Enhanced Fluorescence in a Lens-Less Fiber-Optic Sensor for C-Reactive Protein Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical and Biological Reagents
2.1.1. Reagents
2.1.2. IgG/Anti-IgG Assay
2.1.3. CRP Assay
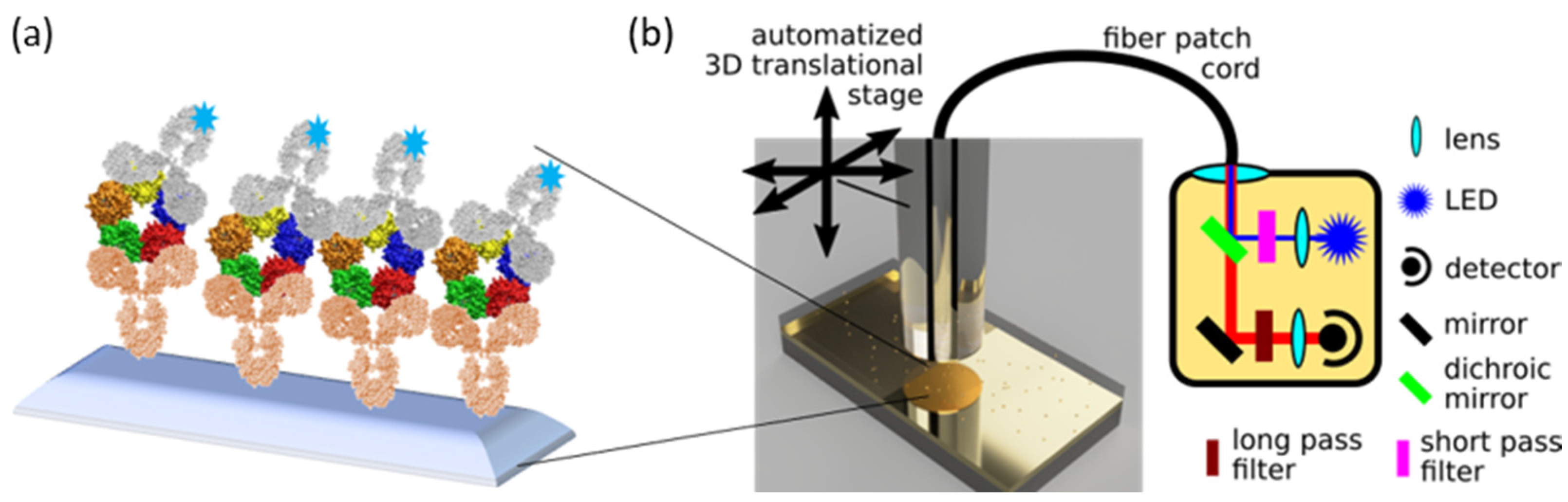
2.2. Fabrication of the Chips
2.2.1. The Chip
2.2.2. Assay Protocol
- functional groups (-COOH) activation by cross-linking chemistry (200 mM EDC and 50 mM NHS) for 30 min;
- covalent immobilization of the capture antibody (IgG or clone C5 as anti-CRP antibody) (0.5 g/mL in PBS) for 60 min;
- washing step with PBS repeated 3 times;
- surface passivation with BSA (0.1% in PBS) for 30 min to prevent nonspecific adsorption onto the surface;
- washing step with PBS repeated 3 times;
- antigen interaction:
- (a)
- anti-mouse-IgG* (concentration range 0–0.1 mg/mL) in PBS for 30 min;
- (b)
- CRP in serum (concentration range 0–0.1 mg/mL) in human serum diluted 1:10 v/v in PBS for 30 min.
- washing step with PBS repeated 3 times;
- detection antibody interaction: Clone C7* (0.01 mg/mL) in PBS for 30 min;
- washing step with PBS repeated 3 times.
