A Metal Organic Framework-Based Light Scattering ELISA for the Detection of Staphylococcal Enterotoxin B
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Apparatus
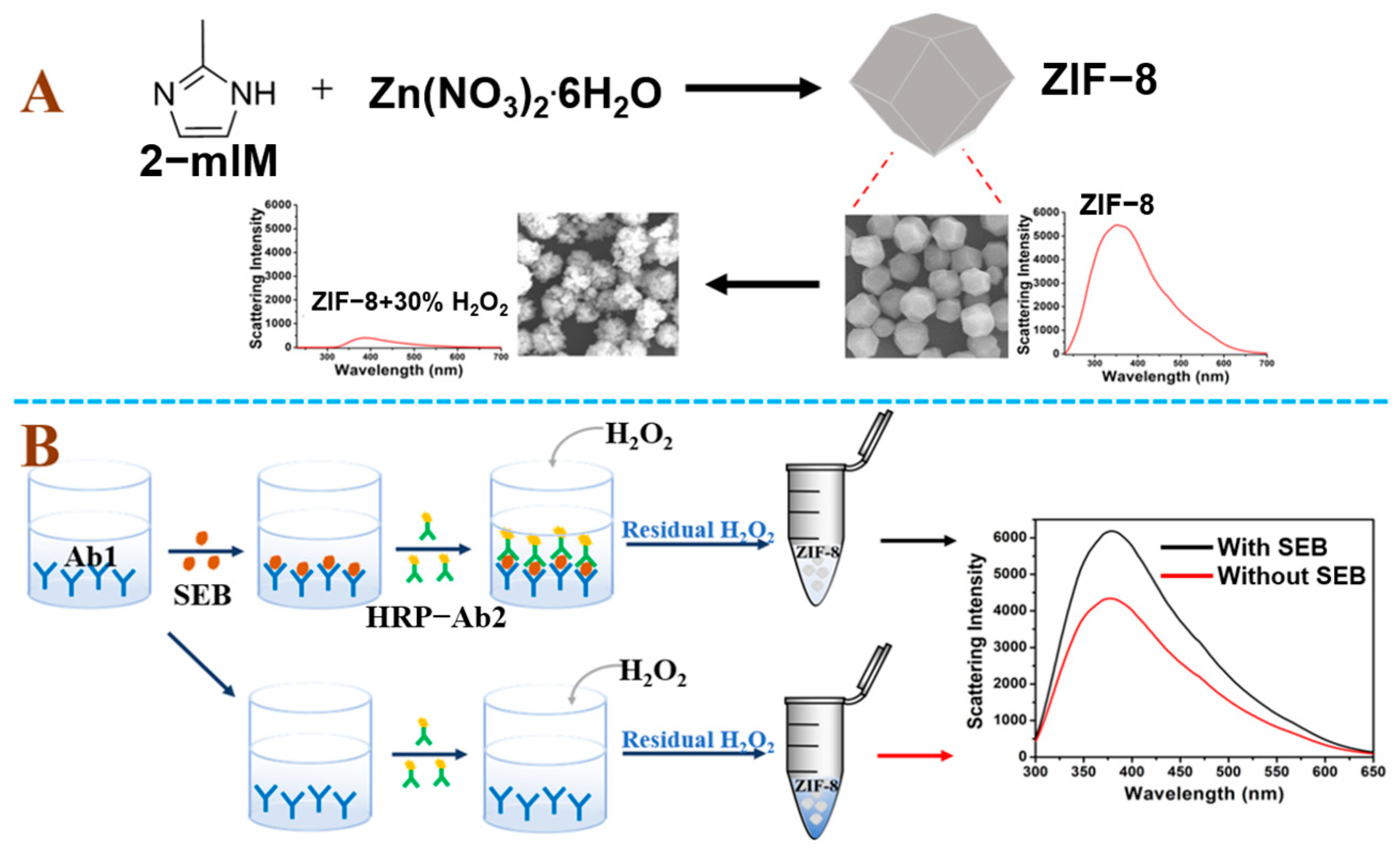
2.2. Synthesis of ZIF-8
2.3. Dark Field Light Scattering Imaging of the Reaction between ZIF-8 and H2O2
2.4. Detection of SEB
3. Results and Discussion
3.1. Principle of the SEB Detection
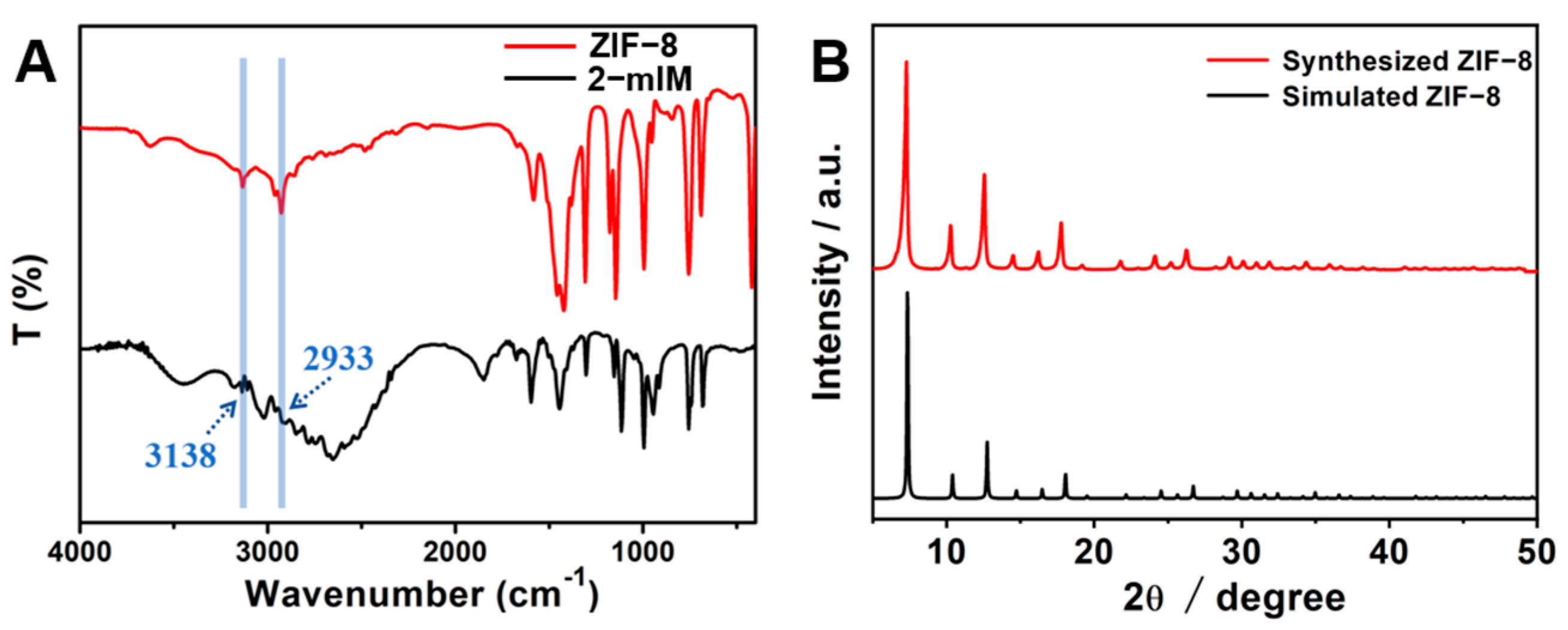
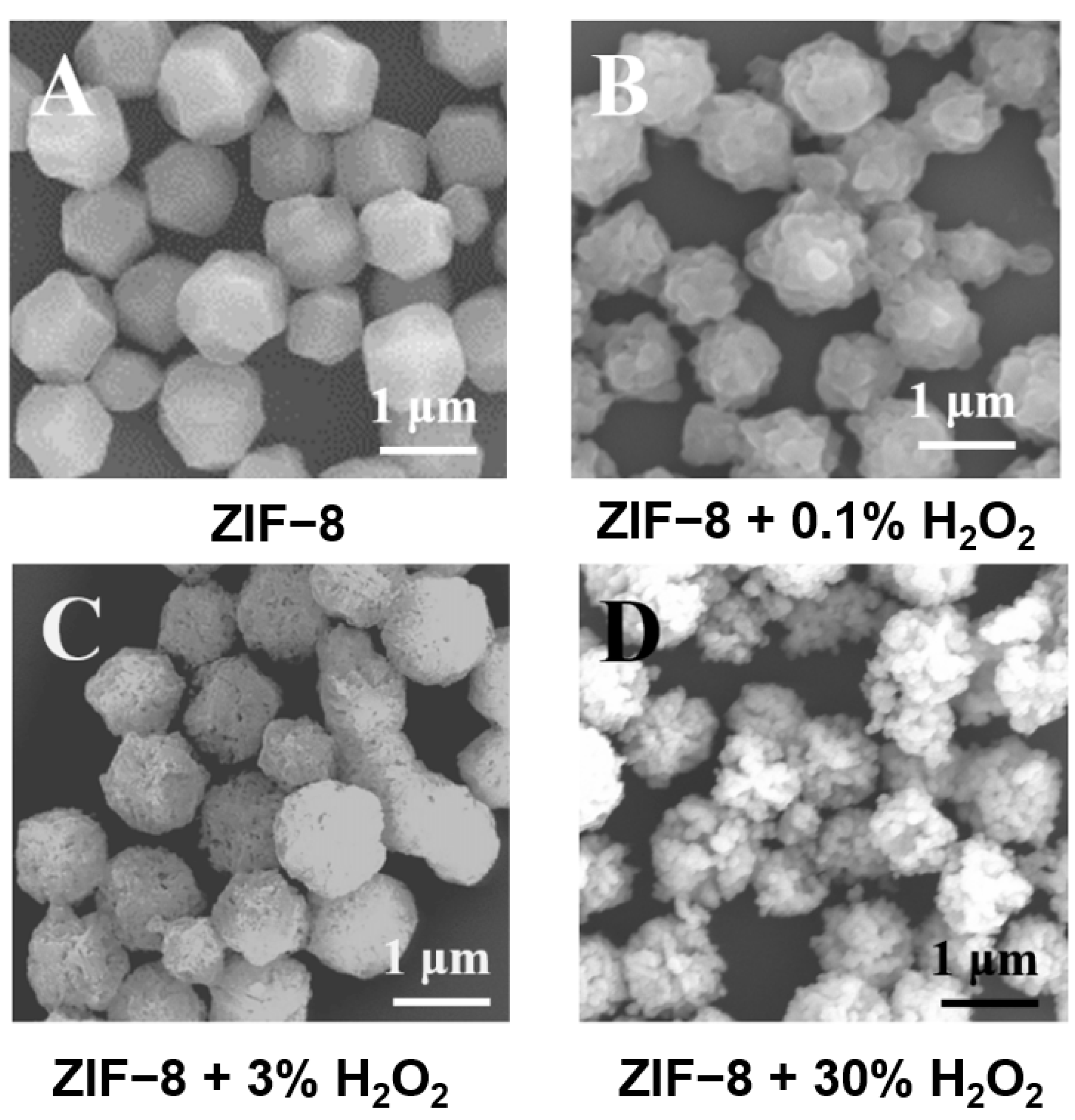
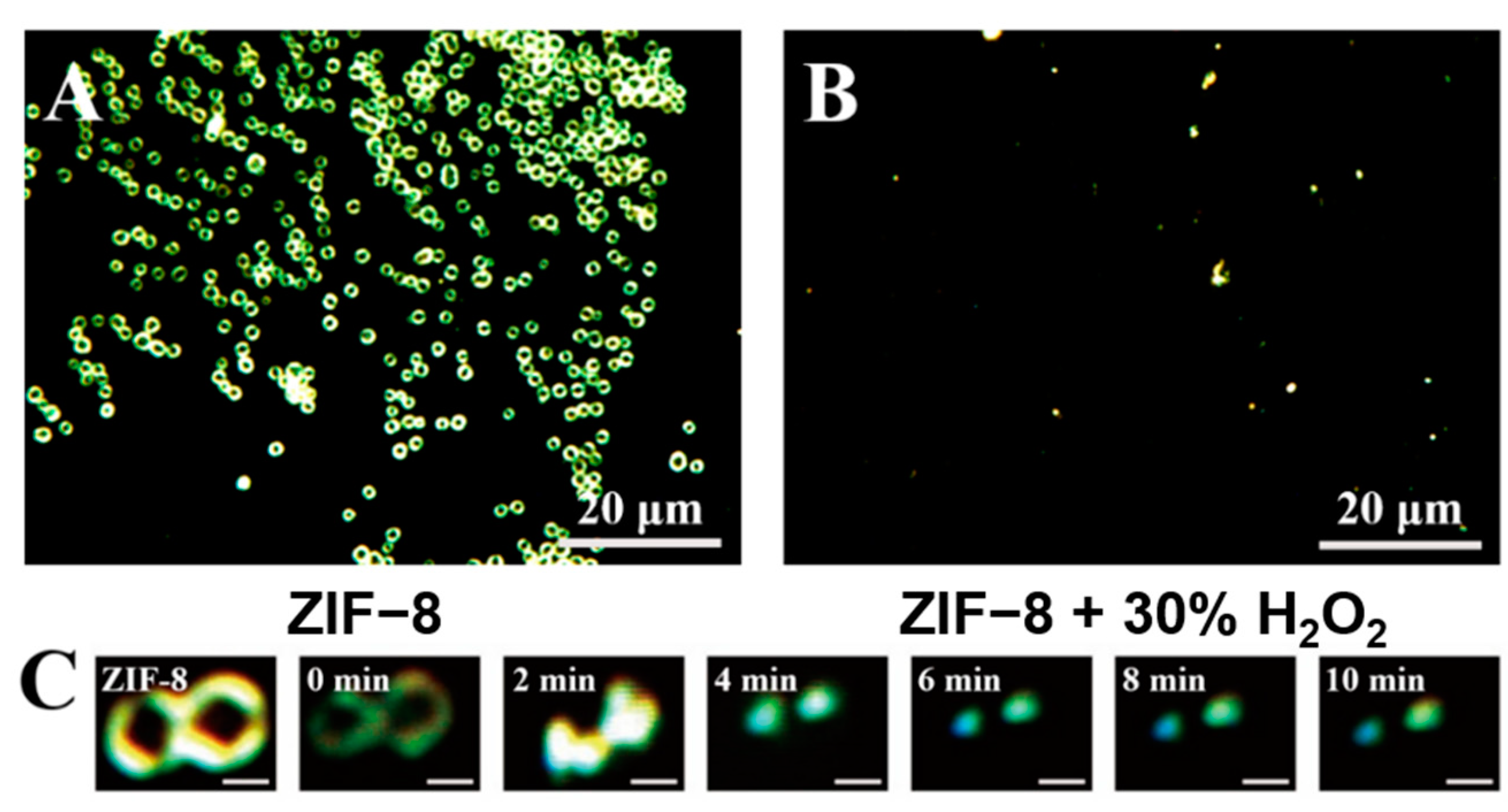
3.2. Characterization of ZIF-8 and Its Reaction with H2O2
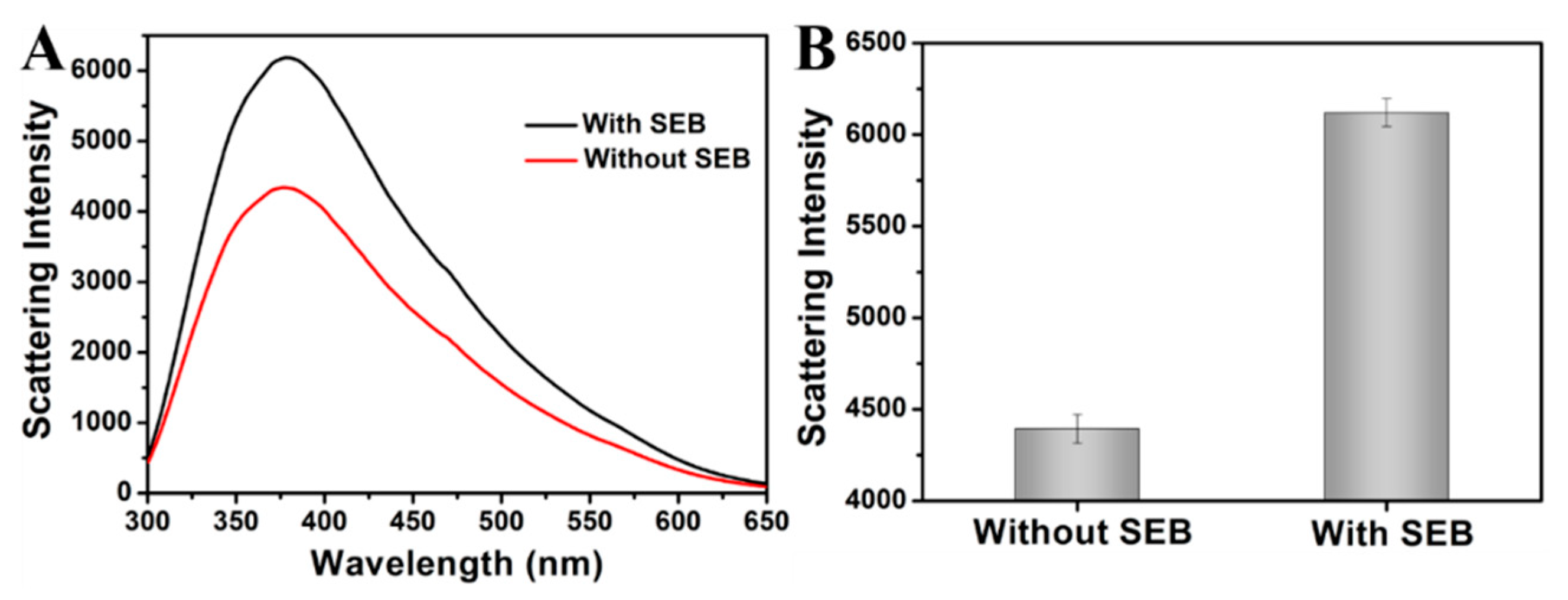
3.3. LS Spectral Characteristics of ZIF-8 for SEB Detection
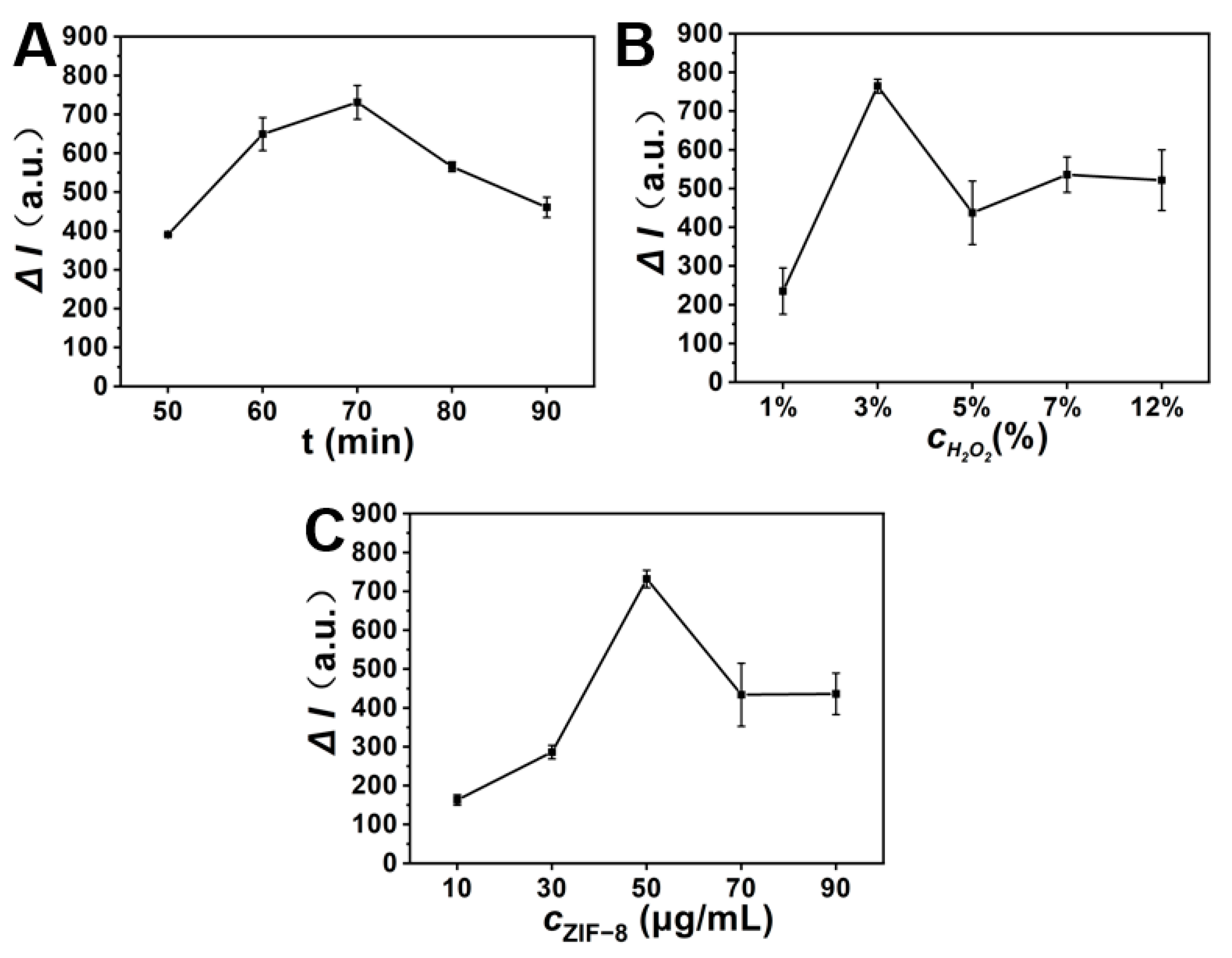
3.4. Optimization of the SEB Detection Conditions
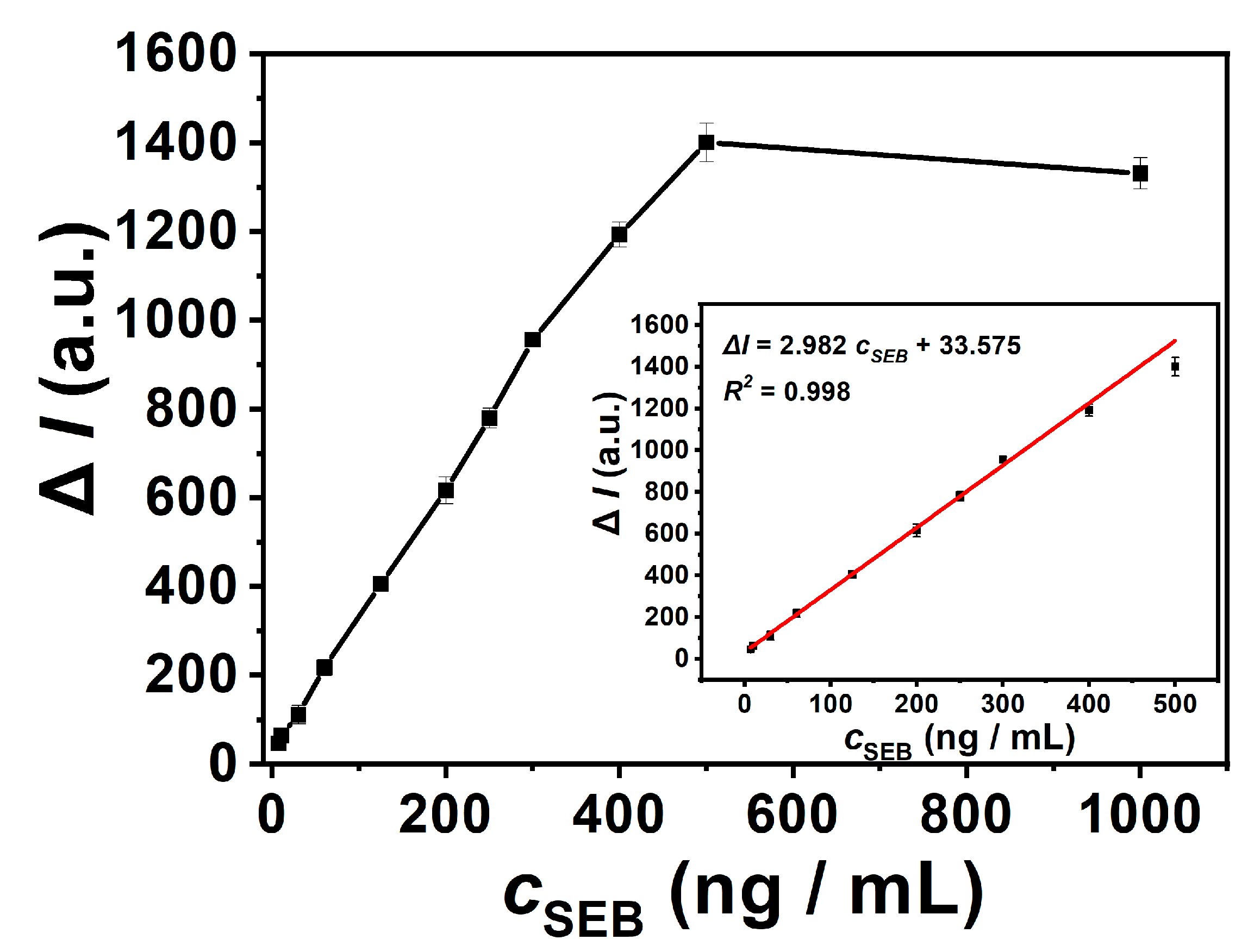
3.5. Analytical Performance of SEB Detection
3.6. SEB Detection in Complex Samples
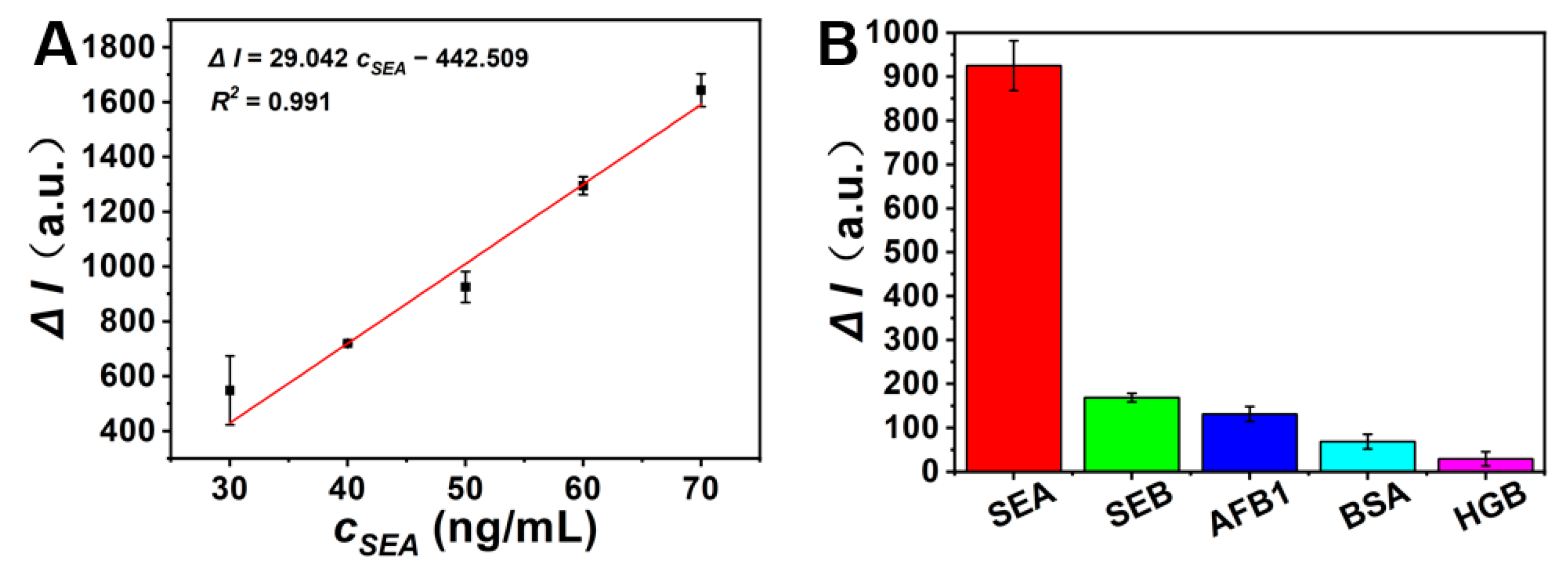
3.7. Staphylococcal Enterotoxin A (SEA) Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mousavi, N.S.; Nasirizadeh, N.; Amani, J.; Halabian, R.; Fooladi, I.A.A. An electrochemical aptasensor for staphylococcal enterotoxin B detection based on reduced graphene oxide and gold nano-urchins. Biosens. Bioelectron. 2019, 127, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Sospedra, I.; Marín, R.; Mañes, J.; Soriano, J.M. Rapid whole protein quantification of staphylococcal enterotoxin B by liquid chromatography. Food Chem. 2012, 133, 163–166. [Google Scholar] [CrossRef]
