1. Introduction
The efforts made to design real-time assays reflect the exponentially growing interest in the fast and easy identification of many biomarkers for several diseases so that many lives can be saved thanks to the detection of a disease at an early stage as well as facilitating the management of more personalized and efficient therapies, aiming to develop and improve personalized medicine. With this in mind, fervent activity is directed towards the advancement of point-of-care (POC) devices [
1]: these tests enable analysis using very small sample volumes with no or little preparation, providing results in shorter times compared to routine lab tests. They offer the additional advantage of being interpreted and implemented without any special equipment or training, while they are administered externally to hospitals and clinics practice, in a home environment. Furthermore, they are cost-effective and can be easily transported. The World Health Organization (WHO) defined the criteria that must be satisfied for an ideal diagnostic system that can be summarized by the ASSURED acronym: affordable, sensitive, specific, user-friendly, rapid, equipment-free and delivered to the end users [
2]. To better understand the relevance of these characteristics, it is necessary to discuss the asymmetries of needs between developed and third-world countries. In developed countries, there is a need for decentralization in order to reduce the volume of presential access to hospitals, freeing resources for properly determined cases. To make it possible, the essential activities of disease screening and monitoring for prevention purposes should be performed at home by the patient themself or by untrained personnel. Access to these data could be provided to medical doctors through remote access, and the interpretation of such a large amount of data could be supported by an existing analytical approach. Proper detection of disease when still in an early stage and risk stratification would be of great help in disease prevention and treatment, improving healthy-aging expectations. Differently, in remote regions, the needs are quite different. Hospitals are insufficient, in numbers and dimensions, and a great number of people have almost no access to any kind of healthcare, even when in great need and in situations of serious illness. Among the many problems that can be claimed as a cause for such a situation, we recall here one of them, which is common to most countries and not depending on the specific government model: the remote geographical location of patients, at large distances from the hospitals, in a rural environment [
3]. Despite having very different natures, these two situations can be both addressed by the employment of new technologies for developing Point-of-Care (POC) systems. New technologies are employed to develop POC devices relying on lab-on-chip (LOC) hardware and remote connections based on Internet-of-Things (IoT) infrastructure. Lab-on-chip devices are detection systems based on microfluidics which integrate multiple procedures of a laboratory on a single chip of only a few square millimeters to a few square centimeters in size, in order to develop ‘from-sample-to-answer’ analytical systems where the input is a rough or low processed sample, and the output is a qualitative or quantitative evaluation of one or more required analytes. The design of an LOC device, thus, will take into account the various steps needed to perform the analysis: fluidic handling, molecular recognition, transduction, sample preparation, and signal amplification [
4]. In addition, LOC devices, including their design and development, have been one of the most important tools in cancer research since their early existence due to the critical importance of clarifying the processes related to cancer, thus improving the speed, accuracy, and accessibility of diagnosis, resulting in earlier detection and better outcomes for patients [
5]. With advancements in sensor technology, the Internet of Medical Things (IoMT) (also referred to as healthcare IoT) is emerging as a next-generation analytical tool, enabling the transition of health systems from a model based on hospitals to caring centred on the patient, by means of its ability to provide access anywhere during the progression of care. The objective of IoT technologies is to create a connection between smart devices incorporating systems, such as processors, sensors, and communication hardware, with each of them providing information and data in a single communication architecture [
6]. Healthcare can be recognized among the main domains of application for IoT, making patients and clinicians closer through medical devices enabling remote access to cumulate, process and transmit medical data over a secure network, thus enhancing the involvement and satisfaction of the patient. In a standard clinical setting, healthcare solutions are provided by an IoT network made up of interconnected devices and dedicated to healthcare analysis, such as patient monitoring and automatic recognition of situations requiring medical assistance [
7]. In addition, HIoT technologies create opportunities to increase the conventional approaches of diagnosis utilized by physicians [
8]. Nowadays, in a further step towards the development of a novel biosensor, the research is focused on combining the innovative properties of plasmonic transducers with advanced microfluidic systems to improve upon the customary techniques and to produce miniaturized, user-friendly, automated, and portable instruments for POC applications, with the capability to perform a label-free and multiplexed (i.e., quantifying several analytes at the same time) analysis. During the last decades, metal nanoparticles have emerged as effective tools to detect different biomolecules with high precision and sensitivity. At the basis of the metal nanoparticles’ properties there are the collective oscillations of the free electrons and the extensive electric-field enhancement when exposed to light, which lead to different light–matter interactions such as localized surface plasmon resonance (LSPR) and surface-enhanced Raman scattering. Significantly, these nanoparticles display clear changes in the optical properties when they change shape as results of growth or etching, or when they aggregate or disaggregate or as results of variations in the dielectric environment around the particle surface. Further, some metal nanoparticles demonstrate enzyme-like activity, producing coloured products from colourless precursor, so that the colour changes generated from their properties represent an outstanding opportunity for colorimetric detection in user-friendly test and devices. In addition, taking advantage of the large electromagnetic field confined around the surface enhancing the light scattering, metal nanoparticles opened the way to surface-enhanced Raman spectroscopy (SERS), a powerful technology to detect molecules with high sensitivity and accuracy down to the molecular level. According to its benefits, a straightforward idea is to integrate SERS analysis based on metal nanoparticles in POC devices able to obtain, in real-time, molecular information for sensing in a low-cost manner. In this scenario, according to real-time sensing arising from their optical properties (colour changes) and their use in SERS spectroscopy enabling powerful readout systems, plasmonic metal nanoparticles have attracted great interest in the design and development of POC devices during recent years. In this work, a critical discussion on the engineering along with the future scenario of POC platforms will be provided: Firstly, the optical properties of plasmonic nanoparticles, the basis of sensing, will be reviewed in
Section 2. The engineering of POC devices will be reviewed starting from
Section 3, where microfluidics will be discussed with a special emphasis on the traditional and most widespread technologies used in commercial POC devices, the lateral flow assay (LFA) and paper-based analytical devices (μPADs). The readout systems will be illustrated in
Section 4, focusing on SERS analysis and smartphone-assisted devices, since they are the most promising technologies for implementation in real analysis. Furthermore, another critical point in the development of POC devices, metal surface functionalization, is reviewed in
Section 5, along with its great potential in multiplexed analysis (
Section 6). In
Section 7, the applications deriving from the combination of various types of engineering such as, for example, LFA with different readout systems will be presented and classified into three categories: smartphone-assisted devices, liquid biopsy, and wearable plasmonic biosensing, according to their practical use. Finally, in
Section 8, the summary of the work along with a critical discussion of the limiting factors of POC technology will be considered along with the future trends in the development of next-generation sensors.
