Abstract
Dynamic detection, as one of the core tenets in tumor diagnosis, relies on specific recognition, rapid reaction, and significant signal output. Circulating tumor cells that carry the most complete information of neoplastic lesions are analytes of interest for sensor designer. To overcome the deficiencies in the use of antibodies, some antibody-like structures were used to integrate chemosensors, such as molecularly imprinted polymers (MIPs) and aptamer conjugates that may perform specific detection of analytes. The sensors inherited from such structurally stable molecular recognition materials have the advantage of being cost-effective, rapid-to-fabricate and easy-to-use, whilst exhibiting specificity comparable to antibody-based kits. Moreover, the fabricated sensors may automatically perform quantitative analysis via digital and microfluidic devices, facilitating advances in wearable sensors, meanwhile bringing new opportunities and challenges. Although most MIPs-based sensors for tumor diagnosis to date have not been practically used in hospitals, with no doubt, material innovation and artificial intelligence development are the driving forces to push the process forward. At the same time, the challenges and opportunities of these sensors moving forward to intelligent and implantable devices are discussed.
1. Introduction
According to worldwide statistics, cancer remains one of the leading threats to human health [1]. It is estimated that 90% of cancer-related deaths are not caused by the primary tumor, but by cancer metastasis closely associated-circulating tumor cells (CTCs) [2]. Specifically, CTCs are cancer cells that shed from tumor lesions into the circulatory system, carrying the complete information of the primary tumor, thereby playing a key role in tumor metastasis [3]. It has been found that CTCs can be detected at an early stage, thus the capture and enrichment of CTCs is primarily important in many regards, such as mechanism exploration, cancer diagnosis and prognosis, drug development, and regenerative medicine [4,5]. Hence, the interest in dynamic capture and release of CTCs, thereby helping to improve patients’ prognosis, prolong survival and improve quality of life.
For sensor development, the dynamic detection of CTCs remains challenging, due to the scarcity (approximately 1–10 CTCs per ml peripheral blood) and heterogeneity of CTCs [6]. Moreover, dynamic capture of CTCs requires a robust and specific cell binding approach to ensure maximum cell collection, which also allows a gentle release to maintain cell viability. Among all affinity ligands for binding interface constitution, the natural antibodies are the most trustworthy, but also the most expensive and fragile [7]. Therefore, researchers spent a long time seeking antibody alternatives in black box until the emerging of aptamer, a single-stranded DNA or RNA obtained after multiple rounds of screening from a random oligonucleotide sequence library artificially synthesized in vitro by using the ligand system evolution technique of exponential enrichment (SELEX) [8]. The aptamers can bind metal ions, carbohydrates, lipids, and proteins, etc., holding the advantages of convenient preparation, good thermal stability, and low immunogenicity. However, the types of aptamers currently available are limited, subject to the discovery of new targets and the exploration of new sequences. To customize antibody-like materials for various targets as desired, the Russian chemist Polyakov observed the “molecular memory” of an aromatic dye in a synthetic silica matrix, which promoted the birth of molecular imprints [9]. Later in a study published in Nature (scientific journal), the group of Mosbach showed that crosslinked polymers synthesized in the presence of a molecular template could bind the target analyte in human serum, exhibiting specificity and selectivity comparable to those of antibodies. Since then, the molecularly imprinted polymers (MIPs) were often termed “artificial antibodies” [10]. Nowadays, some experts in the field of molecular recognition also prepare MIPs on the basis of aptamer, so as to achieve signal amplification strategies. In this paper, we also refer to such antibody-like materials as MIPs.
Currently, the main enrichment techniques include electrostatic adsorption interface, microfluidic devices, and immunomagnetic beads. Particularly, the microfluidic-based enrichment techniques may allow the isolation of peripheral blood mononuclear cells via an automated way, thus have led to the dawn of exciting technological transformation. Unlike the methods that involve unfavorable conditions and harsh environments, some bio-friendly electrochemical sensors, microfluidic devices, and flexible electrode based on aptamers and MIP, have been vigorously validated for accuracy and reproducibility in recent years [11,12]. In this paper, we mainly reviewed the three kinds of recognition elements by evaluating their specific capture performance. Based on these excellent molecular recognition elements, the related detection platforms were studied, with emphasis on the limit of detection (LOD) value and safety for cell viability (Figure 1). Moreover, the challenges and opportunities of these sensors moving forward to intelligent and implantable devices are also discussed.
Figure 1.
Schematic diagram of specific recognition elements-based sensing platforms for accurate, smart and safe capture and detection of CTCs.
2. Specific Recognition Elements for Capturing CTCs
Conventional capture means are mainly based on biospecific identification, and such probes are mainly designed for the recognition of overexpressed antigens possessed by CTCs, such as epithelial cellular adhesion molecule (EpCAM), epidermal growth factor receptor (EGFR), cytokeratin (CK), carcinoembryonic antigen (CEA), prostate antigen (PSA), and the specific tissue-derived markers. Currently, the most widely used and well-established method is the immunomagnetic bead assay. Specifically, immunomagnetic beads that specifically capture CTCs are usually generated by coupling magnetic spheres and antibodies. For instance, the CellSearch system is an immunomagnetic beads-based CTCs detection system, and is the only technology approved by the US Food and Drug Administration (FDA) for baseline CTC counting in recent years [13]. The system uses immunomagnetism to achieve tumor cell isolation, followed by identification and counting of CTCs using immunofluorescence. Thus, the CellSearch system is widely used for the clinical diagnosis of prostate and breast cancers, as well as in the prognostic assessment of tumors. However, this system is unable to detect CTCs that have become fibroblast-like phenotypes, and it is also difficult to achieve nondestructive release after capture.