- Muratovic, A.Z.; Hagstrom, T.; Rosen, J.; Granelli, K.; Hellenas, K.E. Quantitative Analysis of Staphylococcal Enterotoxins A and B in Food Matrices Using Ultra High-Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MS/MS). Toxins 2015, 7, 3637–3656. [Google Scholar] [CrossRef]
- Andjelkovic, M.; Tsilia, V.; Rajkovic, A.; De Cremer, K.; Van Loco, J. Application of LC-MS/MS MRM to Determine Staphylococcal Enterotoxins (SEB and SEA) in Milk. Toxins 2016, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Tiwari, M.; Das, P. Multi format compatible visual and fluorometric detection of SEB toxin in nanogram range by carbon dot-DNA and acriflavine nano-assembly. Sens. Actuators B 2019, 279, 393–399. [Google Scholar] [CrossRef]
- Xiong, X.; Shi, X.; Liu, Y.; Lu, L.; You, J. An aptamer-based electrochemical biosensor for simple and sensitive detection of staphylococcal enterotoxin B in milk. Anal. Methods 2018, 10, 365–370. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Liu, L.; Xu, L.; Kuang, H.; Zhu, J.; Xu, C. Nanoshell-Enhanced Raman Spectroscopy on a Microplate for Staphylococcal Enterotoxin B Sensing. ACS Appl. Mater. Interfaces 2016, 8, 15591–15597. [Google Scholar] [CrossRef] [PubMed]
- Xue, P.; Li, Y.; Zhang, Z.; Fu, A.; Liu, F.; Zhang, X.; Sun, Y.; Chen, L.; Jin, B.; Yang, K. Novel chemiluminescent assay for staphylococcal enterotoxin B. Microchim. Acta 2011, 174, 167–174. [Google Scholar] [CrossRef]
