, 1128, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Tian, Y.; Wang, B.; Guo, Q.; Nie, G. “Signal-on” molecularly imprinting-aptamer electrochemiluminescence platform for ultrasensitive detection of thrombin. Sens. Actuators B-Chem. 2021, 338, 129870. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Amini, M.; Rezaei, B. Molecularly imprinted electrochemical aptasensor for the attomolar detection of bisphenol A. Microchim. Acta 2018, 185, 265. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Alkahtani, S.A.; Alyami, B.A.; El-Wekil, M.M. Dual-recognition molecularly imprinted aptasensor based on gold nanoparticles decorated carboxylated carbon nanotubes for highly selective and sensitive determination of histamine in different matrices. Anal. Chim. Acta 2020, 1133, 58–65. [Google Scholar] [CrossRef]
- Yu, C.; Li, L.; Ding, Y.; Liu, H.; Cui, H. Molecularly imprinted electrochemical aptasensor based on functionalized graphene and nitrogen-doped carbon quantum dots for trace cortisol assay. Analyst 2022, 147, 744–752. [Google Scholar] [CrossRef]
- Huang, Y.; Ye, D.; Yang, J.; Zhu, W.; Li, L.; Ding, Y. Dual recognition elements for selective determination of progesterone based on molecularly imprinted electrochemical aptasensor. Anal. Chim. Acta 2023, 1264, 341288. [Google Scholar] [CrossRef]
- Ali, R.; El-Wekil, M.M. A dual-recognition-controlled electrochemical biosensor for selective and ultrasensitive detection of acrylamide in heat-treated carbohydrate-rich food. Food Chem. 2023, 413, 135666. [Google Scholar] [CrossRef]
- Beiki, T.; Najafpour-Darzi, G.; Mohammadi, M.; Shakeri, M.; Boukherroub, R. Fabrication of a novel electrochemical biosensor based on a molecular imprinted polymer-aptamer hybrid receptor for lysozyme determination. Anal. Bioanal. Chem. 2023, 415, 899–911. [Google Scholar] [CrossRef]
- Rahmati, Z.; Roushani, M. SARS-CoV-2 virus label-free electrochemical nanohybrid MIP-aptasensor based on Ni-3(BTC)(2) MOF as a high-performance surface substrate. Microchim. Acta 2022, 189, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Kan, X.W. Sensitive and selective “signal-off” electrochemiluminescence sensing of prostate-specific antigen based on an aptamer and molecularly imprinted polymer. Analyst 2021, 146, 7693–7701. [Google Scholar] [CrossRef] [PubMed]
- Roushani, M.; Zalpour, N. Impedimetric ultrasensitive detection of trypsin based on hybrid aptamer-2DMIP using a glassy carbon electrode modified by nickel oxide nanoparticle. Microchem. J. 2021, 172, 106955. [Google Scholar] [CrossRef]
- Roushani, M.; Ghanbarzadeh, M.; Shahdost-Fard, F. Fabrication of an electrochemical biodevice for ractopamine detection under a strategy of a double recognition of the aptamer/molecular imprinting polymer. Bioelectrochemistry 2021, 138, 107722. [Google Scholar] [CrossRef] [PubMed]
- Roushani, M.; Rahmati, Z.; Hoseini, S.J.; Fath, R.H. Impedimetric ultrasensitive detection of chloramphenicol based on aptamer MIP using a glassy carbon electrode modified by 3-ampy-RGO and silver nanoparticle. Colloids Surf. B Biointerfaces 2019, 183, 110451. [Google Scholar] [CrossRef]
- Roushani, M.; Nezhadali, A.; Jalilian, Z. An electrochemical chlorpyrifos aptasensor based on the use of a glassy carbon electrode modified with an electropolymerized aptamer-imprinted polymer and gold nanorods. Microchim. Acta 2018, 185, 551. [Google Scholar] [CrossRef]
- Tan, J.; Guo, M.; Tan, L.; Geng, Y.; Huang, S.; Tang, Y.; Su, C.; Lin, C.C.; Liang, Y. Highly efficient fluorescent QDs sensor for specific detection of protein through double recognition of hybrid aptamer-molecular imprinted polymers. Sens. Actuators B Chem. 2018, 274, 627–635. [Google Scholar] [CrossRef]
- Geng, Y.; Guo, M.; Tan, J.; Huang, S.; Tang, Y.; Tan, L.; Liang, Y. A fluorescent molecularly imprinted polymer using aptamer as a functional monomer for sensing of kanamycin. Sens. Actuators B-Chem. 2018, 268, 47–54. [Google Scholar] [CrossRef]
- Duan, N.; Chen, X.; Lin, X.; Ying, D.; Wang, Z.; Yuan, W.; Wu, S. Paper-based fluorometric sensing of malachite green using synergistic recognition of aptamer-molecularly imprinted polymers and luminescent metal–organic frameworks. Sens. Actuators B Chem. 2023, 384, 133665. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).