Unlike antibodies, peptides with specific binding ability to target biomolecules can be obtained by manual screening, nowadays acting as an important affinity ligands in protein–protein interactions [14]. Its unique properties, including small size, high stability and easy-to-functionalize, have drawn intensive attention for CTCs capture applications. For example, Peng et al. developed a HER2 specific peptide that could effectively bind to HER2 overexpressed SKBR3 (human breast cancer cell line) with a capture efficiency of 68.56% [15]. However, the binding capacity of these peptides is still relatively low compared to that of antibodies which may achieve near 100% capture, due to their low density structure.
Inspired by host–guest recognition in nature, artificial antibodies including aptamers origami and MIPs have come to our front [16]. These artificial antibodies have the advantages of easy synthesis, good stability and low price, bringing a wide application prospect in capturing CTCs [17]. Figure 2A shows the aptamer fabrication process, followed by which a 3D structure with ordered arrangement of complementary nucleic acids were generated for effective capture and release of CTCs with detection signal output [18]. To note, aptamer-mediated detection methods could be endowed with signal transduction and amplification functions. Differently, the MIPs not only provide complementary functional groups, but also a complementary space to the target, leading to an increased specificity (Figure 2B) [19]. Moreover, the microenvironment provided by MIPs facilitates the protection of the biological activity of the target, allowing a safe release afterwards, indicating a greater potential in clinical uses. Theoretically, the molecular recognition of MIPs originates from the cavities generated by molecular templates. However, non-imprinted polymers (NIPs) synthesized in the same manner without templates consistently showed significant binding to template molecules. These non-specific “molecular recognitions” can sometimes affect the success or failure of the imprinting process. Therefore, MIPs with the lowest possible non-specific binding are highly desirable. MIP experts have used several strategies to reduce non-specific binding, such as blocking non-cavity regions with polyethylene glycol. However, the molecular recognition mechanism between MIPs and analytes remains unclear. Does MIP produce a conformational change, such as an antibody–antigen interaction, and thus bind a target molecule? This problem persisted until 2013. Bai et al. developed two aptamer-based hydrogel MIPs for the detection of thrombin and platelet-derived growth factors (Figure 2C) [20]. After binding or releasing target proteins, MIPs hydrogels exhibit shrinkage or swelling, which can be quantified by measuring the length of the gel inside the capillary using a mesh magnifier. This finding suggests that MIPs can recognize analytes through dynamic conformational changes similar to natural antibody or receptor recognition mechanisms, suggesting the feasibility of molecular imprinting in the design of synthetic antibodies.
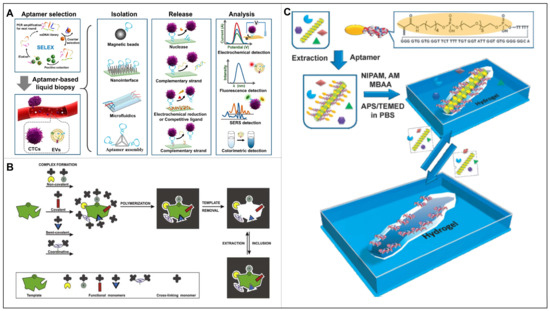
Figure 2.
(A) Schematic diagram for aptamer selection and aptamer-based liquid biopsy. Reproduced with permission from [18]. (B) Consecutive steps of preparing, in different ways, a molecularly imprinted polymer. Reproduced with permission from [19]. (C) Schematic illustration of the bioimprinting process used to create virus responsive super-aptamer hydrogels. Reproduced with permission from [20].
2.1. Aptamer Conjugates
As an antibody-like element, an aptamer is able to target the whole cell via bioengineering means, thereby avoiding false negative results to a great extent with respect to nature antibodies. Therefore, the aptamer technique has been widely used in the capture of CTCs [21,22,23,24]. For example, Wang et al. achieved the capture and identification of EGFR overexpressed human glioblastoma multiforme cells by using an array of anti-EGFR aptamers [25]. Furthermore, since multiple targeting of various biomarkers simultaneously can be accomplished by integrating multiple aptamers into one entity by using some nanostructures or forming dendrimers, leading to improved capture efficiency [26]. Tan et al. developed a platform combining multivalent aptamer nanospheres and a microfluidic device for the isolation of tumor cells from blood (Figure 3A) [27]. Specifically, the gold nanoparticles (AuNPs) that could bind up to 95 aptamers per AuNP were used as scaffold. Compared to individual aptamers, such multivalent aptamer conjugates could significantly enhance the specific molecular recognition, resulting in 39-fold cells attachment. Based on this, Zheng et al. further developed a novel barcode particle that could enable the capture and release of multiple CTCs (Figure 3B) [28]. Specifically, the barcode particles were spherical colloidal crystal clusters with branched amplified aptamer probes which might display specific signal peaks from the coding region. Such barcode particles were successfully applied for simultaneous capture and detection of various CTCs.
Despite advances in selective capture of CTCs by aptamers, it is still challenging to achieve near 100% capture efficiency with a rapid reaction, especially for some samples with ultra-low cell counts. To address this problem, the use of topology to control the spatial distribution of multivalent aptamers could be a potential solution [29]. Fan et al. reported a tetrahedral DNA framework (TDF)-programmed approach to topologically engineer receptor–ligand interactions on cell membranes (Figure 3C) [30]. Specifically, the four vertices of a TDF afford orthogonal anchoring of ligands with spatial organization, based on which they synthesized n-simplexes harboring 1–3 aptamers targeting EpCAM overexpressed tumor cells. Their results showed that the 2-monomorphs with three aptamers were able to significantly increase the binding affinity (~19-fold), while also avoiding cellular endocytosis, thus demonstrating efficient CTCs capture. As a milestone, this work provided a new strategy for the study of affinity–ligand interactions in cancer research.