- Lu, B.; Bai, J.; Zhang, J.; Shen, H.; Wang, M.; Lian, Y.; Gao, Z.; Peng, Y. Iridescent polymeric film with tunable color responses to ultra-trace Staphylococcus aureus enterotoxin B. Sensor. Actuat. B—Chem. 2023, 380, 133318. [Google Scholar] [CrossRef]
- Margalit, Y.; Lu, Y.K.; Top, F.C.; Ketterle, W. Pauli blocking of light scattering in degenerate fermions. Science 2021, 374, 976–979. [Google Scholar] [CrossRef]
- Celiksoy, S.; Ye, W.; Wandner, K.; Kaefer, K.; Sönnichsen, C. Intensity-Based Single Particle Plasmon Sensing. Nano Lett. 2021, 21, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, P.; Song, P.; Kang, B.; Xu, J.J.; Chen, H.Y. The video-rate imaging of sub-10 nm plasmonic nanoparticles in a cellular medium free of background scattering. Chem. Sci. 2021, 12, 3017–3024. [Google Scholar] [CrossRef]
- Wang, J.C.; Wang, Y.S.; Xue, J.H.; Zhou, B.; Qian, Q.M.; Wang, Y.S.; Yin, J.C.; Zhao, H.; Liu, H.; Liu, S.D. An ultrasensitive label-free assay of 8-hydroxy-2’-deoxyguanosine based on the conformational switching of aptamer. Biosens. Bioelectron. 2014, 58, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, T.; Shen, R.; Li, G.; Ling, L. Polymerase Chain Reaction-Dynamic Light Scattering Sensor for DNA and Protein by Using Both Replication and Cleavage Properties of Taq Polymerase. Anal. Chem. 2019, 91, 3429–3435. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Ling, L. Ultrasensitive Detection of HIV DNA with Polymerase Chain Reaction-Dynamic Light Scattering. Anal. Chem. 2018, 90, 13373–13377. [Google Scholar] [CrossRef] [PubMed]
