Abstract
The threat of glyphosate to food safety has attracted widespread attention. Consequently, it is highly urgent to develop a sensitive and accurate method for glyphosate detection. Herein, a turn-on fluorescent method for glyphosate detection using polydopamine-polyethyleneimine (PDA-PEI) copolymer as a fluorescent probe and p-nitrophenylphosphate (PNPP)/alkaline phosphatase (ALP) as a fluorescence quenching system is developed. The PDA-PEI copolymer was prepared by a one-pot method under mild condition, and its fluorescence kept almost unchanged after storing in a refrigerator for one month. ALP catalyzed the hydrolysis of PNPP to p-nitrophenol (PNP) that caused the fluorescence quenching of PDA-PEI copolymer via the inner filter effect. However, glyphosate inhibited ALP activity, thereby preventing the formation of PNP and restoring the fluorescence signal. Under the optimized conditions, the fluorescence of PDA-PEI copolymer depended on glyphosate concentrations ranging from 0.2 to 10 μg/mL with a detection limit of 0.06 μg/mL. Moreover, this assay was applied to detect glyphosate in real samples using the standard addition method. The recoveries were in the range from 88.8% to 107.0% with RSD less than 7.78%. This study provides a novel insight for glyphosate detection and expands the applications of fluorescent copolymer.
1. Introduction
Glyphosate (N-(phosphonomethyl)glycine) is a nonselective, broad-spectrum, and high-efficiency organophosphorus pesticide [1] that has been widely used to control the desirable weeds in global agriculture production. In particular, the development of glyphosate-resistant transgenic crops has contributed to its wide applications [2]. However, the excessive use of glyphosate results in the contamination of soil, air, and environmental water as well as agricultural products. Although its carcinogenicity is controversial, some relevant work has demonstrated that glyphosate shows certain teratogenicity, reproductive toxicity, and mutagenicity [3]. Consequently, the excessive residue of glyphosate represents a risk to the ecological environment, food safety, and human health. Developing sensitive and accurate methods for glyphosate detection is of great importance.
Current methods for glyphosate detection mainly include chromatography [4], capillary electrophoresis [5], electrochemistry [6], photoelectrochemistry [7], and colorimetry [8]. However, these methods are limited by laborious and time-consuming pretreatment or low sensitivity. To address the above problems, fluorimetry has elicited great interest since it offers the advantages of simple and time-saving operation and excellent sensitivity [9,10,11]. The fluorescent methods for glyphosate detection are usually established based on fluorescent probe-Cu2+ (Fe3+) [12] or fluorescent nanomaterials-Au (Ag) nanoparticles, as these ions or metal nanoparticles can cause the fluorescence quenching of those fluorescent nanomaterials, and glyphosate can interact with these ions or nanoparticles, resulting in the fluorescence recovery. For example, Wang et al. have reported a fluorescent assay for glyphosate detection based on carbon dots/Cu2+ [13], finding that Cu2+ quenches the fluorescence of carbon dots and that the introduction of glyphosate interacts with Cu2+, causing the fluorescence recovery. Similarly, various detection platforms based on N-doped silicon quantum dots/Cu2+ [14], carbon dots/Fe3+ [15], CdTe quantum dots/gold nanoparticles [16], and carbon quantum dots/Ag nanoparticles [17] have been successfully applied to detect glyphosate. However, the low selectivity of these methods have undoubtedly limited their practical applications. In view of these cases, many enzyme-based methods on the basis of the inhibition effects of glyphosate on enzymes activity have been proposed. For example, papain-stabilized gold nanoclusters (papain-AuNCs)/tyrosinase/dopamine have been used for glyphosate detection [18]. Tyrosinase catalyzes the oxidation of dopamine to dopamine-chrome, which results in the fluorescence quenching of papain-AuNCs, where glyphosate inhibits the activity of tyrosinase and thereby restores the fluorescence of papain-AuNCs. Similar to the above detection method, glyphosate also presents an inhibition effect on alkaline phosphatase (ALP) activity and causes the fluorescence changes of coordination polymer networks (CPNs) [9]. GMP/Eu/DPA and GMP/Tb CPNs were firstly constructed as a ratiometric probe. ALP quenches the fluorescence of GMP/Tb and enhances the fluorescence emissions of GMP/Eu/DPA. In turn, glyphosate inhibits ALP activity, resulting in the fluorescence enhancement of GMP/Tb and the fluorescence decrease of GMP/Eu/DPA. Based on the above facts, a ratiometric fluorescent method for glyphosate detection has been proposed. Despite the high selectivity of these methods, they still suffer from the limitation of the high cost of these fluorescent materials. Therefore, developing enzyme-based methods for glyphosate detection by utilization of fluorescent materials with low cost is highly desired.
Dopamine (DA), a kind of catecholamine, can be spontaneously oxidized and polymerized to form polydopamine (PDA) under a mild alkaline condition [19]. As an intrinsic fluorescent polymer, PDA has been used to construct fluorescent assays due to its unique physicochemical properties [20,21,22]. However, the low fluorescence intensity of PDA has undoubtedly limited its applications. Consequently, many efforts have been made to enhance its fluorescence intensity, including chemical oxidation [23,24,25] and conjugation with amine- or thiol-containing organic species [26,27,28]. Among them, the conjugation of polyethylenimine (PEI) and DA to form fluorescent PDA-PEI copolymer with enhanced fluorescence intensity has aroused great interest [28]. PEI can not only promote the polymerization of DA but also react with DA, resulting in the fluorescence enhancement of PDA. Despite its easy preparation and high stability, fluorescent PDA-PEI copolymer have been basically used to detect metal ions [29]. Therefore, it is of great importance to expand their applications in the development of fluorescent assays.