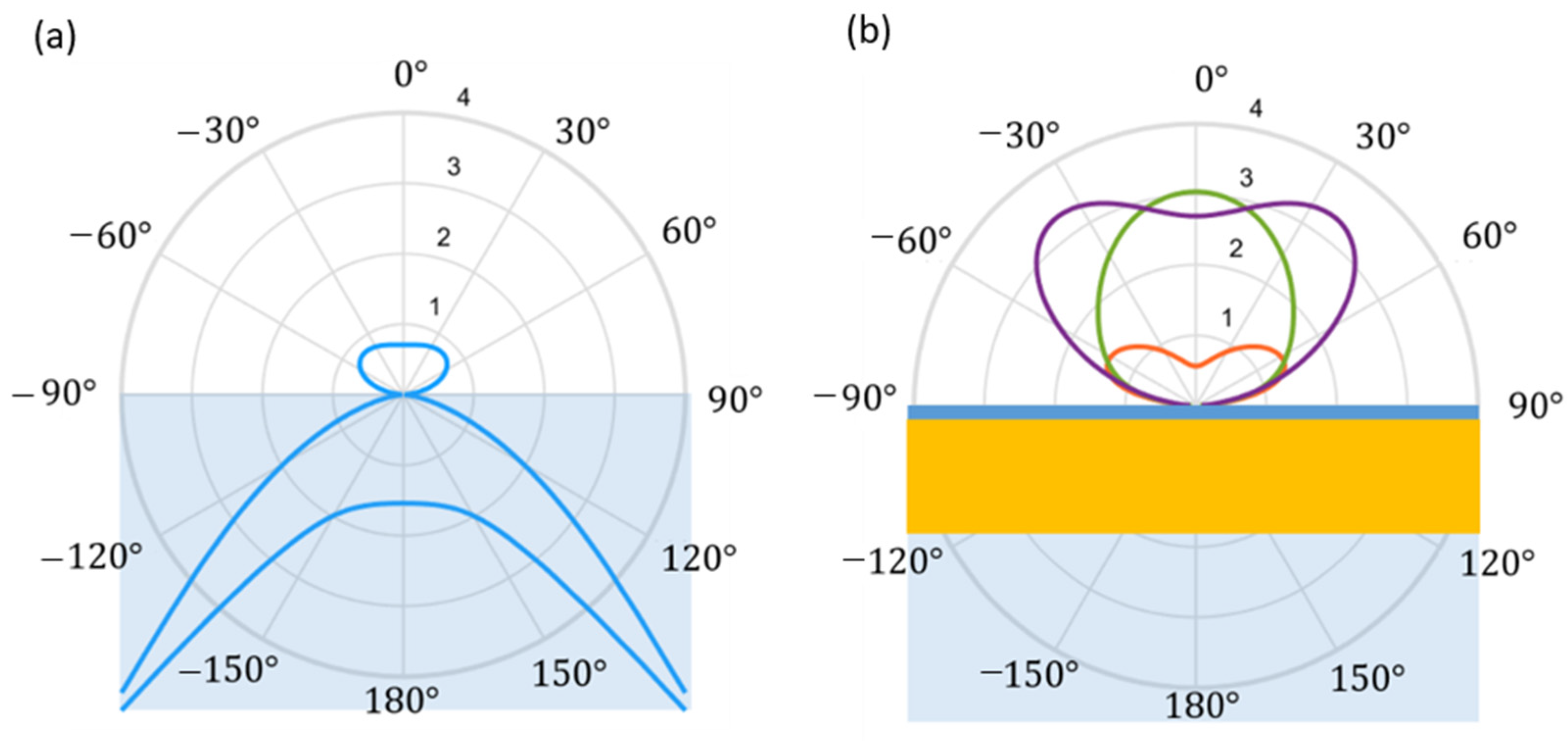
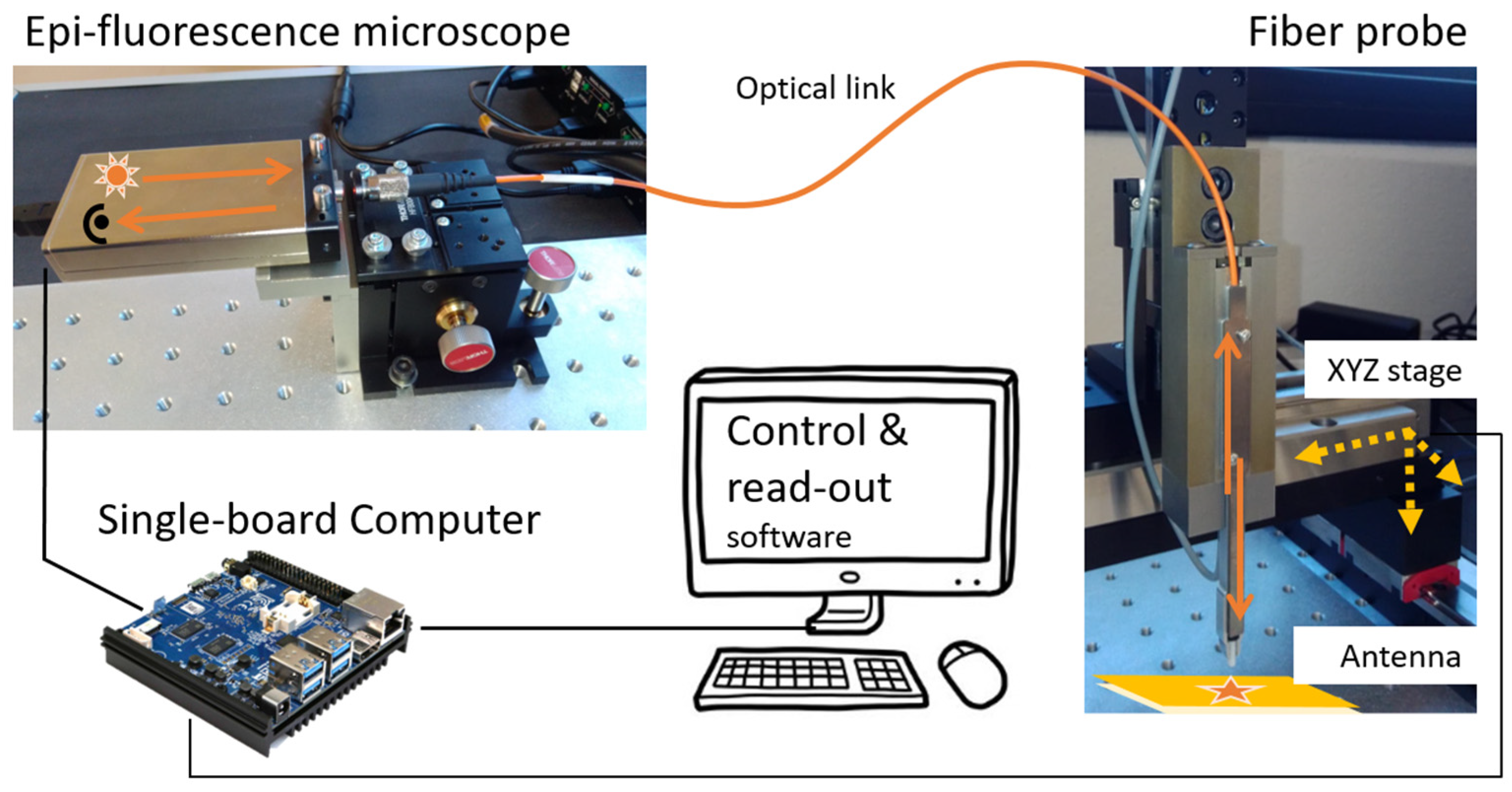
2.3. Sensor Prototype
- a compact epi-fluorescence microscope system (Qiagen Eselog), whose optical scheme is sketched in Figure 2b, which comprises a LED source emitting at 640 nm as excitation source, lenses, and optical filters for the selective collection of fluorescence emitted around 670 nm, and a photodetector for the evaluation of the signal intensity;
- a 3D motorized translation stage with few micrometer repeatability and more than 10 cm scan range in the three directions (Standa 8-0048);
- a multimode optical fiber (400 μm core-size, 0.5 N.A.), terminated on one side with a ceramic ferrule (operating as assay probe), and coupled on the other side to the detection system in free space, through a simple fiber launcher system (see Figure S1 in Supplementary Materials for measurements with 200 μm core-size);
- a single-board mini-computer (Odroid N2+) equipped with a homemade software for the management of the measurement sequences and data storage.