2. Optical Properties of Plasmonic Metal Nanoparticles
When electromagnetic radiation, such as light, interacts with a metal surface, attractive surface excitations are generated. If the free electrons of a metal are coupled with the electromagnetic field of the radiation, thus oscillating in resonance, surface-bound modes, called surface plasmon resonances (SPRs) are originated. Frequently, the SPRs are classified into two categories: propagating surface plasmons (PSPs) and localized surface plasmons (LSPs) [
9]. The first type consists of running surface waves called propagating surface plasmon resonances (PSPRs). Electromagnetic fields are associated with PSPRs: they penetrate the surrounding dielectric environment, thus providing a sensing probe resulting from its sensitivity to changes in a refractive index such as those deriving from biomolecular interactions. To achieve the excitation of PSPRs, when the light interacts with the metal surface, certain conditions must be satisfied, such as polarization, angle, and wavelength, and the most traditional way to obtain an efficient light coupling is to employ a prism-based scheme (i.e., the Kretschmann configuration), even though other approaches are used, such as diffraction grating and waveguide coupling [
10]. For small nanostructures such as metal nanoparticles, boundary and surface effects become important, so that the interaction with the light results in non-propagating collective oscillations of the particle conduction electrons, named localized surface plasmon resonance (LSPR). When light interacts with a metal nanoparticle, electrical charges are accumulated on its surface, thus originating a dipole: the Coulomb attraction between positive and negative charges results in the restoration of forces displacing the electron cloud from the nuclei, with electrical charges starting to oscillate collectively with a unique frequency localized in the UV-visible NIR region of the spectrum [
11]. The position, number and width of such modes are determined by several parameters such as particle size, shape, medium refractive index, surface charge, and interparticle interactions, but also from other additional factors such as surface modification (including the deposition of shells, either of another metal, a semiconductor or a dielectric) and composition (doping, alloying) [
12]. In addition, the electromagnetic fields localized close to the particle surface are largely enhanced, with the enhancement being greater near the surface and quickly decreasing for larger distances. In addition, at the LSPR frequency, the scattering and absorption cross-sections are all strongly enhanced [
13], thus resulting in great interest in sensing techniques such as colorimetric detection or SERS analysis. In fact, LSPR is the source of the brilliant and intense colours displayed from plasmonic metal nanoparticles and have attracted attention since the Lycurgus cup [
14], and recently have been exploited with a growing interest in colorimetric detection. In colorimetric analysis, the event of detection is converted into a colour change, so it is characterized by simplicity, low cost, practicality and the lack of the requirement for bulky and sophisticated instruments because the colour change is revealed by the naked eye. Its intrinsic advantages make it appealing to integrate colorimetric assays into POC devices, and the development of new materials along with new colorimetric strategies has focused enormous efforts in recent years [
15].
3. Microfluidics Technologies
Aiming to integrate nanoplasmonic biosensors into a POC device, the design of the microfluidic and the optical components is crucial. Microfluidic technologies are exploited in order to create POC devices enabling analysis with samples of a very small volume. The devices produced with microfluidic technologies contain channels and chambers with size dimensions of the order of millimeters, so that fluid flow is laminar (namely, without chaotic turbulence), thus controlling the transport and mixing of molecules, thus resulting in a more accurate and sensitive detection [
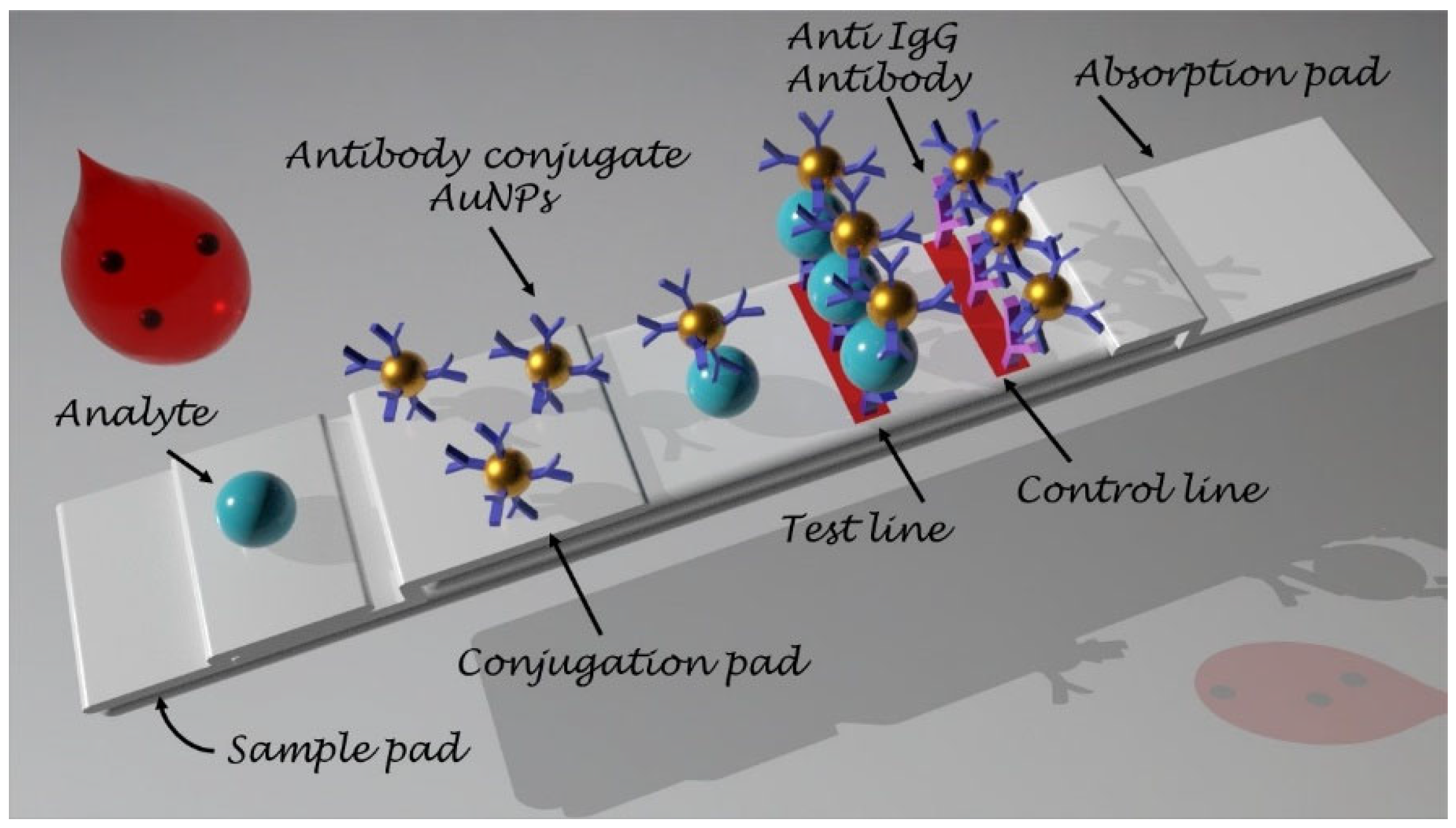
16]. In addition, with microfluidic technologies it is possible to automatically realize the mixing of the sample with the reagent, the washing, and the cleaning and facilitating sensing. Between the devices adopting microfluidics technologies, the one based on capillary flow, the lateral flow assays (LFA), are the most broadly diffused and commercially popular POC tests (such as, for example, the pregnancy test and glucose test kit) and meet the ASSURED criteria. They are self-operating tools consisting of a strip including a sample pad, a conjugate pad, a nitrocellulose (NC) membrane and an adsorbent pad. These components are combined onto an adhesive backing card in such a way that they are sequentially overlapped [
17]. After being introduced onto the sample pad, the sample solution will flow to the downstream components by means of the capillarity, thus performing the test. The next component of the LF strip, the conjugation pad, has been loaded with the micro/nanoparticle coupled to the bio-recognition elements (conjugate): the conjugates will be retained in the conjugation pad during the whole shelf-life of the product, and the analytes will be released from the pad to the NC membrane. In the NC membrane, the analyte molecules migrate until encountering their matching capture components in the test line (T line), where they are immobilized so that a detectable output signal is generated and measured for the analysis. Finally, the analytes that have not been immobilized in the T line are adsorbed in the control line (C line) and the absorbent pad collects the final sample solution, providing the capillary driving force (
Figure 1).