In light of the above facts, herein, PDA-PEI copolymer is prepared and firstly used as fluorescent probe for glyphosate detection. PDA-PEI copolymer prepared by the polymerization of dopamine with PEI under mild condition is endowed with the distinctive advantages of chemical stability, simple synthesis route, and low cost [30]. Due to the large overlap between the absorption spectrum of PNP and the fluorescence excitation spectrum of PDA-PEI copolymer, ALP catalyzes the hydrolysis of p-nitrophenylphosphate (PNPP) to form p-nitrophenol (PNP), which causes the fluorescence quenching of PDA-PEI copolymer through the inner filter effect (IFE). In turn, the inhibition effect of glyphosate on ALP activity prevents the formation of PNP and thereby restores the fluorescence of PDA-PEI copolymer. Therefore, a fluorescent assay with high sensitivity and selectivity for glyphosate detection based on PDA-PEI copolymer is established. This work may provide a novel fluorescent method for glyphosate detection and contribute to expanding the applications of fluorescent copolymer in herbicides detection.
2. Materials and Methods
2.1. Reagents and Instruments
Alkaline phosphatase, dopamine, bovine serum albumin (BSA), tris(hydroxymethyl)methylaminomethane (Tris), acetylcholinesterase (AChE), and MgCl2•6H2O were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lysozyme, pancreatin, and pepsin were supplied by Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Glyphosate, acetamiprid, methylviologen, polyethyleneimine (PEI), p-Nitrophenylphosphate (PNPP), p-nitrophenol (PNP), glucose, fructose, trypsin, and catalase were provided by Aladdin Reagent Company (Shanghai, China). Imidacloprid, thiamethoxam, and clothianidin were obtained from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). All chemicals were of analytical reagent grade and used as received.
Fluorescence measurements were performed using a RF-6000 spectrofluorometer (Shimadzu, Kyoto, Japan). Time-resolved fluorescence decay measurements were collected on a FLS-920 fluorescence spectrophotometer (Edinburgh, UK). UV-vis absorption spectra were recorded using a UV-8000 spectrophotometer (Shanghai Metash Instruments Co., Ltd., Shanghai, China). Transmission electron microscopy (TEM) images were measured using JEM-2100Plus (JEOL Ltd., Tokoy, Japan). Fourier transform infrared (FT-IR) spectroscopy were obtained on a Nicolet iS50 instrument (ThermoScientific, Waltham, MA, USA).
2.2. Preparation of PDA-PEI Copolymer
PDA-PEI copolymer was prepared via a simple one-pot method according to a previous report [19]. First, 50 mg of PEI and 50 mg of dopamine were sequentially introduced into a 100 mL round-bottom flask containing 50 mL of Tris-HCl buffer solution (10 mM, pH = 8.5) and stirred for 12 h at room temperature. Subsequently, the obtained solution was centrifugated at 11,000 rpm/min for 10 min. Then, the supernatant was filtered through a 0.22 µm syringe filter and dialyzed against ultrapure water through a dialysis bag (molecular weight cut-off is 1000 Da) for 4 h. Finally, the PDA-PEI copolymer was collected and stored at 4 °C for subsequent use. As a control, PDA was prepared similarly to the above-mentioned method except for the introduction of PEI.
2.3. Detection Procedure for ALP
In the detection procedure for ALP, 10 μL of various activities of ALP were sequentially introduced into 70 μL of Tris-HCl buffer solution (10 mM, pH = 9.0) containing 10 μL of 1 mM MgCl2 and 10 μL of 40 mM PNPP and incubated at 37 °C for 60 min. Then, 870 μL of ultrapure water and 30 μL of PDA-PEI copolymer were added, mixed thoroughly, and subjected to fluorescence spectral measurements.
2.4. Procedure for Glyphosate Sensing
In the procedure for glyphosate sensing, 10 μL of various concentrations of glyphosate were respectively added to 70 μL of Tris-HCl buffer (10 mM, pH = 9.0) containing 10 μL of 1 mM MgCl2 and 10 μL of 4 U/mL ALP. After reacting at 37 °C for 30 min, 10 μL of 40 mM PNPP was introduced and incubated at 37 °C for another 60 min. Finally, 860 μL of ultrapure water and 30 μL of PDA-PEI copolymer were introduced, mixed uniformly, and subjected to fluorescence spectral measurements.
2.5. Detection Glyphosate in Real Samples
Tap water and lake water samples were respectively obtained from our laboratory and Jiazi Lake of University of Jinan. They were firstly filtered through a 0.22 µm membrane and then diluted 10 times. Apples and pears were purchased from a local supermarket. A total of 5.0 g edible parts of an apple and a pear were firstly chopped and crushed into a homogenate with the introduction of 25 mL Tris-HCl buffer (10 mM, pH = 7.4). After stirring at room temperature for 2 h, the homogenate was centrifugated at 12000 rpm/min for 10 min, and the supernatant was filtered through a 0.22 µm membrane and diluted 100 times. Different concentrations of glyphosate were prepared using the water and food samples, respectively. Detection of glyphosate content in real samples was consistent with the step of detecting glyphosate in buffer solutions except that the glyphosate standard solutions were replaced by the spiked real samples.