2.4. Measurement Protocol and Data Processing
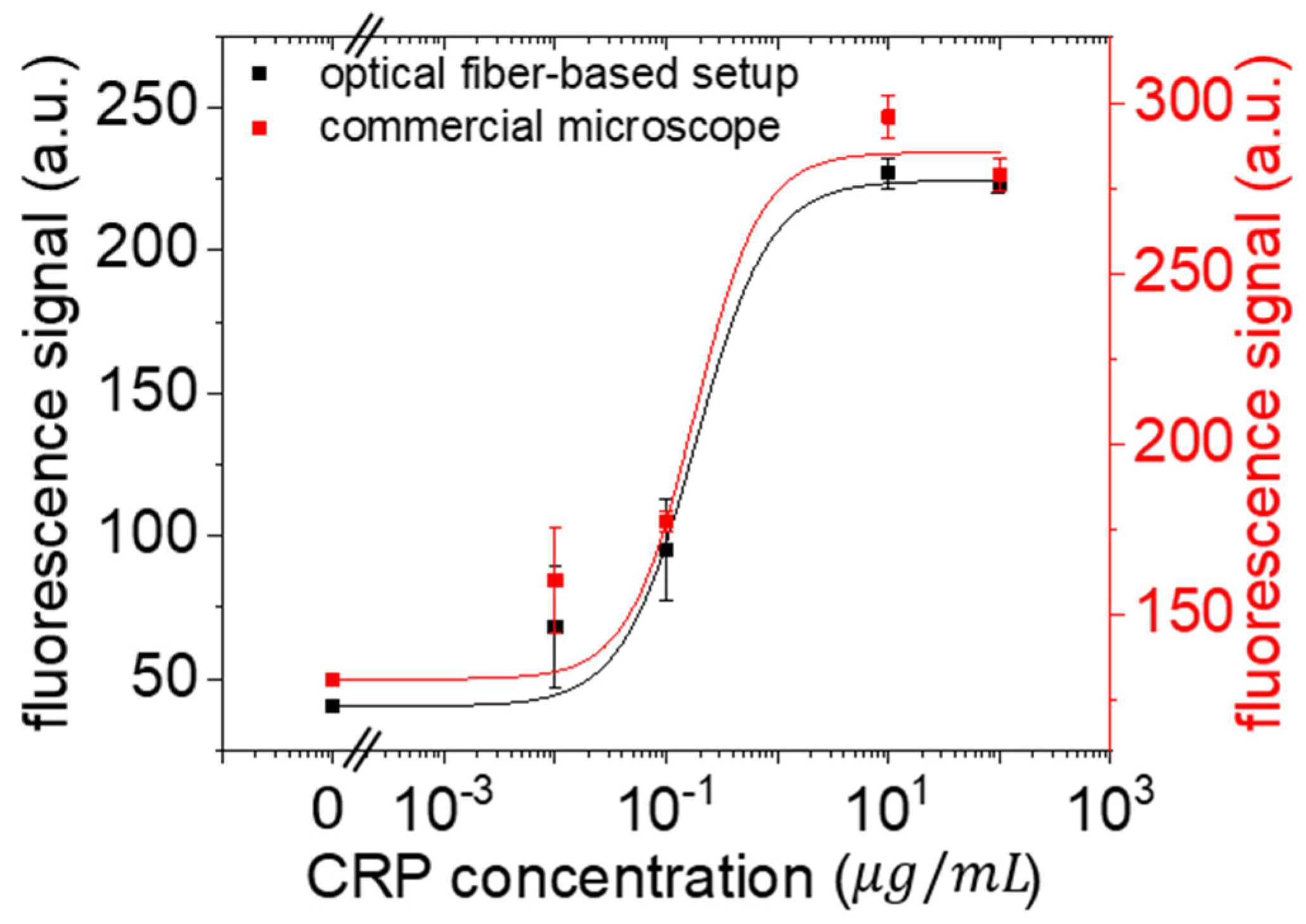
3. Results and Discussion
3.1. Prototype Working Principle
3.2. IgG/Anti-IgG Calibration Curve
3.3. CRP Calibration Curve
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Biosensors for cardiac biomarkers detection: A review. Sens. Actuators B Chem. 2012, 171, 62–76. [Google Scholar]
- Bellagambi, F.G.; Lomonaco, T.; Salvo, P.; Vivaldi, F.; Hangouët, M.; Ghimenti, S.; Biagini, D.; Di Francesco, F.; Fuoco, R.; Errachid, A. Saliva sampling: Methods and devices. An overview. Trends Anal. Chem. 2020, 124, 115781. [Google Scholar]
- Sonker, M.; Sahore, V.; Woolley, A.T. Recent advances in microfluidic sample preparation and separation techniques for molecular biomarker analysis: A critical review. Anal. Chim. Acta 2017, 986, 1–11. [Google Scholar] [PubMed]
- Chandra, P.; Segal, E. Nanobiosensors for Personalized and Onsite Biomedical Diagnosis; eBook; The Institution of Engineering and Technology: Hong Kong, China, 2016. [Google Scholar]
- Stern, E.; Vacic, A.; Rajan, N.K.; Criscione, J.M.; Park, J.; Ilic, B.R.; Mooney, D.J.; Reed, M.A.; Fahmy, T.M. Label-free biomarker detection from whole blood. Nat. Nanotechnol. 2010, 5, 138–142. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Wei, X.; Xue, Y.; Wan, H.; Wang, P. Recent advances in acoustic wave biosensors for the detection of disease-related biomarkers: A review. Anal. Chim. Acta 2021, 1164, 338321. [Google Scholar]
- Campuzano, S.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. New challenges in point of care electrochemical detection of clinical biomarkers. Sens. Actuators B Chem. 2021, 345, 130349. [Google Scholar] [CrossRef]
- Singh, P. SPR biosensors: Historical perspectives and current challenges. Sens. Actuators B Chem. 2016, 229, 110–130. [Google Scholar]
- Damborsky, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar]
- Leung, A.; Shankar, P.M.; Mutharasan, R. A review of fiber-optic biosensors. Sens. Actuators B Chem. 2017, 125, 688–703. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar]
- Hoa, X.; Kirk, A.; Tabrizian, M. Towards integrated and sensitive surface plasmon resonance biosensors: A review of recent progress. Biosens. Bioelectron. 2007, 23, 151–160. [Google Scholar]
- Sepúlveda, B.; Angelomé, P.C.; Lechuga, L.M.; Liz-Marzán, L.M. LSPR-based nanobiosensors. Nano Today 2009, 4, 244–251. [Google Scholar] [CrossRef]