- El-Kurdi, R.; Patra, D. Gold and silver nanoparticles in resonance Rayleigh scattering techniques for chemical sensing and biosensing: A review. Mikrochim. Acta 2019, 186, 667. [Google Scholar] [CrossRef]
- Hu, P.; Huang, C.Z.; Li, Y.F.; Liang, J.; Liu, Y.L.; Fei, L.R.; Xie, J.P. Magnetic Particle-Based Sandwich Sensor with DNA-Modified Carbon Nanotubes as Recognition Elements for Detection of DNA Hybridization. Anal. Chem. 2008, 80, 1819–1823. [Google Scholar] [CrossRef]
- Qi, W.J.; Wu, D.; Ling, J.; Huang, C.Z. Visual and light scattering spectrometric detections of melamine with polythymine-stabilized gold nanoparticles through specific triple hydrogen-bonding recognition. Chem. Commun. 2010, 46, 4893–4895. [Google Scholar] [CrossRef]
- Feng, D.Q.; Liu, G.; Wang, W. A novel biosensor for copper(ii) ions based on turn-on resonance light scattering of ssDNA templated silver nanoclusters. J. Mater. Chem. B 2015, 3, 2083–2088. [Google Scholar] [CrossRef]
- Kohli, R.K.; Davies, J.F. Measuring the Chemical Evolution of Levitated Particles: A Study on the Evaporation of Multicomponent Organic Aerosol. Anal. Chem. 2021, 93, 12472–12479. [Google Scholar] [CrossRef]
- Latimer, P.; Barber, P. Scattering by ellipsoids of revolution a comparison of theoretical methods. J. Colloid Interface Sci. 1978, 63, 310–316. [Google Scholar] [CrossRef]