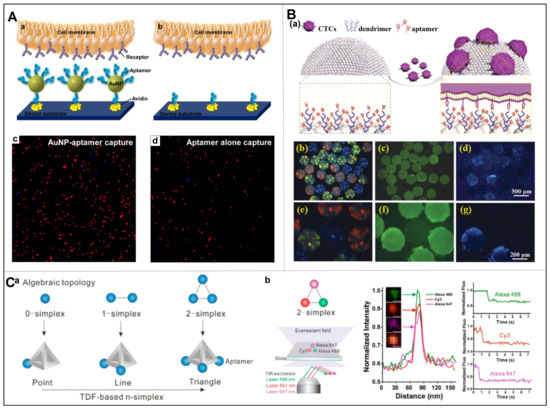
Figure 3.
(A) Platform of multivalent aptamer nanospheres in microfluidic devices for separating tumor cells from blood, as well as the cells capture results of AuNP-aptamer and aptamer alone. (a,b) Enhanced CTCs capture with AuNP-aptamer that could bind toward multiple receptors on the cell membrane. (c,d) Monovalent interaction with aptamer alone to bind receptors on the cell membrane. Reproduced with permission from [27]. (B) Barcode particles capable of capturing and releasing various types of CTC. (a) Enhanced CTC capture with barcode particles that were decorated with dendrimer and DNA aptamers. (b) Optical microscopy image, (c) green fluorescent image, and (d) blue fluorescent image of the barcode particles with captured CTCs. (e–g) Enlarged images of (b–d), respectively. Reproduced with permission from [28]. (C) TDF programming method for the efficient capture of CTCs by receptor-ligand interactions on topologically engineered cell membranes. (a) Representations of algebraic topology (0-simplex indicates one point, 1-simplex indicates a line, and 2-simplex indicates a triangle). (b) Single channel and merged total internal reflection fluorescence microscopy images of the 2-simplex, as well as the corresponding colocalization curves and stepwise photobleaching curves. Reproduced with permission from [30].
2.2. DNA Computation for Smart Recognition
Nowadays, DNAs have emerged as basic elements for multifarious nanostructures (such as branched-DNAs, multifunctional DNA Nanoflowers, aptamer–micelles, etc.), in which smart recognition can be built in due to the unique feature of Watson-Crick base-pairing [31]. Moreover, owning to DNAs’ inherent merits of easy-synthesis, low immunogenicity, and high programmability, the smart recognition has been magically achieved by state-of-the-art DNA computing systems based on Boolean logic [32]. Specifically, the concept of Boolean logic dates back to when George Boole performed logic operations via binary bits, where the “0” value means “false” and the “1” value indicates “1”. Since then, “molecular logic” has drawn substantial interests and obtained burgeoning advances in recent decades. As shown in Figure 4A [33], the binary-encoded molecule can be applied as input stimuli (absence: 0; presence: 1), and afterward, the physical or chemical signals can be displayed as outputs (high: 1; low: 0). Moreover, each kind of “molecular logic” has a unique input–output correlation prototype, following the truth table formulated by George Boole. Later, owning to the milestone work of Adleman who applied DNA to solve Hamilton path problems, the binary-encoded DNAs have been widely used as inputs as well, so as to facilitate the cascade chain growth response and broadcasting signal output.
Aptamers, as special bionic receptors with the ability to selectively bind toward analytes, play an important role in DNA computing systems for the construction of logic gates, i.e., the potential for simultaneous analysis of a series of inputs, and the unique logic of an internal system to decide whether to produce an output or not [34]. A typical example was given by Yoshida et al., where the AND logic gate was constructed by fusion of aptamer 1 and 2 through an 11-nt structure-switching signal aptamer with a fluorescein modification at T32 [35]. Two short strands of DNA partially complementary to the aptamers were modified at their 59 or 39 ends with fluorescent quenchers. In the absence of a ligand, the two quencher moieties exhibit reduced fluorescence in proximity to the fluorophore. When aptamers 1 and 2 bind to their corresponding targets simultaneously, two short strands of DNA modified by the fluorescence quencher were released, resulting in enhanced fluorescence. On the contrary, the OR logic gate was designed by partially modifying the AND gate by only using the fluorescent group and the quenching group at the ends of the two short-strand DNAs. When the target corresponding to one of the aptamers appeared, a certain DNA strand disassociated from the complex, resulting in fluorescence-enhancement. Based on this work, Chen et al. developed a DNA logic gate with dual-anchored neighboring aptamers for the accurate identification of CTCs (Figure 4B) [36]. The authors designed an “AND” DNA logic gate that could combine multiple biomarkers recognition with signal amplification. When two specific tumor-associated proteins were simultaneously overexpressed on the surface of CTCs, an “AND” logic gate was launched, inducing rolling circle amplification (RCA) reactions and a large amount of signal. Even in complex cell mixtures with small numbers of target cells, the signal of low abundance targets could be captured. Thus, DNA logic gates can directly and accurately detect and analyze CTCs in blood, presenting overall information when multiple targets of interest are presented, indicating a great significance for convenient and intelligent biomedical analysis of CTCs.