Generally, the conventional microfluidics devices contain channels created by etching or molding into PDMS, silicone or glass substrates, or other polymers or plastics [
18]; however, diagnostics devices can also be produced by patterned paper, obtaining the so-called microfluidic paper-based analytical devices (μPADs). In μPADs systems, sheets of paper are patterned into hydrophilic channels bounded by hydrophobic barriers. Using μPADs systems, the process of the analysis is facilitated by the microchannels patterned on paper substrates: the samples and buffer solutions that must be analysed can flow naturally within the microchannels as a result of the natural hydrophilicity of paper fibers, so that external driving sources are not required. The production of μPADs devices is based on technologies that employ chemical modification or physical deposition techniques to change the material properties of the cellulose matrix [
19]. The most conventional methods to manufacture μPADs include wax printing, photolithography, inkjet printing, laser cutting, polydimethylsiloxane (PDMS) stamping and printing, thermal embossing, hydrophobic silylation, and paper folding. The μPADs systems combine the advantages of paper—such as the low cost, the biocompatibility, the low weight and the removal of the need for external support equipment such as pumps or power because the movement of the fluid is controlled mainly by capillary forces and evaporation—with that of microfluidics analysis—such as the requirement of samples of a small volume and the rapidity. In 1949, Muller and Clegg [
20] produced, for the first time, paraffin-patterned paper containing microfluidic channels, considered also the first paper-based assay, but it was the work of Whitesides’ group at Harvard University [
21] which really introduced the μPADs systems to the scientific world. Since then, microfluidic paper-based analysis has experienced a growing interest and an evolution, representing a breakthrough in cost-effective and portable POC diagnostics and a promising alternative to traditional microfluidics devices.
5. Surface Functionalization
The bioactive area, the place where analytes are recognized by specific receptors, is one of the critical components of a sensor, affecting the signal and, as consequence, the limit of detection, the sensitivity, and the overall performance of the device. In light of its importance, when producing a biosensor, one of the most challenging steps is the conjugation of the sensor’s active surface with the biological receptors in order to maximize the signal that is proportional to (i) the percentage of the surface occupied by the capturing elements (coating of the bioactive area); (ii) the effectiveness of the binding event; and (iii) the selectivity between the inert area and the bioactive area [
31]. In these circumstances, during the design and development of a biosensor, several details must be considered, particularly the capacity of retaining the native conformation of the biorecognition element after immobilization, so that its natural activity is preserved, along with the facility to access the analyte molecules and prevention of non-specific absorption onto the solid substrate. In addition, in the case of a plasmonic sensor, a relevant detail to be considered is the decay length, the distance to the metal surface, which can be decreased exponentially [
32], so that the maximum signal is reached when the molecules are attached directly to the particle surface. From this perspective, it is important to immobilize the receptors in such a way that the binding event takes place relatively close to the plasmonic surface, thus ensuring the signal readout. As a consequence of the rapid decay length, furthermore, at low concentrations, the detection of analytes could be challenging because a molecule will scarcely adsorb or be in the immediate surroundings of the plasmonic surface. One of the central points at the time of immobilizing a biomolecule onto a sensor surface is to achieve its proper orientation, namely, the appropriate exposure of the binding sites to the sample where analytes must be detected, because the effectiveness of recognition is stronger, and, also, the orientation affects the density of the receptors on the surface, thus assuring a higher recognition specificity and an improved sensitivity of the sensor. Along with the orientation, optimal immobilization also takes into account stability and activity while performing the analysis, the packing density and the exclusive binding onto the sensing area. Over recent years, several strategies have been developed to immobilize receptors on the sensor surface and, broadly speaking, they can be divided into two main categories: non-covalent and covalent immobilization [