3. Results
3.1. Characterization of PDA-PEI Copolymer
TEM was firstly used to characterize the morphologies of PDA-PEI copolymer. As shown in Figure 1A, PDA-PEI copolymer was well dispersed with an average size of 110 nm in diameter. FT-IR spectrum was utilized to identify its functional groups (Figure 1B). The peak at 3433 cm−1 originated from the stretching vibrations of N-H and O-H. The peaks at 2922 and 2852 cm−1 were assigned to the stretching vibration of C-H. The peak at 1631 cm−1 was attributed to C-N vibration. The above results have demonstrated that PDA-PEI copolymer contained amino and hydroxyl groups. Then, its optical properties were further evaluated. PDA-PEI copolymer was yellow and showed green fluorescence under a UV lamp (Figure 1C). When excited at 380 nm, PDA-PEI copolymer exhibited an emission peak centered at 526 nm. The fluorescence of PDA-PEI copolymer was excitation wavelength-dependent. Its fluorescence intensity reached the maximum when excited at 380 nm (Figure 1D). Differently from PDA-PEI copolymer, PDA and PEI displayed weak fluorescence (Figure S1A). The above results suggested that PEI was helpful to enhance the fluorescence intensity of PDA. Finally, the stability of PDA-PEI copolymer was studied by testing its fluorescence intensity every five days. After storing in a refrigerator for one month, the fluorescence intensity of PDA-PEI copolymer stayed almost unchanged (Figure S1B), indicating the high stability of the as-prepared PDA-PEI copolymer.
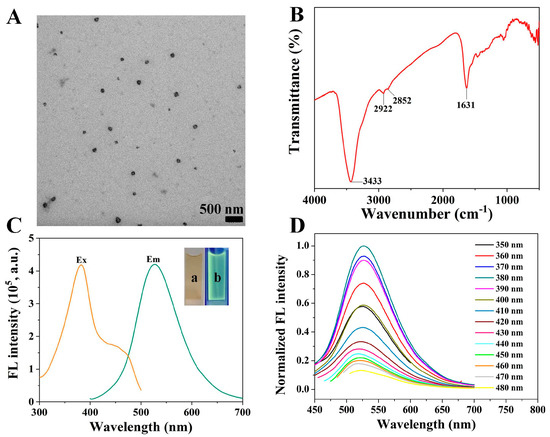
Figure 1.
TEM image (A), FT-IR spectrum (B), and fluorescence excitation and emission spectra of PDA-PEI copolymer (C). Inset shows the images of PDA-PEI copolymer taken under sunlight (a) and a UV lamp excited at 365 nm (b). (D) Fluorescence emission spectra of PDA-PEI copolymer under different excitation wavelengths.
3.2. ALP Detection Based on PDA-PEI Copolymer
To demonstrate the feasibility of PDA-PEI copolymer in ALP detection, either PNPP or ALP was firstly individually introduced to PDA-PEI copolymer solution. The introduction of ALP showed negligible effect on the fluorescence of PDA-PEI copolymer (Figure 2A). The addition of PNPP caused a fluorescence decrease of PDA-PEI copolymer. Moreover, its fluorescence decreased gradually with the increased concentrations of PNPP (Figure S2A). However, its fluorescence decreased dramatically after the simultaneous introduction of PNPP and ALP. Taking the above facts into consideration, PDA-PEI copolymer could be utilized for ALP detection.
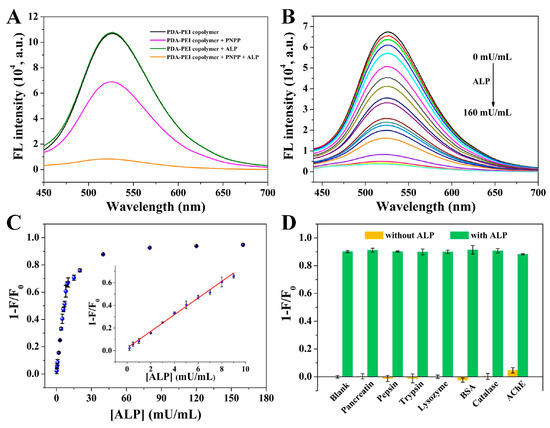
Figure 2.
(A) Fluorescence spectra of PDA-PEI copolymer (1), (1)-PNPP, (1)-ALP, and (1)-PNPP-ALP. (B) Fluorescence responses of PDA-PEI copolymer/PNPP after the introduction of various activities of ALP. From top to bottom, the ALP activities are 0, 0.2, 0.5, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 40, 80, 120, and 160 mU/mL. (C) The relationship between 1 − F/F0 and ALP activities. Inset shows the corresponding calibration plot between 1 − F/F0 and ALP activities. (D) Fluorescence responses of PDA-PEI copolymer-PNPP after the individual addition of pancreatin, pepsin, trypsin, lysozyme BSA, catalase, and AChE or the simultaneous addition of each with ALP. The concentrations of these interfering substances and ALP were 4 μg/mL. Error bars demonstrate the standard deviation of three independent measurements.
Prior to testing its use for ALP detection, several factors (including PNPP concentration, incubation time of PNPP with ALP, and the volume of PDA-PEI copolymer) were optimized to sensitively detect ALP. The value of 1 − F/F0 was used as the criterion to optimize detection conditions, where F0 and F, respectively, represented the fluorescence of this assay at 526 nm before and after the addition of ALP. The effect of PNPP concentration was firstly studied. As shown in Figure S2A, the value of F0 decreased gradually with the increasing concentration of PNPP, while the value of F firstly decreased step by step and then stayed almost unchanged when the concentration of PNPP was higher than 0.4 mM. As a result, when the concentration of PNPP was 0.4 mM, the value of 1 − F/F0 was the largest (Figure S2B). Then, the impact of incubation time of PNPP with ALP was investigated. With the extension of incubation time, the value of F0 stayed almost constant, while the value of F decreased gradually and then remained almost unchanged when the incubation time was longer than 60 min (Figure S2C). As displayed in Figure S2D, the value of 1 − F/F0 increased gradually and stayed constant when the incubation time was longer than 60 min (Figure S2D). Finally, the volume of PDA-PEI copolymer was studied. Both the value of F0 and F increased step by step with the increasing volume of PDA-PEI copolymer (Figure S2E). As demonstrated in Figure S2F, the value of 1 − F/F0 reached the maximum when the volume of PDA-PEI copolymer was 30 μL. Consequently, 0.4 mM of PNPP, incubation time of 60 min, and 30 μL of PDA-PEI copolymer were utilized for ALP detection.