The surface of cell membranes is composed of thousands of chemical compounds that play important roles in cell growth, proliferation, and signaling. In diagnosis, we pay the most attention to cell surface receptor proteins because they participate in the communication between cells and play an important role in the energy/substance transfer between cells and the environment. In order to achieve a more accurate diagnosis and treatment of diseases, the properties of individual cells can be more accurately identified by analyzing multiple highly expressed membrane proteins of cells. Assembled logic devices based on multiple DNA aptamers are one of the most promising strategies for single-cell identification. Therefore, You et al. developed a general platform to facilitate the identification of higher-order multicellular surface markers (Figure 4C) [37]. The system could evaluate truth values (0 and 1) based on operations as AND, OR, and NOT. Through the DNA cascade reaction, they built in a series of aptamer-driven Boolean logic operations into the system, which was ultimately able to identify the expression levels of multiple cell membrane markers. Specifically, the platform was composed of two types of components, which were short oligonucleotide tags attached to specific aptamers (serving as barcodes), and fluorophore- or therapeutic-labeled ssDNA or dsDNA (serving as barcode reader and actuator), respectively. While aptamers specifically bound toward their targeting cell membrane, tags and actuators would perform logical operations via bind, dissociate, negate. For example, in the “AND” DNA logic gate, the cX*/cY* reporter DNA duplex was partially complementary to the remaining toehold region. When cY* was replaced by the invading strand X*, the substituted cY* subsequently hybridizes with the invading strand Y* to form another new duplex Y*/cY*. In their study, target labeling was only achieved when both barcode labels were attached to the cell surface, i.e., when both inputs were true. More specifically, the ON signal (output 1) was only possible when both sets of aptamer-targeted antigens were present on the cell surface.
Since typical logic gates exhibit low speed for molecular proximity effect, due to the freely diffusible components in solution, Peng et al. constructed an aptamer-based 3D DNA logic gate nanomachine, where all recognition and computational modules were integrated in a single structure to target overexpressed cancer cell biomarkers with specific recognition (Figure 4D) [38]. Specifically, they designed a DNA triangular prism (TP) structure as a substrate for subsequent functionalization. On this basis, three functional toes extended from its top surface and two sides. On the top, a reporter toe consisting of F/S/R conjugate was loaded; while on the bottom, two separate recognition toes were loaded. Each recognition toe hybridized to its respective piece of complementary DNA (cDNA) in the absence of target cells. However, in the presence of tumor cells expressing the two biomarkers of interest, the bottom recognition toes simultaneously released cDNA as output, and reporter toes were then opened after undergoing a DNA strand displacement reaction for outputs.
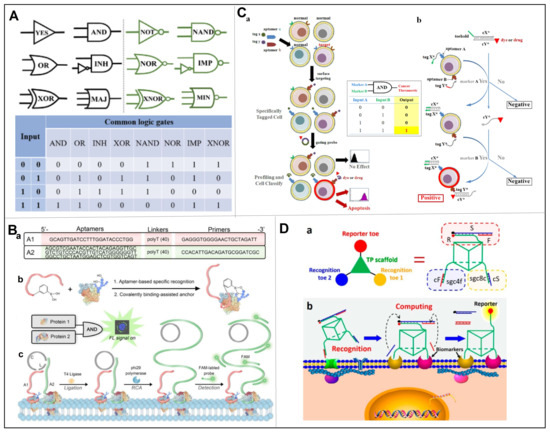
Figure 4.
(A) Basic logic gates corresponded truth tables reproduced with permission from [33]. (B) Schematics of the DNA logic gate based on dual-anchored proximity aptamers for identification of target cells. (a) Sequences of A1 and A2 aptamers, respectively. (b) Recognition of the target tumor-associated protein by the dual-anchored proximity aptamer. (c) The operation of the DNA logic gate on the surface of target cells. Reproduced with permission from [36]. (C) Schematics of the cell-surface logic gates with symbols, truth tables, and experimental schemes of toehold based strand displacement “AND” gate. (a) Example of two-input “AND” gate: tagged aptamer probes were incubated with various cells, among which only the target cell expressed both membrane receptors (blue and red); after washing, dye or drug-labeled probe was added, and the final cellular fluorescence signal or cellular viability could be detected. (b) Toehold-based strand displacement “AND” gate, where tag is named X or Y, meanwhile cX or cY indicates the complementary sequence, and * represents positive signal. Reproduced with permission from [37]. (D) Working principles of 3D DNA nanomachine for cell-surface recognition and computing. (a) Structure of the DNA nanomachine consisted of reporter toe, recognition toe 1, recognition toe 2 and DNA TP scaffold. (b) Nanomachine for targeted cell surface computing: two aptamers recognized the corresponding membrane proteins, thus releasing the complementary cS and cF, leading to the fluorescence signal in reporter toe to “ON”. Reproduced with permission from [38].
2.3. Molecular Imprints
Since MIPs hold imprinted cavities that are complementary to the three-dimensional shape and interaction sites of the template molecule, these MIPs have drawn lots of focus in areas such as chemosensing, isolation, and catalysis [39,40,41]. However, traditional MIPs were always restricted due to their potential damage to cells and non-conductive properties [42]. Owning to recent advances in material science and nanotechnology, some cell-imprinted materials are expected to enable sensitive adhesion and bio-friendly capture of CTCs for further detection. With respect to antibody–antigen interactions, the layered micro/nanostructures, i.e., the cell-imprinted interfaces using aptamer as functional monomers, can also be used to modulate cell adhesion behavior. In view of this, Liu et al. proposed a cell-imprinted hydrogel with site-directed modification of aptamers (APT-CIH) for specific capture of CTCs (Figure 5) [43]. Specifically, they designed the imprinted sites toward the target cells by using a whole cell imprinting technique, after which the aptamers were site-directed and modified within the imprinted sites via a trifunctional cleavable cross-linker. Using this APT-CIH, SMMC-7721 cells were captured with significant efficiency of 94.7 ± 0.9%, when 105 cells were incubated on the hydrogels. On the contrary, few SMMC-7721 cells bound onto the APT-NIH, with the capture efficiency of only 16.4 ± 0.4%, indicating the limited ability of non-imprinted hydrogel to capture the target cells. Moreover, the number of proliferated SMMC-7721 cells released from APT-CIH showed an exponential increasing trend with respect to the time, similar to that of the untreated cells, indicating that the released cells had unaffected viability and proliferation capacity, demonstrating their ability for the downstream analysis.