- Al-Chalabi, S.A.M.; Jones, A.R. Light scattering by irregular particles in the Rayleigh-Gans-Debye approximation. J. Phys. D Appl. Phys. 1995, 28, 1304. [Google Scholar] [CrossRef]
- Ling, J.; Li, Y.F.; Huang, C.Z. Visual Sandwich Immunoassay System on the Basis of Plasmon Resonance Scattering Signals of Silver Nanoparticles. Anal. Chem. 2009, 81, 1707–1714. [Google Scholar] [CrossRef]
- Ling, J.; Huang, C.Z.; Li, Y.F.; Zhang, L.; Chen, L.Q.; Zhen, S.J. Light-scattering signals from nanoparticles in biochemical assay, pharmaceutical analysis and biological imaging. TrAC 2009, 28, 447–453. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Bustamante, C.; Collings, P.J.; Giannetto, A.; Gibbs, E.J. Porphyrin Assemblies on DNA as Studied by a Resonance Light-Scattering Technique. J. Am. Chem. Soc. 1993, 115, 5393–5399. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Li, D.S.; Bu, X.; Feng, P. Metal-Organic Frameworks for Separation. Adv. Mater. 2018, 30, e1705189. [Google Scholar] [CrossRef] [PubMed]
- Terzopoulou, A.; Nicholas, J.D.; Chen, X.Z.; Nelson, B.J.; Pane, S.; Puigmarti-Luis, J. Metal-Organic Frameworks in Motion. Chem. Rev. 2020, 120, 11175–11193. [Google Scholar] [CrossRef]
- Troyano, J.; Carne-Sanchez, A.; Avci, C.; Imaz, I.; Maspoch, D. Colloidal metal-organic framework particles: The pioneering case of ZIF-8. Chem. Soc. Rev. 2019, 48, 5534–5546. [Google Scholar] [CrossRef]
- Saliba, D.; Ammar, M.; Rammal, M.; Al-Ghoul, M.; Hmadeh, M. Crystal Growth of ZIF-8, ZIF-67, and Their Mixed-Metal Derivatives. J. Am. Chem. Soc. 2018, 140, 1812–1823. [Google Scholar] [CrossRef]