Under the optimized experimental conditions, the fluorescence emission spectra of PDA-PEI copolymer in the presence of 0.4 mM PNPP after the introduction of various activities of ALP were recorded. As shown in Figure 2B, the fluorescence intensity of PDA-PEI copolymer decreased gradually with the increasement of ALP activity. Conversely, the value of 1 − F/F0 increased step by step and stayed almost unchanged when ALP activity was higher than 40 mU/mL (Figure 2C). A linear relationship between the value of 1 − F/F0 and ALP activities within the dynamic range from 0.2 to 9 mU/mL was obtained. The working equation was 1 − F/F0 = 0.023 + 0.074 [ALP] (mU/mL) (R2 = 0.990). The detection limit was calculated to be 0.16 mU/mL based on the signal-to-noise ratio of 3, which was comparable or even lower than the limit found using previously reported methods, as shown in Table S1 [31,32,33,34,35,36,37,38]. The selectivity and anti-interference ability of this method were further investigated. The individual introduction of these possible interfering substances caused no obvious fluorescence changes compared to blank solution, whereas the individual introduction of ALP or simultaneous introduction of ALP with these interfering substances resulted in significant decrease of fluorescence intensity (Figure 2D). These results undoubtedly verified the high selectivity and good anti-interference ability of this method for ALP detection.
3.3. Analytical Performance for Glyphosate Determination
Glyphosate, a typical organophosphorus pesticide, could inhibit the activity of ALP. As a consequence, this assay was further used to determine glyphosate. As exhibited in Figure 3A, the introduction of glyphosate showed no obvious effect on the fluorescence of PDA-PEI copolymer-ALP or the PDA-PEI copolymer-PNPP system. However, the quenched fluorescence of PDA-PEI copolymer by PNPP and ALP could recover after the addition of glyphosate. The above results have verified the feasibility of this assay for glyphosate detection.
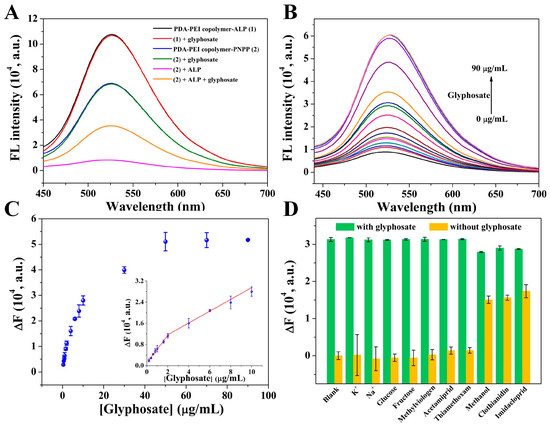
Figure 3.
(A) Fluorescence spectra of PDA-PEI copolymer-ALP (1), (1)-glyphosate, PDA-PEI copolymer-PNPP (2), (2)-glyphosate, (2)-ALP, and (2)-ALP-glyphosate. (B) Fluorescence emission spectra of PDA-PEI copolymer-PNPP-ALP after the addition of different concentrations of glyphosate. From bottom to top, the glyphosate concentrations are 0, 0.2, 0.4, 0.6, 0.8, 1, 1.6, 2, 4, 6, 8, 10, 30, 50, 70, and 90 μg/mL. (C) Scatter diagram of the value of ∆F versus glyphosate concentration. Inset is the linear curve of ∆F versus glyphosate concentration. (D) Fluorescence responses of this assay against K+, Na+, glucose, fructose, methylviologen, acetamiprid, thiamethoxam, methanol, clothianidin, and imidacloprid in the absence and presence of glyphosate. The concentrations of these pesticides and carbohydrates were 10 μg/mL, and the concentrations of K+ and Na+ were 100 μg/mL. Error bars demonstrate the standard deviation of three independent measurements.
The analytical performance of this assay was affected by some critical factors, such as the incubation time of ALP with glyphosate and the pH of the buffer solution. Therefore, these testing conditions were systematically optimized before its application in glyphosate detection. The value of ∆F (F − F0) was used to screen the detection conditions, where F0 and F represented the fluorescence intensity of PDA-PEI copolymer-PNPP-ALP at 526 nm before and after the addition of glyphosate. The effect of the incubation time of glyphosate with ALP was firstly investigated. As obviously shown in Figure S3A, the value of both F and F0 changed little with the increasing incubation time. The value of ∆F increased gradually with the incubation time, ranging from 10 to 60 min (Figure S3B). However, the value of ∆F increased slowly when the incubation time was longer than 30 min. Taking high sensitivity and short detection time into consideration, the incubation time was fixed at 30 min. Then, the effect of buffer solution pH on F and F0 was studied (Figure S3C). The value of F0 decreased gradually with the increasing pH value of the buffer solution, while the value of F stayed almost unchanged. The value of ∆F increased with the increasing pH of the buffer solution, and the change between pH 9 and 9.5 was not significant (Figure S3D). As a result, the incubation time of 30 min and pH 9.0 buffer solution were used in the next experiment.