33]. The non-covalent immobilization is based on the absorption of biomolecules by means of forces, such as Van-der-Waals forces, hydrophobic interactions and electrostatic or ionic bonds; however, despite the simplicity of the approach, it is not advisable because the control over the biomolecule orientation is limited, resulting in the low performance of the sensing device. Covalent immobilization includes the modification of the surface to integrate reactive groups such as hydroxy, thiol, carboxy, or amino groups which will then be exploited to attach the bioreceptors. This approach typically requires two steps: (a) the formation of an appropriate self-assembled monolayer (SAM) of bifunctional alkane thiols onto the metal surface operating as a linker and (b) the following cross-linking between the functional thiols and the complementary functional group on the analyte molecule. The choice of the thiol group (-SH) is the most widespread in the case of gold surfaces: the -SH groups strongly attach to the gold surface, thus spontaneously forming an SAM, also exploiting the electrostatic and hydrophobic interactions between the carbon chains. Other approaches are based on the interaction between a biomolecule, acting as a linker, and a second molecule employed for the detection of the analyte, enabling a highly specific and powerful recognition mechanism (bioaffinity interactions). The most popular example of a bioaffinity-mediated immobilization technique utilizes the streptavidin–biotin complex: in this case, streptavidin is immobilized onto the substrate, whereas a biotinylated moiety is attached to the receptor molecule. Despite the further steps required for the chemical modification of the receptor with biotin and to attach streptavidin, this approach provides a highly oriented and stable layer with improved specificity. Another approach is based on the bioaffinity interactions among antibodies and protein A or G produced in bacteria: such proteins are able to selectively bind the Fc portions of Abs, thus assuring the correct orientation and resulting in the proper orientation onto the metal surface, thus guaranteeing high recognition of antigens as well as high purity and long-term stability. In addition, a further approach employs the DNA-directed recognition by means of a single strained DNA (ss-DNA) able to selectively bind an antibody. Other strategies for the immobilization of receptors have emerged with the progresses in molecular chemistry and bioengineering, such as calixarenes, DNA-mediated coupling, or recombinant antibody fragments; however, they are less frequently utilized. Many biological systems showing an exceptional affinity and specificity towards their corresponding analyte molecules have been employed to develop sensing devices [
34]. As an example, aptamers (RNA oligonucleotide sequences or single-strained DNA (ss-DNA) with the ability to bind, in a specific manner, an analyte possessing an affinity comparable to that of antibodies) are frequently used as receptors, but their production is labour-intensive and time-consuming, although their engineering is easier and more suitable compared to that of antibodies [
35]. Sugars are less commonly used, but sometimes could be very selective, but they need to be modified before being anchored to the surface. Recently, molecularly imprinted polymers (MIPs) are emerging as recognition elements: they are affinity polymers provided with specific binding sites designed with the proper shape, size and orientation of the functional groups for the selective trapping of the target molecule, offering an interesting alternative for their affordability and robustness [
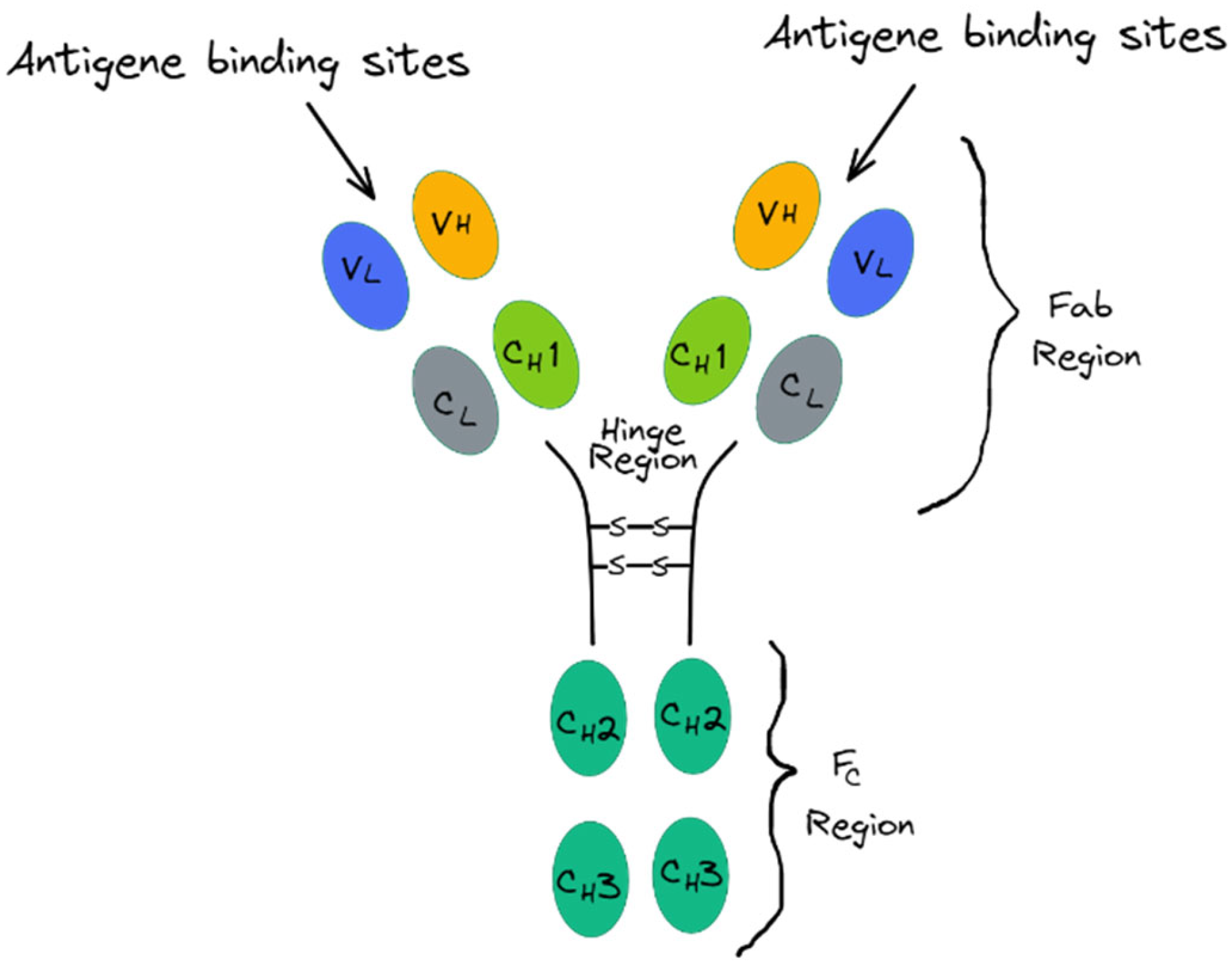
36]. Antibodies, or immunoglobulins (Abs), are the biological recognition elements most frequently used in the development of a biosensor: they are large glycoproteins of about 150 KDa which are part of the immunoglobulin (Ig) superfamily produced by the immune system to recognize external antigens in order to neutralize them and induce an additional immune response [
37,
38]. The bone structure of an antibody includes two identical light chains and two heavy chains connected through disulfide bonds in the hinge region to form a complex quaternary Y-shaped structure [
39]. The light chains are formed by two identical light domains: a constant domain (C
L) and a variable domain (V
L). In contrast, according to the type of heavy chain, antibodies can be classified into five distinct classes—IgA, IgD, IgE, IgG, and IgM—which are different in structure and function. IgA, IgD and IgG have three constant domains (C
H) and one variable (V
H), whereas the IgE and IgM have four constant domains and one variable [
40]. The whole quaternary structure of the antibody can be divided into three fragments: the first two, recognized as Fab portions, are responsible for the antigen recognition and are localized at each tip of the Y (corresponding to the “harms” of the antibody) and contain one light chain, one variable heavy domain and one constant heavy domain (V
L, C
L, V
H and C
H) [
41]. The remaining fragment is the fragment crystallizable region (F
C) and is located at the base of the Y structure: it comprises the rest of both heavy chains and is responsible for the interactions between the antibody and other members of the immune system [
42]. Finally, the smallest fragment of an antibody functioning in antigen-binding activities is the single-chain fragment variable (scFv) which is composed of the variable regions of the light chain (V
L) and heavy chain (V
H) linked by a flexible peptide to generate a single-chain protein with an affinity for its antigen comparable to that of the parental mAb [
43] (
Figure 2).