- He, L.Z.; Huang, G.N.; Liu, H.X.; Sang, C.C.; Liu, X.X.; Chen, T.F. Highly bioactive zeolitic imidazolate framework-8–capped nanotherapeutics for efficient reversalofreperfusion-induced injury in ischemic stroke. Sci. Adv. 2020, 6, 9751. [Google Scholar] [CrossRef]
- Hu, X.; Liu, X.; Zhang, X.; Chai, H.; Huang, Y. One-pot synthesis of the CuNCs/ZIF-8 nanocomposites for sensitively detecting H(2)O(2) and screening of oxidase activity. Biosens. Bioelectron. 2018, 105, 65–70. [Google Scholar] [CrossRef]
- Carraro, F.; Williams, J.D.; Linares-Moreau, M.; Parise, C.; Liang, W.; Amenitsch, H.; Doonan, C.; Kappe, C.O.; Falcaro, P. Continuous-Flow Synthesis of ZIF-8 Biocomposites with Tunable Particle Size. Angew. Chem. Int. Ed. Engl. 2020, 59, 8123–8127. [Google Scholar] [CrossRef]
- Lee, Y.R.; Jang, M.S.; Cho, H.Y.; Kwon, H.J.; Kim, S.; Ahn, W.S. ZIF-8: A comparison of synthesis methods. Chem. Eng. J. 2015, 271, 276–280. [Google Scholar]
- Hu, P.; Zhuang, J.; Chou, L.Y.; Lee, H.K.; Ling, X.Y.; Chuang, Y.C.; Tsung, C.K. Surfactant-directed atomic to mesoscale alignment: Metal nanocrystals encased individually in single-crystalline porous nanostructures. J. Am. Chem. Soc. 2014, 136, 10561–10564. [Google Scholar] [CrossRef]