Under the optimal experimental conditions, the fluorescence responses of this assay to different concentrations of glyphosate were studied. The fluorescence signal increased gradually with the continuing addition of glyphosate (Figure 3B). Consequently, ∆F increased gradually within the glyphosate concentration range of 0.2–50 μg/mL and stayed almost constant when its concentration was higher than 50 μg/mL (Figure 3C). A linear response was obtained in the glyphosate concentration range of 0.2–2 μg/mL (∆F = 1096.60 + 5157.98 [glyphosate] (μg/mL), R2 = 0.986). Another good linear relationship between ∆F and glyphosate concentration in the range of 2–10 μg/mL (∆F = 7192.10 + 2232.18 [glyphosate] (μg/mL), R2 = 0.986) was obtained. The detection limit was estimated to be 0.06 μg/mL, which was comparable or lower than other methods shown in Table 1 [1,9,11,14,15,17,39,40]. Moreover, the detection limit was much lower than the maximum residue level of glyphosate in drinking water (0.9 μg/mL) set by the World Health Organization [41]. Additionally, the low cost and high stability of fluorescent PDA-PEI copolymer, simple and time-saving detection procedure, makes this fluorescent assay more attractive.

Table 1.
Comparison of the analytical performances of different methods for glyphosate detection.
Then, the selectivity and anti-interference ability of this assay were investigated by evaluating the fluorescence responses of the PDA-PEI copolymer-PNPP-ALP system toward metal ions, carbohydrates, organochlorine pesticides (methylviologen), and neonicotine pesticides (acetamiprid, thiamethoxam, clothianidin, and imidacloprid) both in the absence and presence of glyphosate. As depicted in Figure 3D, a remarkable fluorescence response toward glyphosate was observed, while no obvious fluorescence changes were found after the introduction of metal ions, carbohydrates, methylviologen, acetamiprid, and thiamethoxam. Fluorescence changes caused by clothianidin and imidacloprid were ascribed to their preparation in methanol medium since their fluorescence changes were similar to that of methanol. The above results demonstrated the high selectivity of this assay for glyphosate detection. Then, the anti-interference ability of this assay toward glyphosate detection was further investigated by the simultaneous addition of glyphosate with these interfering substances. It could be clearly observed that the fluorescence responses toward glyphosate stayed almost unchanged even when these interfering substances existed, demonstrating the good anti-interference ability of this assay.
3.4. Determination of Glyphosate in Actual Samples
The high selectivity and good anti-interference ability of this assay demonstrated the great potential of this assay in practical applications. To verify the practicality of this fluorescent assay, detection of glyphosate in water and fruits samples was carried out based on a standard addition method. The real samples were spiked with known quantities of glyphosate (1 μg/mL and 10 μg/mL) and analyzed according to the established fluorescent assay. The recoveries of glyphosate in the spiked samples were in the range from 88.8% to 107.0%, with RSD ranging from 2.57% to 7.78% (Table 2). The results have revealed that this assay was reliable for detection of glyphosate in real samples.

Table 2.
The results for detection of glyphosate in real samples based on the standard addition method.
3.5. Mechanism of This Assay for Glyphosate Detection
PNPP as ALP substrate could be hydrolyzed to PNP, inducing the increase of absorbance at 400 nm accompanied with the decrease of absorbance at 310 nm (Figure S4A). However, ALP activities were inhibited by glyphosate, causing a decrease of absorbance at 400 nm and an increase of absorbance at 310 nm (Figure S4B). The fluorescence of PDA-PEI copolymer was progressively decreased with the increasing concentration of PNP (Figure 4A). Consequently, the decreased fluorescence of PDA-PEI copolymer was attributed to PNP. The absorption spectrum of PNP largely overlapped with the fluorescence excitation spectrum of PDA-PEI copolymer (Figure 4B), indicating that the fluorescence quenching of PDA-PEI copolymer caused by PNP may be attributed to IFE. Then, time-resolved fluorescence spectra were explored to further verify the quenching mechanism. Figure 4C shows that the fluorescence lifetime of PDA-PEI copolymer stayed almost unchanged before and after the introduction of PNP, suggesting the IFE of PNP on PDA-PEI copolymer. Finally, the IFE-based fluorescence quenching mechanism was further confirmed by the excitation wavelength-dependent fluorescence quenching spectrum of PDA-PEI copolymer. As exhibited in Figure 4D, the excitation wavelength-dependent fluorescence quenching spectrum was similar to the absorption spectrum of PNP. Consequently, it was reasonable to deduce that the fluorescence quenching of PDA-PEI copolymer by PNP could be attributed to IFE [42]. Based on the above facts, a fluorescent assay for glyphosate detection was constructed on the basis of the IFE between PNP and PDA-PEI copolymer and the inhibition effect of glyphosate on ALP activity (Scheme 1).
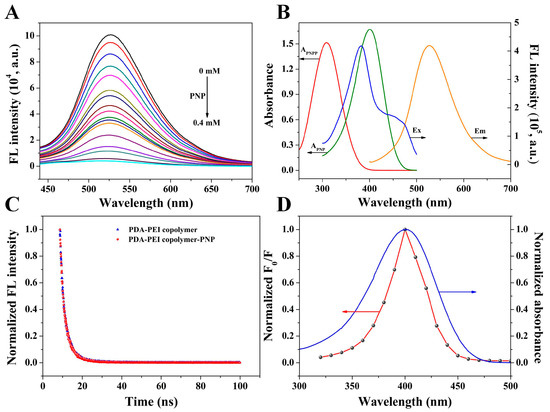
Figure 4.