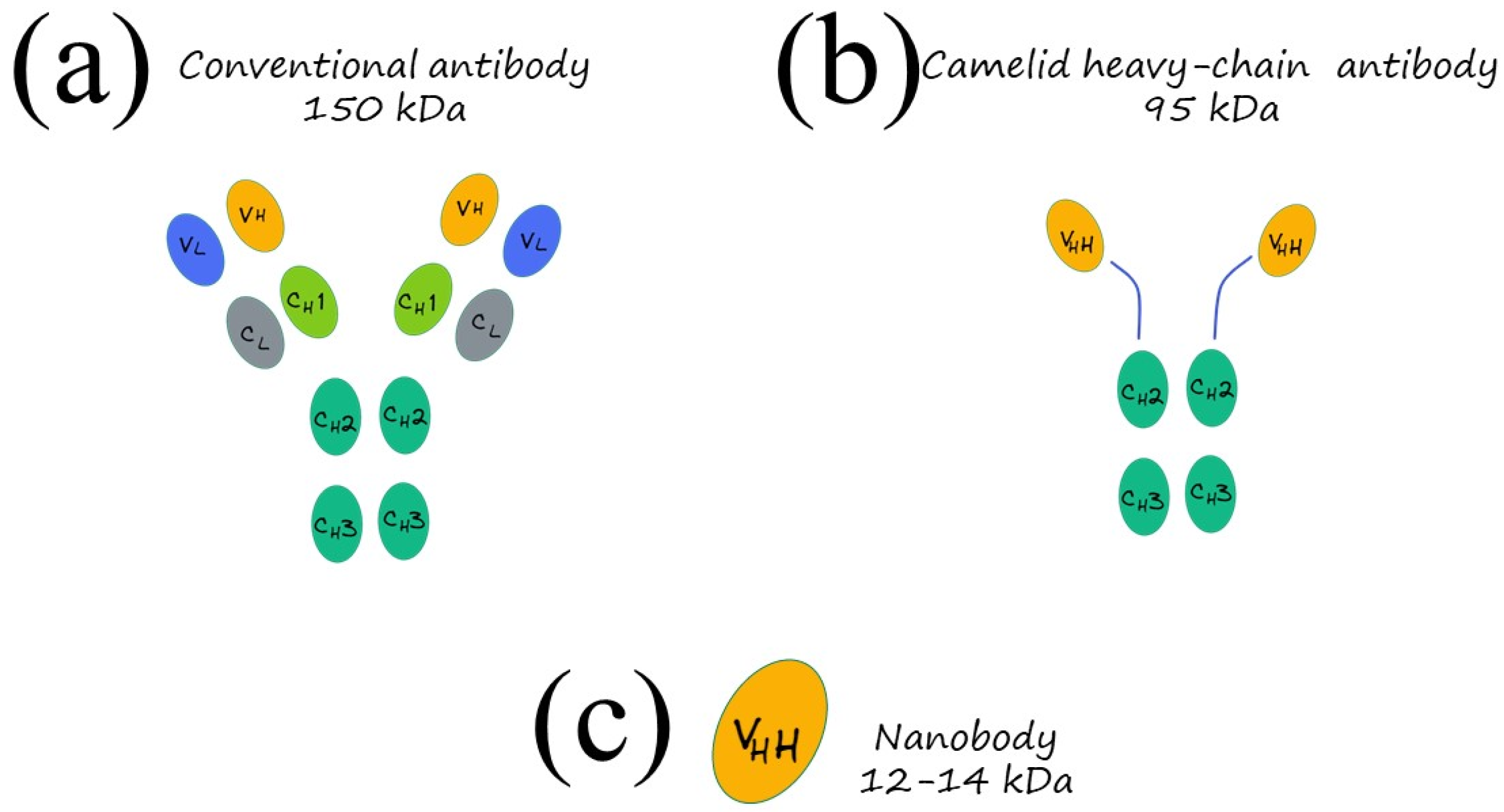
Immunoassays are based on the specific antigen–antibody interactions and the detectable signal produced by the changes in a substrate to qualitatively and quantitatively determine molecules: compared to standard instrumental analysis, they offer several advantages such as low cost, simple pre-treatment with no need for trained professionals nor complex equipment, rapid response and, more significantly, they have high sensitivity and specificity, thus offering great potential as a POC device. There are several formats for immunoassays, including the most famous, enzyme-linked immunosorbent assay (ELISA), chemiluminescent immunoassay (CLIA), quantum dot fluorescence immunoassay, colloidal gold immunoassay, microarray-based immunoassay and many others; however, all of them rely on the specific binding between the antibody recognition sites and antigenic determinants. Within that framework, it is extremely important to design specific antibodies with a high affinity for the analytes, to improve the specificity. In standard immunoassays, polyclonal and monoclonal antibodies are usually exploited, but in recent years, a new type of antibody, recognized as “nanobodies,” is emerging as an appealing alternative to conventional antibodies and ideal basic components for a broad assortment of sensing devices and tests to be used in medical, biotechnological, and environmental fields [
44]. Nearly 30 years ago, in 1993, heavy-chain antibodies (HCAbs), naturally lacking light chains, were casually discovered in the peripheral blood of
Camelus dromedarius by the Belgian immunologist Hamers and his colleagues, paving the way for a new type of antibody, the nanobodies. A heavy-chain-only antibody consists of two heavy-chain constant regions (CH2 and CH3) while lacking the first constant CH1 domain, a hinge region, and a heavy-chain variable domain (VHH), preserving the antigen-binding capacity of the antibody [
45]. The molecular mass of the HCAbs is about 95 kDa while their variable antigen-binding domains (VHH) have a molecular mass of 12–14 kDa with a prolate shape with dimensions of 4 nm × 2.5 nm × 3 nm (
Figure 3)
In the VHH domain is included the strong affinity towards its related antigen and the full potential to bind the antigen, so it is considered the smallest naturally occurring fragment with intact antigen-binding activity. The small size, in the low nanometre range, was the stimulus for introducing, in 2003, the term “nanobody” as a trademark of the company Ablynx, which has subsequently been utilised as a general tag for those proteins. They are distinguished by superior properties, compared to conventional antibodies—such as, for example, elevated stability—expressed in various forms: tolerance to increased temperatures; prolonged shelf life; resistance to several harsh conditions, such as exposure to different pH values from physiological, elevated pressure; proteolytic degradation; and the fact that chemical denaturants hardly affect their antigen-binding ability. The efficient ability to refold after thermal, physical and chemical denaturation is the basis of the nanobodies’ robustness. Within the framework region 2 in nanobodies, named FR2, four hydrophobic and highly conserved amino acids are replaced by smaller and/or more hydrophilic amino acids, thus increasing solubility in aqueous solutions. The simple monomeric structure and the lack of post-translational changes make it possible to express nanobodies in milligram quantities per litre of culture in shake flasks by means of microbial expression systems such as
Escherichia coli,
Saccharomyces cerevisiae and
Pichia pastoris, thus enabling industrial mass production at a low cost and the availability of purified antibodies with homogeneous properties in sufficient amounts [
46]. In addition, nanobodies possess an improved ability to identify antigens compared to conventional antibodies arising from their unique structure and small size, which make it possible to bind sites that are difficult to reach by conventional antibodies [
38]. As a result of their extraordinary properties, nanobodies, in recent years, have emerged as a powerful tool to design and develop next-generation immunoassays. The interest in the use of nanobodies to develop sensing platforms is only in its early stage, so much still needs to be explored in this field. In the work of Simões [
47], an effective and simple strategy for the spontaneous self-assembly of nanobodies on gold surfaces was proposed, which has remarkable potential to be employed to develop sensing platforms. Firstly, the model nanobody NbVCAM1, targeting the vascular cell adhesion molecule-1 (VCAM1), was strategically engineered at its C-terminal, at the opposite end of the binding antigen pocket, with an alkyne-modified cysteine with the capacity to absorb on gold surfaces. As a result of the incorporation of the modified cysteine, nanobodies can be directly immobilized on gold sensors, forming a monolayer that was then investigated using many complementary surface-analysis techniques such as three-dimensional (3D) Orbitrap secondary ion mass spectroscopy (3D OrbiSIMS), time-of-flight secondary ion mass spectrometry (ToF-SIMS) and circular dichroism (CD). The obtained results indicated that the activity and structure of nanobodies are maintained upon the immobilization and confirmed the arrangement in a stable, homogeneous and well-packed monolayer onto the gold surface. In addition, experimental and theoretical results confirmed the high proportion of well-oriented nanobodies led to a strong ability to bind antigens, thus paving the way for the design and development of robust, reliable and stable sensing platforms for the sensing of biomolecules. While nanobodies (Nbs) technology and the immune-sensing area are currently undergoing considerable progress, the functionalization of AuNPs with Nbs is still in its early phase; thus, aiming to take further steps forward in this direction, Goossens [