- Hammond, J.L.; Bhalla, N.; Rafiee, S.D.; Estrela, P. Localized surface plasmon resonance as a biosensing platform for developing countries. Biosensors 2014, 4, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Chiavaioli, F.; Santano Rivero, D.; Del Villar, I.; Socorro-Leránoz, A.B.; Zhang, X.; Li, K.; Santamaría, E.; Fernández-Irigoyen, J.; Baldini, F.; van den Hove, D.L.A.; et al. Ultrahigh sensitive detection of Tau protein as Alzheimer’s biomarker via microfluidics and nanofunctionalized optical fiber sensors. Adv. Photonics Res. 2022, 3, 2200044. [Google Scholar]
- Chiavaioli, F.; Janner, D. Fiber optic sensing with lossy mode resonances: Applications and perspectives. J. Light. Technol. 2021, 39, 3855–3870. [Google Scholar]
- Altug, H.; Oh, S.-H.; Maier, S.A.; Homola, J. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 2022, 17, 5–16. [Google Scholar]
- Taitt, C.R.; Anderson, G.P.; Ligler, F.S. Evanescent wave fluorescence biosensors. Biosens. Bioelectron. 2005, 20, 2470–2487. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, H.; Anderson, A.S.; Grace, W.K.; Grace, K.M.; Hartman, N.; Martinez, J.S.; Swanson, B.I. Waveguide-based biosensors for pathogen detection. Sensors 2009, 9, 5783–5809. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-enhanced Raman spectroscopy for in vivo biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar]
- Li, P.; Long, F.; Chen, W.; Chen, J.; Chu, P.K.; Wang, H. Fundamentals and applications of surface-enhanced Raman spectroscopy–based biosensors. Curr. Opin. Biomed. Eng. 2020, 13, 51–59. [Google Scholar] [CrossRef]
- Guo, Y.; Li, H.; Reddy, K.; Shelar, H.S.; Nittoor, V.R.; Fan, X. Optofluidic Fabry–Pérot cavity biosensor with integrated flow-through micro-/nanochannels. Appl. Phys. Lett. 2011, 98, 041104. [Google Scholar]
- Wu, H.; Huang, H.; Bai, M.; Liu, P.; Chao, M.; Hu, J.; Hao, J.; Cao, T. An ultra-low detection-limit optofluidic biosensor based on all glass Fabry-Perot cavity. Opt. Express 2014, 22, 31977–31983. [Google Scholar] [PubMed]
- Checcucci, S.; Lombardi, P.; Rizvi, S.; Sgrignuoli, F.; Gruhler, N.; Dieleman, F.B.; Cataliotti, F.S.; Pernice, W.H.; Agio, M.; Toninelli, C. Beaming light from a quantum emitter with a planar optical antenna. Light Sci. Appl. 2017, 6, e16245. [Google Scholar]
- Li, J.; Salandrino, A.; Engheta, N. Shaping light beams in the nanometer scale: A Yagi-Uda nanoantenna in the optical domain. Phys. Rev. B 2007, 76, 245403. [Google Scholar]
- Heykel, A.; Mahboub, O.; Bonod, N.; Devaux, E.; Popov, E.; Rigneault, H.; Ebbesen, T.W.; Wenger, J. Bright unidirectional fluorescence emission of molecules in a nanoaperture with plasmonic corrugations. Nano Lett. 2011, 11, 637–644. [Google Scholar]