(A) Fluorescence emission spectra of PDA-PEI copolymer upon introduction of various concentrations of PNP. From top to bottom, PNP concentrations are 0, 0.001, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.125, 0.15, 0.2, 0.3, and 0.4 mM. (B) Absorption spectra of PNPP and PNP, fluorescence excitation, and emission spectra of PDA-PEI copolymer. (C) The lifetime of PDA-PEI copolymer in the absence and presence of 1.0 mM PNP. (D) The relationship between the normalized F0/F and different excitation wavelengths and the absorption spectrum of PNP, where F0 and F represented the fluorescence of PDA-PEI copolymer in the absence and presence of 0.1 mM PNP.
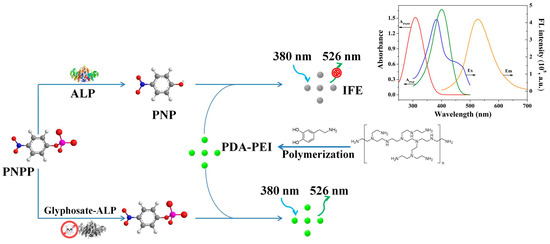
Scheme 1.
Detection principle of PDA-PEI copolymer-based fluorescent assay.
4. Conclusions
In summary, a fluorescent assay for glyphosate detection was successfully established on the basis of the quenching effect of PNP on PDA-PEI copolymer via IFE and the inhibition effect of glyphosate on ALP activity, where PNP was obtained from the hydrolysis of PNPP catalyzed by ALP. The established assay was successfully applied to detect glyphosate in real samples with satisfactory results. It exhibited several distinctive advantages, such as the facile preparation procedure, low cost, and high stability of the fluorescent PDA-PEI copolymer, simple and time-saving detection procedure with high sensitivity, excellent selectivity, and good anti-interference ability. This study offered a new insight for glyphosate detection and expanded the application of PDA-PEI copolymer in pesticide detection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/chemosensors11070398/s1. Figure S1. (A) Fluorescence spectra of 0.2 mg/mL PDA-PEI copolymer, PDA and PEI. (B) The stability of PDA-PEI copolymer. Figure S2. The impacts of PNPP concentration (A), incubation time of PNPP with ALP (C), the volume of PDA-PEI copolymer (E) on the fluorescence intensity of PDA-PEI QDs copolymer in the absence (F0 value) and presence of ALP (F value). The relationship between PNPP concentration (B), incubation time of PNPP with ALP (D), the volume of PDA-PEI copolymer (F) and 1-F0/F. Figure S3. The effects of incubation time of glyphosate with ALP (A) and the pH value of buffer solution (B) on glyphosate detection. The change trend of ∆F at different incubation time (C) and different pH values of buffer solution (D). Figure S4. UV-vis absorption spectra of PNPP in the absence and presence of various activities of ALP (A) and PNPP-ALP before and after the introduction of various concentrations of glyphosate (B). Table S1. Comparison the analytical performances of different methods for ALP detection.
Author Contributions
P.N., conceptualization, investigation, and writing-original draft; S.L., methodology, formal analysis, writing–review and editing; Y.L., writing–review and editing, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32202145, 22172063), the Young Taishan Scholars Program (tsqn201812080), and the Natural Science Foundation of Shandong Province (ZR2022QB192, ZR2019YQ10, and ZR2020QB008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The experimental data are available upon reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sawetwong, P.; Chairam, S.; Jarujamrus, P.; Amatatongchai, M. Enhanced selectivity and sensitivity for colorimetric determination of glyphosate using Mn-ZnS quantum dot embedded molecularly imprinted polymers combined with a 3D-microfluidic paper-based analytical device. Talanta 2021, 225, 122077. [Google Scholar] [CrossRef] [PubMed]
- Wiwasuku, T.; Boonmak, J.; Burakham, R.; Hadsadee, S.; Jungsuttiwong, S.; Bureekaew, S.; Promarak, V.; Youngme, S. Turn-on fluorescent probe towards glyphosate and Cr3+ based on Cd(II)-metal organic framework with Lewis basic sites. Inorg. Chem. Front. 2021, 8, 977–988. [Google Scholar] [CrossRef]
- Luo, D.; Huang, X.; Liu, B.; Zou, W.; Wu, Y. Facile colorimetric nanozyme sheet for the rapid detection of glyphosate in agricultural products based on inhibiting peroxidase-like catalytic activity of porous Co3O4 nanoplates. J. Agric. Food Chem. 2021, 69, 3537–3547. [Google Scholar] [CrossRef]
- Zelaya, I.A.; Anderson, J.A.H.; Owen, M.D.K.; Landes, R.D. Evaluation of spectrophotometric and HPLC methods for shikimic acid determination in plants: Models in glyphosate-resistant and -susceptible crops. J. Agric. Food Chem. 2021, 59, 2202–2212. [Google Scholar] [CrossRef]