Similarly, Gao et al. prepared a cell-imprinted bionic interface that could effectively recognize and selectively capture targeted MCF-7 cells [44]. In their work, the specific cavities were prepared by template cell imprinting via soft lithography onto polydimethylsiloxane. Following the template removal, the natural antibody anti-EpCAM was introduced onto the cell-imprinted interfaces. Lastly, synergistic effects of the natural and plastic antibodies endowed the cell-imprinted surface with the ability of specific recognition of MCF-7 cells, superior to that of non-imprinted substrates (Flat), and better than streptavidin-modified interface and nothing modified interface (CIP). It is well known that the biggest problem of MIPs comes from the embedding of template molecules and the unsatisfactory cross-selectivity. To address the issue of template embedding, the imprinting of whole cells may provide solutions by controlling the thickness of the polymer layer. Moreover, the creation of multivalent binding sites may improve the selectivity of MIPs.
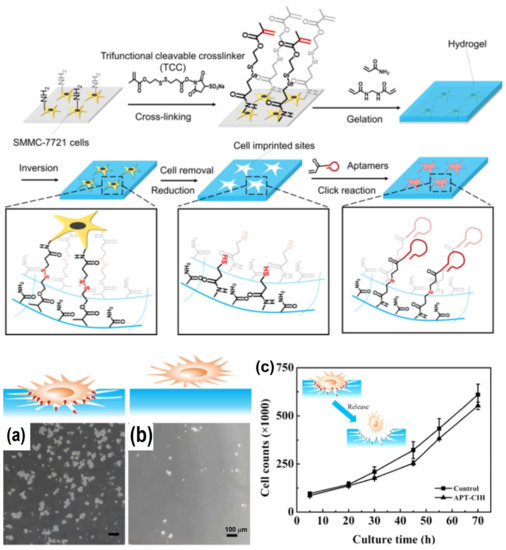
Figure 5.
Preparation of cell-imprinted hydrogel for CTCs capture. (a,b) SMMC-7721 cells on APT-CIH and NIH, respectively. (c) Proliferation curve of SMMC-7721 cells released from APT-CIH and untreated control. Reproduced with permission from [43].
2.4. Magnetic MIPs for Dynamic Recognition
Although many examples of molecular imprints have been reported for dynamic recognition of analytes, it remains a challenge to overcome the functional decline over time caused by the time-consuming and laborious nature of additional centrifugation [45]. Different from diffusion-controlled capture and release, magnetic nanoparticles (MNPs) supported MIPs may bring more efficient enrichment and separation due to a series of superior physical and chemical properties [46]. For instance, their strong magnetic force makes them easy to enrich and separate in magnetic fields or to orientate and move for localization, while the large surface-area-to-volume ratio of MNPs exhibit magical binding efficiency for molecular recognition. Moreover, the low toxicity makes them environmentally and biologically friendly, thus highly applicable [47,48,49].
Usually, magnetic molecularly imprinted polymers (MMIPs) are prepared by embedding MNPs that respond to external magnetic fields into MIPs or by coating them on the surface of MIPs. This way, the synthesized MMIPs not only have the high selectivity and affinity for the target molecules, but also have the superparamagnetic properties for subsequent isolation. Since the identified and adsorbed target analytes can be efficiently separated from the mixture by an external magnetic field without additional centrifugation or filtration, these MMIPs can be recycled many times [50]. Recently, Kuhn et al. prepared a porous MMIP network that could selectively bind target antibiotics, leading to the efficient removal and monitoring of harmful antibiotic contaminants in food [51]. Specifically, they selected vinyl silane-modified magnetic nanoparticles to form a porous polymer network with high binding capacity for erythromycin and ciprofloxacin, respectively. As a result, a good binding capacity of 75% and 68% have been achieved and remained after four cycles. For biomacromolecules, Chen et al. developed a novel MMIPs for selective extraction of glycoproteins at physiological pH by using phenylboronic acid-functionalized magnetic graphene oxide as substrate (Figure 6A) [52]. The synthesized MMIPs exhibited not only a good binding ability for template glycoproteins, but also a fast adsorption/elution rate in the magnetic field. On the other hand, the low pKa value (pKa = 6.5) of 2,4-difluoro-3-formyl phenylboronic acid-modified magnetic graphene oxide as a support matrix for glycoprotein immobilization allowed the polymer to be used at physiological pH, demonstrating the reliable application of this material.
As well known, the manufacturing process of MMIPs has significant flaws. During polymerization, MNPs were usually dispersed directly in the polymer precursor solution, which could lead to non-uniform distribution of magnetism or even non-entrapment. Moreover, it is difficult to prepare uniform and controllable shells of MIPs on the surface of magnetic nanoparticles. To address this issue, An et al. successfully fabricated a controllable core–shell MMIP based on konjac glucomannan, combining a controlled core–shell strategy with polysaccharide graft copolymerization for the selective recognition and enrichment of trichlorfon (Figure 6B) [53]. Particularly, the shell thickness of the core/shell layer structure could be efficiently regulated by the dose of the polymer precursor over a wide range. The adsorption isotherm of this MMIP could be fitted with Langmuir equation with a maximum adsorption capacity of 22.04 mg/g. At the end, the environmentally friendly MMIPs have been successfully applied in the detection of trichlorfon in real samples, contributing to the enrichment and separation of pesticide residues in food samples.