- Attri, P.; Kim, Y.H.; Park, D.H.; Park, J.H.; Hong, Y.J.; Uhm, H.S.; Kim, K.N.; Fridman, A.; Choi, E.H. Generation mechanism of hydroxyl radical species and its lifetime prediction during the plasma-initiated ultraviolet (UV) photolysis. Sci. Rep. 2015, 5, 9332. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, K.; Li, X.; Liu, Z.; Liu, Y.; Xiao, R.; Wang, S. Highly sensitive detection of three protein toxins via SERS-lateral flow immunoassay based on SiO(2)@Au nanoparticles. Nanomedicine 2022, 41, 102522. [Google Scholar] [CrossRef]
- Jang, J.H.; Kim, S.; Kim, S.G.; Lee, J.; Lee, D.G.; Jang, J.; Jeong, Y.S.; Song, D.H.; Min, J.K.; Park, J.G.; et al. A Sensitive Immunodetection Assay Using Antibodies Specific to Staphylococcal Enterotoxin B Produced by Baculovirus Expression. Biosensors 2022, 12, 787. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Huang, H.; Peng, Y.; Wang, J.; Gao, Z. Rapid determination of Staphylococcus aureus enterotoxin B in milk using Raman spectroscopy and chemometric methods. J. Raman Spectrosc. 2022, 53, 709–714. [Google Scholar] [CrossRef]

| Test Methods | LOD | Ref. |
|---|---|---|
| Spectral method | 0.5 ng/mL | [5] |
| SERS-lateral flow immunoassay | 0.05 ng/mL | [36] |
| Enzyme-linked immunosorbent assay | 0.38 ng/mL | [37] |
| Raman spectroscopy and chemometric methods | 0.2 ng/L | [38] |
| Light scattering ELISA | 0.69 ng/mL | This work |
| Sample | Added (ng/mL) | Found (ng/mL) Mean a ± SD b | Recovery (%, n = 3) | RSD (%, n = 3) |
|---|---|---|---|---|
| 400 | 403.5 ± 8.46 | 98.9–103.2 | 8.21 | |
| Orange juice | 250 | 251.3 ± 12.3 | 96.9–106.2 | 4.91 |