48] elaborated a strategy to obtain stable AuNPs functionalized with nanobodies (Nb-AuNP). Using three different types of well-characterized nanobodies (BCII10 binding β-lactamase II of Bacillus cereus, cAbLys3 able to identify the hen-egg white lysozyme and a GFP binder), the practicability of functionalised AuNPs with Nbs through physical absorption, along with the possibility of applying the rules of antibodies absorption to gain further insights into the mechanism for the production of stable Nb-AuNP, were investigated. When developing a sensing system, colloidal stability is a key point as well as the functionality of the biological molecules. In the case of monoclonal antibodies (mAbs), the larger size enables the formation of repulsive barriers between particles by means of electrostatic hindrance; however, due to their reduced size, in the case of Nbs, the driving repulsive force is their charge. The absorption of antibodies produces stable particle complexes only when the negative charge of Nbs is negative and the values of the solution ionic strength are low. In such circumstances, no unfolding mechanism occurs at the interface with gold, so that the produced Nb colloids are functional units with great potential to be employed in immune assays. On this basis, engineering Nbs in such a way as to realize an optimal electrostatic configuration by integrating strong binding residues, such as cysteine, and then creating Nb-AuNP colloids through physical absorption has been proposed as a general strategy for the development of a successful sensing system. One of the early approaches in the use of AuNPs conjugated with nanobodies as a sensing device was presented in the work of Alsadig [
49]: an advanced biosensing platform was created through a combination of a mixed self-assembled monolayer (SAM) of thiolated single-stranded DNA (ssDNA) and bio-repellent molecules, indicated as top-terminated oligo-ethylene glycol (TOEG6) and the DNA-directed immobilization (DDI) of DNA–protein conjugates. One of the primary challenges related to the immobilization of protein onto a solid substrate concerns the preservation of the protein functionality by preventing alterations in their structure during the immobilization progress, along with the steric hindrance of the recognition sites. In that direction, the DNA-directed immobilization (DDI) of DNA-conjugated proteins, resulting from the combination of short ss-DNA oligos self-assembling with the sequence-specific hybridization of complementary DNA strands conjugated with definite proteins and/or proteins binders, emerges as a versatile tool to transform a DNA microarray into a protein microarray exploiting Watson–Crick hybridization while retaining the protein structure. Unlike covalent binding, when biomolecules are anchored by means of DDI onto a surface, they completely preserve their functionality, as well as the steric hindrance being reduced by employing DNA strands as spacers between the substrate and the biomolecules of interest, thus resulting in a broader opportunity for molecular recognition. In light of implementing the DDI approach to create sensing platforms, AuNPs were functionalized with an optimized quantity of ssDNA molecules in order to prevent the effects of steric hindrance and to ensure the active loading of the DNA–protein conjugates. To avoid the absence of the specific binding of protein or small molecules, the thiolated ssDNA moieties were mixed with bio-repelling TOEG6 molecules (ss-DNA/TOEG6@AuNPs). In a successive step, the potential of the ssDNA-modified AuNPs in the context of cancer-associated antigen sensing was investigated employing the complementary ssDNA fragments conjugated with the C8 nanobody fragment optimized for the recognition of a specific biomarker extracellular domain of the human epidermal growth factor receptor 2 (HER2-ECD), a transmembrane protein belonging to the family of the epidermal growth factor receptors (EGFRs) and overexpressed in 15–30% of breast-cancer patients. To detect the hybridization of the ss-DNA/TOEG6@AuNPs with the C8-labelled DNA target, the changes in the refractive index, induced by the biorecognition event occurring close to the AuNPs’ surface, were evaluated by means of a novel technique. Employing a conventional LSPR detection technique, the sensitivity is too low to identify the binding of ECD-HER2 to the functional nanoparticles, so that, to assure the enhanced sensitivity of the test, an approach based on a miniaturized gel electrophoresis chip integrated with online thermal lens spectroscopy (MGEC-TLS) was employed as a sensing system, thus obtaining a sensitivity 100-fold higher than the standard SPR resulting from the acquisition of quantitative information through the relation of the immunocomplex biological identification with the mobility of the particle. The time required by the conjugates to arrive at the measurement point was measured and, as expected, increases with the quantity of HER2-ECD, thus making it possible to relate the concentration of loaded HER2-ECD to the time of MGEC-TLS signal, namely, the signal registered when the sample passes through the measurement point. The changes in the refractive index were amplified by the presence of the gel matrix and are associated with the temperature alterations generated by the energy-to-heat conversion of light adsorption at the LSPR wavelength, thus obtaining a system with enhanced sensitivity combined with ease of preparation and reduced costs, with potential to be used in contexts of low or limited resources (
Figure 4).