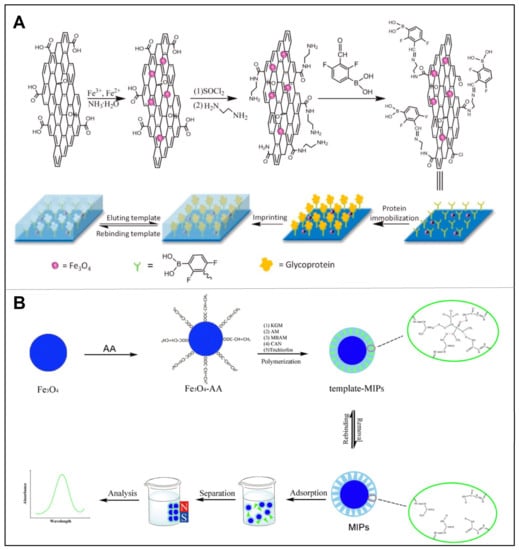
Figure 6.
(A) Schematic illustration for preparation of a novel type of MMIP for selective extraction of glycoprotein at physiological pH. Reproduced with permission from [52]. (B) Preparation of core/shell magnetic MIPs and their application. Reproduced with permission from [53].
3. Integrated Sensors for Dynamic Detection of CTCs
Currently, chip integration technology at the macro level has promoted the development of electrochemical sensors, microfluidic systems, and wearable devices. Therefore, the integrated sensors are playing an important role in the detection of circulating tumor cells, further promoting the era of precision medicine towards the direction of smaller, faster, and simpler. In this section, the applications of integrated sensors for dynamic detection of CTCs are reviewed, with a focus on their recyclability, as well as cell viability after capture and release, so as to point out the way to go for intelligent and popular diagnostic technology.
3.1. Electrochemical Sensors
Electrochemical sensing technology is an integral part of modern analytical chemistry, especially for medical testing, due to its excellent sensitivity (LOD as low as fM level) and real-time response rate (as quick as ms level) [54,55]. For CTCs detection, since the adhesion of cells to the electrode interface changes the impedance of the electrode, the electrochemical impedance spectroscopy (EIS) may correctly reflect the relevant biological information [56]. Based on this, Gao et al. developed an on-site electrochemical sensor that realized controlled capture and release of MCF-7 cells by introducing a host-guest based redox response on an indium tin oxide (ITO) [57]. To note, they have for the first time constructed an electrochemical system for reversible capture-and-release and immediate detection of cancer cells and the cell viability after one time capture and release remained to be over 86%. For quantitative analysis, Shen et al. designed a novel label-free electrochemical sensor that could specifically collect and detect MCF-7 cells using EpCAM aptamers-hybridized probes on gold electrodes (Figure 7A) [58]. Under stable external conditions of 10 μM of MCH, 100 nM of capture probe, 120 min of incubation time of the capture probe, and 60 min of cell incubation time, the linear detection range covered 30–106 cells/mL-1 with a LOD of 10 cells/mL. Moreover, the gold sensor could be reused eight times without affecting cell activity.
As the most commonly used molecular recognition elements, aptamers show unique advantages in detection of low CTCs containing blood samples, by implementing signal amplification strategies [59,60]. Ou et al. developed a sandwich-type sensor to analyze MCF-7 cells, so as to determine cell viability and detect high selectivity of the sensor (Figure 7B) [61]. Specifically, a tetrahedral DNA nanostructure was simultaneously hybridized with AS1411 and MUC1 that specifically target MCF-7 cell surface biomarkers, and then immobilized on the surface of gold electrodes. Upon cell capture on the functionalized gold electrode, the metal organic framework PCN-224-based oxidase mimics might recognize the captured MCF-7 cells and mediate the oxidation reaction for differential pulse voltammetry (DPV) signal output. Their sensor was successfully used to detect 20–107 cells/mL with LOD of 6 cells/mL. Furthermore, the captured cells could be released by electrochemical reductive desorption via disrupting the gold-thiol bond, resulting in 93% of the cells to retain their original viability. Particularly, Yang et al. explored a photoelectrochemical (PEC) sensor for MCF-7 detection by measuring changes in photocurrent intensity (Figure 7C) [62]. Specifically, Fe3O4 nanospheres and Cu2O nanoparticles were used for efficient magnetic capture and output signal amplification, respectively. The Cu2O-nucleic acid aptamer probe binds specifically to MUC1 overexpressed on the surface of MCF-7 cells, where Cu2O NPs competitively absorb excitation light, thereby amplifying the detection response. This PEC biosensor was successfully applied to detect 3 to 3000 cell mL-1 MCF-7 cells with an LOD of 1 cell mL-1. Owning to the inherent pairing property of nucleic acids, aptamers-based electrochemical sensors have great application perspectives in in cancer diagnosis [63,64].
MIPs, also called artificial antibodies, show incomparable stability for the preparation of electrochemical sensors, while maintaining antibody-like affinity and specificity [65,66,67]. An interesting example was provided by Eersels et al., where cells were imprinted onto a PDMS surface by forming polyurethane. Upon the binding of cells, the heat transfer resistance would increase for quantification analysis (Figure 7D) [68]. This study reported specific binding of cells to the blotting lumen with a LOD of 28,930 cells/mL for this sensor. Lv et al. report a study combining the near infrared (NIR) photothermal effects of gold nanorods (GNR) and thermally responsive hydrogel substrates for the capture and release of highly active EpCAM-positive cells. Hydrogel substrates are prepared by imprinting target cancer cells onto GNR pre-embedded gelatin hydrogels followed by antibody modification. Gelatin can be rapidly solubilized at 37 °C to recover large numbers of CTCs, or site-specific release of individual CTCs can be achieved by NIR-mediated photothermal activation of the embedded GNR. The temperature-responsive release of cancer cells has a 95% viability rate and the photothermal selective release of cancer cells has a 90% viability rate [69].