- Soltani, N.; Agio, M. Planar antenna designs for efficient coupling between a single emitter and an optical fiber. Opt. Express 2019, 27, 30830–30841. [Google Scholar]
- Soltani, N.; Rabbany Esfahany, E.; Druzhinin, S.; Schulte, G.; Müller, J.; Sledz, F.; Flatae, A.M.; Butz, B.; Schönherr, H.; Markešević, N.; et al. Scanning planar Yagi-Uda antenna for fluorescence detection. J. Opt. Soc. Am. B 2021, 38, 2528. [Google Scholar]
- Soltani, N.; Esfahany, E.R.; Druzhinin, S.I.; Schulte, G.; Müller, J.; Butz, B.; Schönherr, H.; Agio, M.; Markešević, N. Biosensing with a scanning planar Yagi-Uda antenna. Biomed. Opt. Express 2022, 13, 539–548. [Google Scholar]
- Chiavaioli, F.; Zubiate, P.; Del Villar, I.; Zamarreño, C.R.; Giannetti, A.; Tombelli, S.; Trono, C.; Arregui, F.J.; Matias, I.R.; Baldini, F. Femtomolar detection by nanocoated fiber label-free biosensors. ACS Sens. 2018, 3, 936–943. [Google Scholar] [CrossRef]
- Kumari, S.; Moirangthem, R.S. Development of cost-effective plasmonic biosensor using partially embedded gold nanoparticles for detection of immunoglobulin proteins. Mater. Res. Express 2018, 5, 025031. [Google Scholar]
- Yu, C.; Irudayaraj, J. Quantitative evaluation of sensitivity and selectivity of multiplex nanoSPR biosensor assays. Biophys. J. 2007, 93, 3684–3692. [Google Scholar] [CrossRef]
- Baldini, F.; Bolzoni, L.; Giannetti, A.; Kess, M.; Krämer, P.M.; Kremmer, E.; Porro, G.; Senesi, F.; Trono, C. A new procalcitonin optical immunosensor for POCT applications. Anal. Bioanal. Chem. 2009, 393, 1183–1190. [Google Scholar] [PubMed]
- Chiavaioli, F.; Gouveia, C.A.J.; Jorge, P.A.S.; Baldini, F. Towards a uniform metrological assessment of grating-based optical fiber sensors: From refractometers to biosensors. Biosensors 2017, 7, 23. [Google Scholar] [PubMed]
- Mocak, J.; Bond, A.M.; Mitchell, S.; Scollary, G. A Statistical Overview of Standard (IUPAC and ACS) and New Procedures for Determining the Limits of Detection and Quantification: Application to Voltammetric and Stripping Techniques. Pure Appl. Chem. 1997, 69, 297–328. [Google Scholar]
- Buchegger, P.; Preininger, C. Four assay designs and on-chip calibration: Gadgets for a sepsis protein array. Anal. Chem. 2014, 86, 3174–3180. [Google Scholar] [PubMed]
- Clyne, B.; Olshaker, J.S. The C-reactive protein. J. Emerg. Med. 1999, 17, 1019–1025. [Google Scholar] [PubMed]
- Faix, J.D. Biomarkers of sepsis. Crit. Rev. Clin. Lab. Sci. 2013, 50, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, N.; Mohanty, S.; Miller, C.S.; Langub, M.C.; Floriano, P.N.; Dharshan, P.; Ali, M.F.; Bernard, B.; Romanovicz, D.; Anslyn, E.; et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip 2005, 5, 261–269. [Google Scholar]