- Munoz, R.; Guevara-Lara, A.; Santos, J.L.M.; Miranda, J.M.; Rodriguez, J.A. Determination of glyphosate in soil samples using CdTe/CdS quantum dots in capillary electrophoresis. Microchem. J. 2019, 146, 582–587. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, L.; Shen, C.; Wang, C.; Hu, X.; Wang, G. An electrochemical sensor on the hierarchically porous Cu-BTC MOF platform for glyphosate determination. Sens. Actuators. B Chem. 2019, 283, 487–494. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, L.; Wang, C.; Hu, X.; Liu, Y.; Wang, G. Sensitive detection of glyphosate based on a Cu-BTC MOF/g-C3N4 nanosheet photoelectrochemical sensor. Electrochim. Acta 2019, 317, 341–347. [Google Scholar] [CrossRef]
- Hamedpour, V.; Sasaki, Y.; Zhang, Z.; Kubota, R.; Minami, T. Simple colorimetric chemosensor array for oxyanions: Quantitative assay for herbicide glyphosate. Anal. Chem. 2019, 91, 13627–13632. [Google Scholar] [CrossRef]
- Qu, F.; Wang, H.; You, J. Dual lanthanide-probe based on coordination polymer networks for ratiometric detection of glyphosate in food samples. Food Chem. 2020, 323, 126815. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, J.; Liu, S.; Yang, J.; Zhang, H.; Yan, J.; Hu, X. Fluorescent carbon dots for glyphosate determination based on fluorescence resonance energy transfer and logic gate operation. Sens. Actuators B Chem. 2017, 242, 545–553. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, J.; Chen, X.; Yang, W.; Pei, H.; Hu, N.; Li, Z.; Suo, Y.; Li, T.; Wang, J. The simultaneous detection and removal of organophosphorus pesticides by a novel Zr-MOF based smart adsorbent. J. Mater. Chem. A 2018, 6, 2184–2192. [Google Scholar] [CrossRef]
- Sun, F.; Yang, L.; Li, S.; Wang, Y.; Wang, L.; Li, P.; Ye, F.; Fu, Y. New fluorescent probes for the sensitive determination of glyphosate in food and environmental samples. J. Agr. Food Chem. 2021, 69, 12661–12673. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bi, Y.; Gao, J.; Li, Y.; Ding, H.; Ding, L. Carbon dots based turn-on fluorescent probes for the sensitive determination of glyphosate in environmental water samples. RSC Adv. 2016, 6, 85820–85828. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Huo, D.; Ji, Z.; Ma, Y.; Yang, M.; Luo, H.; Luo, X.; Hou, C.; Lv, J. A turn-on fluorescent nanoprobe based on N-doped silicon quantum dots for rapid determination of glyphosate. Microchim. Acta 2020, 187, 341. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, X.; Lan, S.; Zhang, C.; Hou, C.; He, Q.; Huo, D. A turn-on fluorescent sensor based on carbon dots from Sophora japonica leaves for the detection of glyphosate. Anal. Methods 2020, 12, 4130–4138. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, Y.; Luo, Y.; Shen, F.; Sun, C. Efficient fluorescence resonance energy transfer between oppositely charged CdTe quantum dots and gold nanoparticles for turn-on fluorescence detection of glyphosate. Talanta 2014, 125, 385–392. [Google Scholar] [CrossRef]
- Wang, L.; Bi, Y.; Hou, J.; Li, H.; Xu, Y.; Wang, B.; Ding, H.; Ding, L. Facile, green and clean one-step synthesis of carbon dots from wool: Application as a sensor for glyphosate detection based on the inner filter effect. Talanta 2016, 160, 268–275. [Google Scholar] [CrossRef]
- Hong, C.; Ye, S.; Dai, C.; Wu, C.; Chen, L.; Huang, Z. Sensitive and on-site detection of glyphosate based on papain-stabilized fluorescent gold nanoclusters. Anal. Bioanal. Chem. 2020, 412, 8177–8184. [Google Scholar] [CrossRef]
- Zhong, Z.; Jia, L. Room temperature preparation of water-soluble polydopamine-polyethyleneimine copolymer dots for selective detection of copper ions. Talanta 2019, 197, 584–591. [Google Scholar] [CrossRef]
- Xiao, T.; Sun, J.; Zhao, J.; Wang, S.; Liu, G.; Yang, X. FRET effect between fluorescent polydopamine nanoparticles and MnO2 nanosheets and its application for sensitive sensing of alkaline phosphatase. ACS Appl. Mater. Interfaces 2018, 10, 6560–6569. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, S.; Chen, X.; Liu, X.; Wang, Z.; Li, Y. Recent developments in polydopamine fluorescent nanomaterials. Mater. Horiz. 2020, 7, 746–761. [Google Scholar] [CrossRef]
- Xue, Q.; Cao, X.; Zhang, C.; Xian, Y. Polydopamine nanodots are viable probes for fluorometric determination of the activity of alkaline phosphatase via the in situ regulation of a redox reaction triggered by the enzyme. Microchim. Acta 2018, 185, 231. [Google Scholar] [CrossRef]
- Kong, X.J.; Wu, S.; Chen, T.T.; Yu, R.Q.; Chu, X. MnO2-induced synthesis of fluorescent polydopamine nanoparticles for reduced glutathione sensing in human whole blood. Nanoscale 2016, 8, 15604–15610. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, K.; Wang, L.; Zhou, K.; Zeng, J.; Gao, D.; Xia, Z.; Fu, Q. Redox modulation of polydopamine surface chemistry: A facile strategy to enhance the intrinsic fluorescence of polydopamine nanoparticles for sensitive and selective detection of Fe3+. Nanoscale 2018, 10, 18064–18073. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, C.; Lu, Y.; Wei, G.; Ye, G.; Sun, T.; Chen, J. Microplasma electrochemistry controlled rapid preparation of fluorescent polydopamine nanoparticles and their application in uranium detection. Chem. Eng. J. 2018, 344, 480–486. [Google Scholar] [CrossRef]