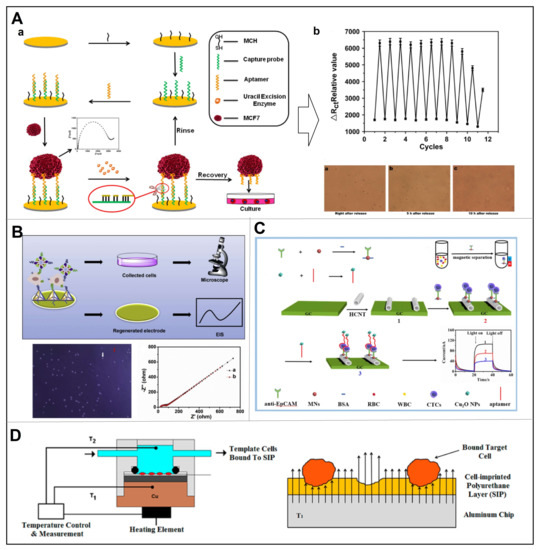
Figure 7.
(A) Sensor fabrication process (a), as well as cycling experiment results and cell viability microscope images (b). Reproduced with permission from [58]. (B) Schematic illustration of the sensor for cell release, with the microscope image of trypan blue staining of collected cells (the red arrow points to the dead cell, the white arrow points to the living cell), as well as EIS of (a) bare electrode and (b) the regenerated electrode. Reproduced with permission from [61]. (C) Schematic representation of CTC measurement in whole blood based on MNs isolation of CTCs and Cu2O-aptamer as signal probe. Reproduced with permission from [62]. (D) Schematics of the measuring setup and illustration of cell binding to the binding cavities of the SIP, thereby blocking the heat transfer from the chip to the liquid. Reproduced with permission from [68].
Prospects analysis. Moving towards a more flexible direction, so as to facilitate the development of wearable devices, researchers begun to focus on graphene film. This flexible nanostructured film can provide biomimetic cellular microenvironment and has high capture efficiency [70,71]. Owing to its large surface area and easy-to-modify property, Wu et al. found that grapheme-covered glassy carbon electrode could effectively accelerate electron transfer and enhance the detection signal. Moreover, when the grapheme was integrated with anti-EpCAM antibodies, the captured CTCs could be traced by aptamer- or MIP-conjugated QD nanoparticles, thereby exhibiting significant electronic signals [72]. Recently, Wang et al. prepared a ZnO nanorod arrays-based sensor, where the ZnO nanorods were used as a vertical biological interface to enhance the interaction between cancer cells and the electrode. Specifically, the ZnO nanorods were modified by aminopropyl triethoxysilane (APTES) and glutathione (GA), followed by coupling with the sambucus nigra agglutinin (SNA), leading to specific recognition of sialic acid (SA) on the cell surface that brought in good cell trapping ability [73]. Inspired by this work, we believe that MIPs can be endowed with dual functions, i.e., forming nanoarrays and SA targeting, which is an interesting topic worth exploring. For the preparation of MIPs-based sensors toward accurate, smart, and safe, we still have a long way to go (as summarized in Table 1).

Table 1.
Performance comparison of different recognition element-based electrochemical sensors.
Table 1.
Performance comparison of different recognition element-based electrochemical sensors.
| Receptor | Target CTC | LOD | Recycle | Dynamic | Ref. |
|---|---|---|---|---|---|
| Antibody | Hep3B cells | 5 cells/mL | 6 | yes | [72] |
| Aptamer | MCF-7 cells | 1–10 cells/mL | 8 | yes | [58,61,62] |
| MIP | MCF-7 cells | 28,930 cells/mL | >2 | yes | [68] |
3.2. Microfluidic Devices
Recently, researchers started to detect CTCs in a more dynamic way so as to facilitate final release and collection. To achieve this goal, microchannel devices have drawn a great attention due to the large surface-to-volume property and hydrodynamic advantages, which in turn increases the chances of capture events and automated collection [74]. Moreover, combining nanomaterials with microchannels is expected to improve the capture efficiency, sensitivity and solve the problem of non-destructive release of CTCs. The integration of nanomaterials into microfluidic chips is therefore a promising method for the dynamic detection of CTCs. In this section, the performance of microfluidic devices combined with two types of nanoparticles (i.e., magnetic nanoparticles and Au nanoparticles) were reviewed, and an immobilized strategy was highlighted for CTCs isolation [75].
3.2.1. Microfluidic Chip-Combined with Magnetic Nanoparticles
As well-known, immunomagnetic separation has received considerable attention for the capture of CTCs, due to the rapid magnetic response, high surface area, good suspension properties, and biocompatibility. A good example is provided by Yu et al., who achieved effective capture and identification of multiple types of CTCs on immunomagnetic nanospheres (IMNs) (Figure 8A) [76]. To note, superparamagnetic iron oxide nanospheres were functionalized with polymeric ionic liquid (PIL) and subsequently coated with anti-EpCAM nanospheres. The IMNs showed outstanding cell capture efficiency (>95%) and specificity when multiple EpCAM-high expressing CTCs alone were used for the experiments. As the structure of PIL is rich in carboxyl groups, the efficiency of coupling the two is enhanced by the chemical binding of the carboxyl group to the amino group, resulting in more target cells adhering to the IMNs.