- Wolf, M.; Juncker, D.; Michel, B.; Hunziker, P.; Delamarche, E. Simultaneous detection of C-reactive protein and other cardiac markers in human plasma using micromosaic immunoassays and self-regulating microfluidic networks. Biosens. Bioelectron. 2004, 19, 1193–1202. [Google Scholar]
- Baldini, F.; Carloni, A.; Giannetti, A.; Porro, G.; Trono, C. An optical PMMA biochip based on fluorescence anisotropy: Application to C-reactive protein assay. Sens. Actuators B Chem. 2009, 139, 64–68. [Google Scholar] [CrossRef]
- Algarra, M.; Campos, B.B.; Gomes, D.; Alonso, B.; Casado, C.M.; Arrebola, M.M.; Diez de los Rios, M.J.; Herrera-Gutiérrez, M.E.; Seller-Pérez, G.; Esteces da Silva, J.C.G. Thiolated DAB dendrimer/ZnSe nanoparticles for C-reactive protein recognition in human serum. Talanta 2012, 99, 574–579. [Google Scholar] [CrossRef] [PubMed]

| Instrument | LOD (μg/mL) IgG/Anti-IgG | LOD (μg/mL) CRP in Serum | LOQ (μg/mL) IgG/Anti-IgG | LOQ (μg/mL) CRP in Serum |
|---|---|---|---|---|
| Commercial equipment | 0.14 | 0.005 | 0.46 | 0.013 |
| Lens-less prototype | 0.03 | 0.0015 | 0.1 | 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteso, V.; Lombardi, P.; Chiavaioli, F.; Majumder, P.; Colautti, M.; Howitz, S.; Cecchi, P.; Baldini, F.; Giannetti, A.; Toninelli, C. Enhanced Fluorescence in a Lens-Less Fiber-Optic Sensor for C-Reactive Protein Detection. Chemosensors 2023, 11, 448. https://doi.org/10.3390/chemosensors11080448
Esteso V, Lombardi P, Chiavaioli F, Majumder P, Colautti M, Howitz S, Cecchi P, Baldini F, Giannetti A, Toninelli C. Enhanced Fluorescence in a Lens-Less Fiber-Optic Sensor for C-Reactive Protein Detection. Chemosensors. 2023; 11(8):448. https://doi.org/10.3390/chemosensors11080448
Chicago/Turabian StyleEsteso, Victoria, Pietro Lombardi, Francesco Chiavaioli, Prosenjit Majumder, Maja Colautti, Steffen Howitz, Paolo Cecchi, Francesco Baldini, Ambra Giannetti, and Costanza Toninelli. 2023. "Enhanced Fluorescence in a Lens-Less Fiber-Optic Sensor for C-Reactive Protein Detection" Chemosensors 11, no. 8: 448. https://doi.org/10.3390/chemosensors11080448
APA StyleEsteso, V., Lombardi, P., Chiavaioli, F., Majumder, P., Colautti, M., Howitz, S., Cecchi, P., Baldini, F., Giannetti, A., & Toninelli, C. (2023). Enhanced Fluorescence in a Lens-Less Fiber-Optic Sensor for C-Reactive Protein Detection. Chemosensors, 11(8), 448. https://doi.org/10.3390/chemosensors11080448