| 60 | 60.9 ± 5.0 | 91.9–106.5 | 2.10 | |
| 400 | 397.2 ± 36.73 | 92.0–109.6 | 9.25 | |
| Fresh milk | 250 | 250.5 ± 16.61 | 93.4–106.7 | 6.63 |
| 60 | 60.3 ± 0.97 | 98.6–101.4 | 0.16 | |
| 400 | 369.0 ± 12.24 | 90.2–95.8 | 3.32 | |
| Milk powder | 250 | 257.0 ± 3.73 | 101.5–104.4 | 1.45 |
| 60 | 59.5 ± 4.08 | 94.7–107.0 | 6.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, K.; Tian, L.; Luo, Y.; Li, Q.; Chen, X.; Zhan, L.; Li, Y.; Huang, C.; Zhen, S. A Metal Organic Framework-Based Light Scattering ELISA for the Detection of Staphylococcal Enterotoxin B. Chemosensors 2023, 11, 453. https://doi.org/10.3390/chemosensors11080453
Mao K, Tian L, Luo Y, Li Q, Chen X, Zhan L, Li Y, Huang C, Zhen S. A Metal Organic Framework-Based Light Scattering ELISA for the Detection of Staphylococcal Enterotoxin B. Chemosensors. 2023; 11(8):453. https://doi.org/10.3390/chemosensors11080453
Chicago/Turabian StyleMao, Kai, Lili Tian, Yujie Luo, Qian Li, Xi Chen, Lei Zhan, Yuanfang Li, Chengzhi Huang, and Shujun Zhen. 2023. "A Metal Organic Framework-Based Light Scattering ELISA for the Detection of Staphylococcal Enterotoxin B" Chemosensors 11, no. 8: 453. https://doi.org/10.3390/chemosensors11080453
APA StyleMao, K., Tian, L., Luo, Y., Li, Q., Chen, X., Zhan, L., Li, Y., Huang, C., & Zhen, S. (2023). A Metal Organic Framework-Based Light Scattering ELISA for the Detection of Staphylococcal Enterotoxin B. Chemosensors, 11(8), 453. https://doi.org/10.3390/chemosensors11080453