- Chen, M.; Wen, Q.; Gu, F.; Gao, J.; Zhang, C.C.; Wang, Q. Mussel chemistry assembly of a novel biosensing nanoplatform based on polydopamine fluorescent dot and its photophysical features. Chem. Eng. J. 2018, 342, 331–338. [Google Scholar] [CrossRef]
- Tang, L.; Mo, S.; Liu, S.G.; Liao, L.L.; Li, N.B.; Luo, H.Q. Synthesis of fluorescent polydopamine nanoparticles by Michael addition reaction as an analysis platform to detect iron ions and pyrophosphate efficiently and construction of an IMPLICATION logic gate. Sens. Actuators B Chem. 2018, 255, 754–762. [Google Scholar] [CrossRef]
- Liu, M.; Ji, J.; Zhang, X.; Zhang, X.; Yang, B.; Deng, F.; Li, Z.; Wang, K.; Yang, Y.; Wei, Y. Self-polymerization of dopamine and polyethyleneimine: Novel fluorescent organic nanoprobes for biological imaging applications. J. Mater. Chem. B 2015, 3, 3476–3482. [Google Scholar] [CrossRef]
- Zhao, C.; Zuo, F.; Liao, Z.; Qin, Z.; Du, S.; Zhao, Z. Mussel-inspired one-pot synthesis of a fluorescent and water-soluble polydopamine-polyethyleneimine copolymer. Macromol. Rapid Comm. 2015, 36, 909–915. [Google Scholar] [CrossRef]
- Lin, Z.Y.; Xue, S.F.; Chen, Z.H.; Han, X.Y.; Shi, G.; Zhang, M. Bioinspired copolymers based nose/tongue-mimic chemosensor for label-free fluorescent pattern discrimination of metal ions in biofluids. Anal. Chem. 2018, 90, 8248–8253. [Google Scholar] [CrossRef]
- Lin, T.; Liu, S.; Huang, J.; Tian, C.; Hou, L.; Ye, F.; Zhao, S. Multicolor and photothermal dual-mode assay of alkaline phosphatase based on the UV light-assisted etching of gold nanorods, Anal. Chim. Acta 2021, 1181, 338926. [Google Scholar] [CrossRef]
- Xiang, J.; Wang, X.; Lin, A.; Wei, H. In situ exsolution of noble-metal nanoparticles on perovskites as enhanced peroxidase mimics for bioanalysis. Anal. Chem. 2021, 93, 5954–5962. [Google Scholar]
- Wu, T.; Ma, Z.; Li, P.; Liu, M.; Liu, X.; Li, H.; Zhang, Y.; Yao, S. Colorimetric detection of ascorbic acid and alkaline phosphatase activity based on the novel oxidase mimetic of Fe-Co bimetallic alloy encapsulated porous carbon nanocages. Talanta 2019, 202, 354–361. [Google Scholar] [CrossRef]
- Ye, K.; Niu, X.; Song, H.; Wang, L.; Peng, Y. Combining CeVO4 oxidase-mimetic catalysis with hexametaphosphate ion induced electrostatic aggregation for photometric sensing of alkaline phosphatase activity. Anal Chim Acta 2020, 1126, 16–23. [Google Scholar] [CrossRef]
- Ni, P.; Chen, C.; Jiang, Y.; Zhang, C.; Wang, B.; Lu, Y.; Wang, H. A fluorescent assay for alkaline phosphatase activity based on inner filter effect by in-situ formation of fluorescent azamonardine, Sens. Actuators B: Chem. 2020, 302, 127145. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Z.; Rao, H.; Xue, X.; Luo, M.; Xue, Z.; Lu, X. A two fluorescent signal indicator-based ratio fluorometric alkaline phosphatase assay based on one signal precursor. Chem. Commun. 2021, 57, 4444–4447. [Google Scholar] [CrossRef]
- Zhan, Y.; Yang, S.; Chen, L.; Zeng, Y.; Li, L.; Lin, Z.; Guo, Z.; Xu, W. Ultrahigh efficient FRET ratiometric fluorescence biosensor for visual detection of alkaline phosphatase activity and its inhibitor, ACS Sustain. Chem. Eng. 2021, 9, 12922–12929. [Google Scholar]
- Wang, M.; Zhou, X.; Cheng, L.; Wang, M.; Su, X. Lysozyme-functionalized 5-methyl-2-thiouracil gold/silver nanoclusters for luminescence assay of alkaline phosphatase. ACS Appl. Nano Mater. 2021, 4, 9265–9273. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, H.; Li, Z.; Pang, J.; Lin, T.; Guo, L.; Fu, F. Colorimetric sensing of glyphosate in environmental water based on peroxidase mimetic activity of MoS2 nanosheets. J. Nanosci. Nanotechnol. 2017, 17, 5730–5734. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, Z.; Hao, J.; Yang, W.; Tang, J. A simple label free colorimetric method for glyphosate detection based on the inhibition of peroxidase-like activity of Cu(II). Sens. Actuators B Chem. 2016, 228, 410–415. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, L.; Sharma, A.S.; Chen, M.; Chen, Q. A system composed of polyethylenimine-capped upconversion nanoparticles, copper(II), hydrogen peroxide and 3,3′,5,5′-tetramethylbenzidine for colorimetric and fluorometric determination of glyphosate. Microchim. Acta 2019, 186, 835. [Google Scholar] [CrossRef]
- Tang, D.; Zhang, J.; Zhou, R.; Xi, Y.N.; Hou, X.; Xu, K.; Wu, P. Phosphorescent inner filter effect-based sensing of xanthine oxidase and its inhibitors with Mn-doped ZnS quantum dots. Nanoscale 2018, 10, 8477–8482. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).