A colorimetric and photothermal dual-mode immunochromatographic test strip (KNb-DITS) based on the determination of nanobodies for
Salmonella Typhimurium (
S. Typhimurium) was presented in the work of Zhang [
50], which provides an innovative path and ideas for foodborne pathogen detection and, more broadly, for immunosensing POC devices. Encouraged by their intrinsic advantages, nanobodies were integrated in colorimetric/photothermal sensing probes. Firstly, flower-like three-dimensional (3D) K
0.27MnO
2·0.54 H
2O (KMO nanoflowers) were synthesized using hydrothermal methods and then decorated with AuNPs; successively, the KMO@Au composites were effectively conjugated with anti-
S. Typhimurium Nb9 was used to form the colorimetric/photothermal-KMO@Au@Nb9 sensing probes. As in classical tests, the strip is composed of a sample pad, nitrocellulose (NC) membrane, absorbent pad, and PVC backing. The test line (TL) and the control line (CL) are placed onto the NC membrane and are made up of capture anti-
S. Typhimurium pAb and anti-HA tag mAb. In the presence of the target analyte
S. Typhimurium, an interaction with the capture anti-
S. Typhimurium pAb of the TL is established, so that a KMO@Au@Nb9-
S. Typhimurium complex is formed, which can be directly observed by the naked eye and the colour analysed using software. In addition,
S. Typhimurium can be detected in a photothermal mode: the TL line is irradiated with a laser of 808 nm (radiation in the NIR region of the spectrum) and temperature and images collected by means of a portable infrared camera and processed for a quantitative analysis. Using the KNb-DITS sensor, it is possible to easily perform not only visual qualitative detection with the naked eye, but also quantitative detection using both colorimetric and photothermal analysis. The colorimetric detection is able to detect the
S. Typhimurium with a LOD of 10
4 CFU mL
−1, whereas in photothermal mode the obtained LOD is 10
3 CFU mL
−1, thus providing a new direction for advanced POC devices.
6. Multiplexed Analysis
The detection of different analytes at the same time enables the performance of accurate diagnostics about the health of an individual, so that personalised therapy could be designed, which also offers the additional advantages of lowering the analysis costs and significantly simplifying the diagnostic workflow [
51]. The benefits arising from the analysis performed in a single run make the multiplexed detection an intriguing alternative to other approaches. Despite the promising advantage of supporting future advances in personalized medicine, multiplexed detection still presents some challenges, making its full implementation in real diagnostics difficult [
52]. One of the main issues is to generate a sufficient number of signals that are each distinguishable from the other by means of the detection instrument in order to distinguish the individual analytes. This means that when the signals can be more easily distinguishable and their total number is greater, better detection could be obtained. Within a biological sample, the concentrations of analytes may differ by an order of magnitude, such that the additional need concerning the linear range and the detection sensitivity is introduced. In addition, it is necessary to reduce the time and the costs of the analysis, as well as to minimize errors resulting from the complexity of the operations, so it is critical to simplify the overall detection process as much as possible. In light of effective clinical implementation, several strategies have been designed to face the challenges arising in multiplexed detection, along with different approaches to analysis. During recent years, plasmonic nanoparticles have attracted increasing interest for their unique properties; they have proven to be a promising resource in the multiplexed detection of analytes [
53]. In fact, by means of different synthetic techniques [
54], researchers are able to control various parameters such as size, shape, composition, surface modification, surface charge and so on; it is possible to produce the desired optical response such as position and shape of the LSPR peak in the UV-visible spectrum. Given the possibility of synthesizing plasmonic nanoparticles with tailored optical properties, it is easy to figure out how including various nanoparticles generating different signals in a unique assay enables the detection of multiple analytes in a single run. On these bases, several approaches have been designed to exploit plasmonic metal nanoparticles in multiplexed detection. Spectroscopic methods are emerging as analytical techniques for the multiplexed detection of analytes, due to their remarkable advantages, such as ultrasensitive detection down to the level of the single molecule and the ability to perform the measurements in a biological environment without the need for preparing the sample in advance. Despite this great potential, there are still some limitations that must be overcome to effectively implement SERS technology in multiplexed analysis for daily medical diagnostics. One of the main difficulties is the number of the analytes that can be detected simultaneously in a multiplexed analysis: when increasing the number of molecules to be revealed, competing signals are likely to arise, so that the vibrational modes of different molecules are likely to overlap, thus making the interpretation of the spectra difficult if not nearly impossible. A way to solve the complexity of the SERS spectra of multiple analytes is to apply chemometric algorithms, such as principal component analysis (PCA), partial least-squares regression (PLS) and hierarchical cluster analysis (HCA) [
55]. The multiplexed detection of analytes could be performed in one of two modalities: the first is the direct one involving the use of nanoparticles with bare surfaces to obtain enhanced Raman signals, whereas the second is the indirect modality, where SERS tags provide the signal and confirm only indirectly the presence of the analyte. The elevated quantity of salt, the low analyte concentration, typical of the physiological environment, and voiding of results in the nanoparticle aggregation make the reproducibility and quantitative identification of the target limited, and, in this context, indirect detection becomes increasingly important along with the concept of the SERS tag (also called encoded SERS nanoparticles) [
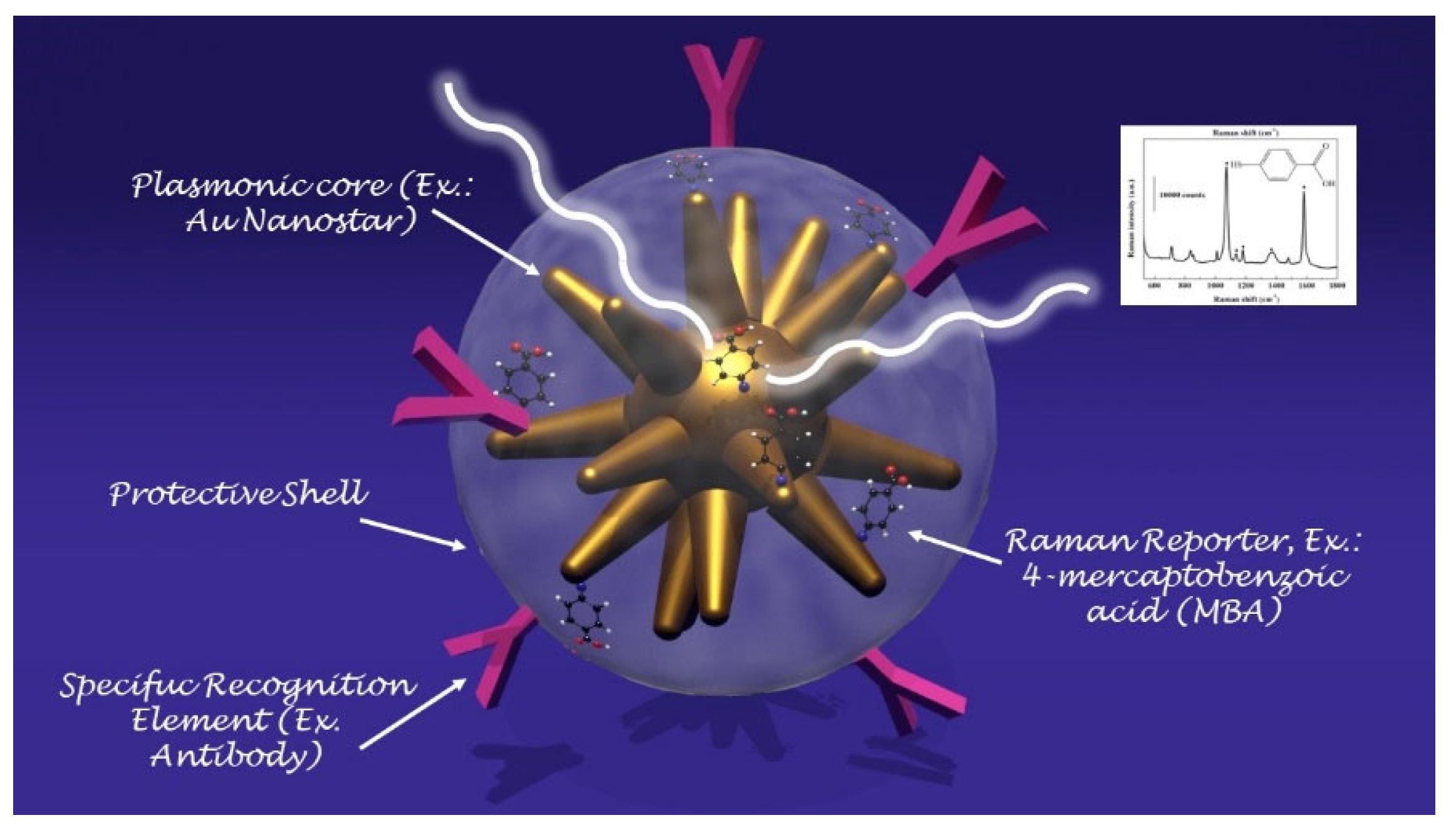
56]. A SERS tag is a probe created to recognize, in a selective way, the target molecule by means of the targeting moieties designed with high specificity towards the analyte molecules (such as, for example, antibody, polymers or antibodies) and to generate the SERS signal. Even though the characteristics strongly rely on the specific applications for which the tag has been designed, the SERS tags share some common characteristics: (a) the presence of a plasmonic core generating the electromagnetic field required for the enhancement of the Raman signal; (b) the addition of a Raman reporter molecule (SERS code) which originates the unique molecular fingerprint arising from the encoded particle; and (c) a protecting layer to stabilize the nanoparticle against aggregation and to provide improved biocompatibility to the particle surface [
57]. In
Figure 5, a schematic of a SERS nanotag is depicted.
On this basis, it can be easily figured out how multiplexed assays can be produced by designing different SERS nanotags generating different signals and successively integrating them in a unique test. The production of SERS nanotags originating non-overlapping signals is an effective strategy for the multiplex detection of analytes so that their integration into advanced POC devices is beneficial to the rapid determination of biomarkers in complex fluids such as blood or saliva. The optical properties of plasmonic metal nanoparticles have also been exploited to perform multiplexed analysis in other multiplexed technologies: for example, when designing LFA devices, the most straightforward and direct method to realize multiplexed detection is creating an array of parallel test lines (TLs) on the detection zone, where every single TL is responsible for the identification of a different analyte. The majority of analyses adopting this kind of multiplexed detection are able to determine up to three analytes [
58]. One of the critical drawbacks with multiplexed analysis with LFA devices is the reduced length of the detection zone, limiting the number of TLs that can be included in the assay. To insert more TLs, the test strip should be extended, which exponentially increases the assay time. Alternatively, the detection of different analytes within a single TL, as long as it maintains simplicity in the production and in the results read out, could be performed by designing a single test line with two or more different bioreceptors conjugating the analytes. When using a single TL to perform a multiplexed analysis, various labels generating signals that can be easily distinguished, such as, for example, different plasmonic nanoparticles, must be designed and exploited. In view of taking better advantage of the test area provided by lateral flow strips, TLs have been replaced with microfluidic arrays, where a microarray is an ordered arrangement of various assay units into a single device [
59].
8. Summary and Outlook
This work outlined the growing interest in developing POC platforms based on plasmonic sensing platforms to create devices with great potential to be implemented as next-generation diagnostic tools. In order to gain a better understanding of the novel sensing transducers, firstly, the optical properties of plasmonic metal nanoparticles were reviewed, and then the key issues of the POC devices were discussed. It was illustrated that the microfluidics of the system is a critical point to integrate the plasmonic transducer into a POC device and the attention was focused on LFA and μPADs, for their simplicity arising from the lack of a need for external forces such as micropumps and their low-cost. In addition, it was illustrated how common technologies, such as smartphones, and analytical methodologies, such as SERS, are considered intriguing readout systems in view of their ability to provide quantitative analysis, a key point for timely and highly precise diagnostics. For an accurate prognosis and treatment evaluation, the determination of multiple biomarkers at the same time is central so the strategies developed for the multiplexed analysis were also discussed. Finally, several applications of POC devices were reported, such as in liquid biopsy, wearable sensors and smartphone-assisted devices. The objective of the work is a critical discussion of the progress achieved in developing POC devices: while the interest in new nanomaterials such as hybrid composites of plasmonic metal nanoparticles and 2D nanomaterials creates new possibilities for the creation of new sensing transducers, the route towards their effective implementation as commercial biosensing platforms still has limitations that must be overcome. One of the critical challenges in the development of POC devices, for example, are the SERS readers: they are not very user-friendly, need long time to acquire data, and have a high cost. In addition, the results obtained from SERS are not easily understood, so that, for applications in the real world where the analysis is performed by untrained people, it is necessary to develop a software able to accurately and reliably process, interpret, and report the results in a way that they are simple to understand. In this scenario, some work must be performed to develop handheld a Raman spectrometer which gives a simple-to-understand readout to be integrated into POC devices. Still, despite SERS technology having great potential, its use in clinical diagnostics is not approved because of the reproducibility of SERS substrates and the complexity of the equipment, which limits a great number of applications. Efforts are directed towards the creation of a “closed case” system for the final user, where the sample to be determined is introduced and analysed and the results generated on a quantified, reliable and simple readout system; this will enable the widespread use of this technology as commercial biosensing platforms. In addition, another one of the main challenges to face in the development of the next generation of biosensing diagnostic devices is their integration into eHealth systems and, in this context, the technological developments in the field of Internet of medical things (IoMT) are becoming more important; these offer wireless-based operation and the connectivity of POC devices with health experts and medical centres. During recent years, POC platforms have also been integrated with machine-learning (ML) algorithms, such as artificial intelligence (AI), resulting in a new model of making decisions based on data analytics. Firstly, the AI sensors analyse the sample using portable instruments which are then able to automatically digitalize the detected biomarker concentration. Successively, on the basis of the acquired data, the disease spectrum can be predicted by means of algorithms such as classification, cluster, and pattern, to make decisions. In this context, AI is proving to be a promising tool for providing medical practitioners a further possibility in terms of personal treatment and observations of their outcomes. The use of health analytics, such as IoMT and AI, thus, contributes to personalised care for the active management and prevention of disease risk, improving the wellness of the patient. The greatest breakthrough of the next generation of diagnostic tools will be the realization of so-called P6 medicine (personalized, predictive, preventive, participatory, psycho cognitive, and public), and, ultimately, also a contribution to the achievement of cancer chronology, as a further step in the fight against the disease.