Once the cells are labeled with magnetic particles, it is difficult to separate the magnetic bead-cell aggregates from the mixture of bare magnetic beads, as the magnetic field attracts both. Therefore, to separate the magnetic beads from the magnetic bead-cell aggregates, Lin et al. used immunomagnetic separation techniques to extract magnetic beads and bead-bound cancer cells from test samples by injecting particles suspended into microchannels, in which impedance measurements were performed at multiple frequencies simultaneously [77]. Since the magnetic bead-cell aggregates show a larger diameter, with respect to bare cells and bare beads, this manifests itself as a larger peak intensity. Therefore, the electrical impedance can be used to differentiate between cells bound to magnetic beads and bare cells/beads. Recently, Lee et al. designed an automated motion-controlled microfluidic system for one-step cell enrichment (Figure 8B) [78]. On this device, a micro-mixer generated multiple vortex currents to enhance the interaction between CD45 antibody-coupled magnetic nanoparticles (MNP) and leukocytes (WBCs), and a magnetic sorter placed at the top and bottom of the microchannel removed MNP-coated WBCs. After specific removal of WBCs, over 85% of MCF-7 cells can be obtained from the blood sample. Moreover, Shi et al. developed a wavy-herringbone (wavy-HB) structured microfluidic device in which anti-EpCAM antibody-coated MNP was immobilized on the wavy-HB surface under an external magnetic field [79]. High yields (92%) was obtained in whole blood samples using this wavy-HB structured device.
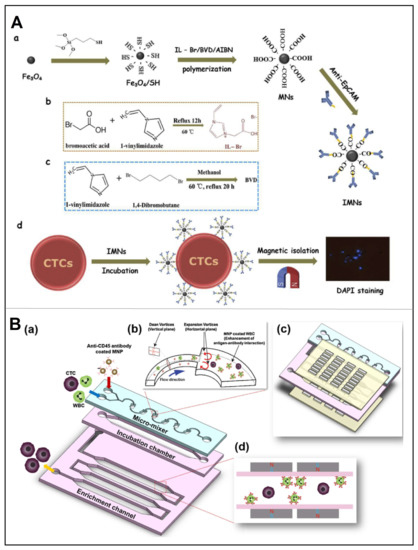
Figure 8.
(A) Representation of CTCs captured by PIL-decorated IMNs. (a) Preparation of anti-EpCAM conjugated IMNs. (b) Synthesis of IL-Br. (c) Synthesis of 1,4-butanediyl-3,3′-bis-l-vinylimidazolium bromide (BVD). (d) Application of the PIL-decorated IMNs for CTCs capturing and isolation. Reproduced with permission from [76]. (B) Schematic diagram of a μ-MixMACS chip for one-step CTCs isolation. (a) Structure of the chip: a microfluidic mixer, an incubation chamber and a magnetic activated cell sorter (MACS). (b) In the microfluidic mixer, the interaction between CD45-conjugated MNPs and WBCs was enhanced. (c) Two magnetic sorters were placed at the top and bottom of the microfluidic mixer, respectively. (d) The μ-MixMACS chip was used to capture the MNP-coated WBCs via magnetic activation, thus eluting CTCs through the outlet. Reproduced with permission from [78].
3.2.2. Microfluidic Chip-Combined with Au Nanoparticles
In addition to magnetic nanoparticles, gold nanoparticles have also been explored for the capture of CTCs during a long time. Due to the simple synthesis and modification process, gold nanoparticles (AuNPs) are commonly used to assemble various aptamers, so as to improve the efficiency of CTCs capture [80].
Recently, Park et al. developed sulfhydryl ligands-based AuNPs on herringbone microchannels to effectively isolate cancer cells from peripheral blood [81]. Specifically, the functionalized AuNPs were immobilized within herringbone microchannels, thereby the coated anti-EpCAM antibodies could promote specific CTCs binding with a high capture efficiency and low non-specific binding. Besides, Song et al. designed a deterministic lateral displacement (DLD)-patterned microfluidic chip, mimicking the multivalent tentacles of octopus by aptamer-functionalized AuNPs (Figure 9A) [82]. Compared to the monovalent aptamer-modified chip, the capture efficiency of their device was improved more than 3-fold, due to the synergistic interactions of AuNP-aptamer and EpCAM. The strong interaction between the S atom and Au surface follows the acid-base interaction between hard and soft to form the Au-S bond, which can be readily disrupted by excess biocompatible thiol molecules, without compromising cell viability, thereby enabling effective release for further analysis [83]. In this experiment, the AuNP-aptamer interface achieved 80% capture efficiency, and maintained 96% cell viability through the sulfhydryl exchange reaction, which was fully compatible with downstream mutation detection and CTCs culture.
Inspired by the high-efficiency preying mechanism performed by the multiplex tubing feet and endoskeletons of sea urchins [84], Zhang et al. designed a super-efficient biomimetic single-CTC recognition platform by conjugating dual-multivalent-aptamers (DMAs) Sgc8 and SYL3C onto AuNPs to form a sea urchin-like nanoprobe (sea urchin-DMA-AuNPs) (Figure 9B) [85]. Aptamers Sgc8 and SYL3C selectively bind to the biomarker proteins PTK7 and EpCAM expressed on the surface of CTCs. Compared with the single-aptamer nanoprobe/-interface, the binding efficiency of the dual-aptamer nanoprobe was increased twice (Kd < 0.35 nM). At the flow rate of 60 μL min-1, the separation rate of single CTC was 93.6%, and the measurement efficiency of single CTC was 73.8 ± 5.0%. Moreover, their strategy ensured the operation and detection of a single CTC in 100 μL whole blood within 1 h.
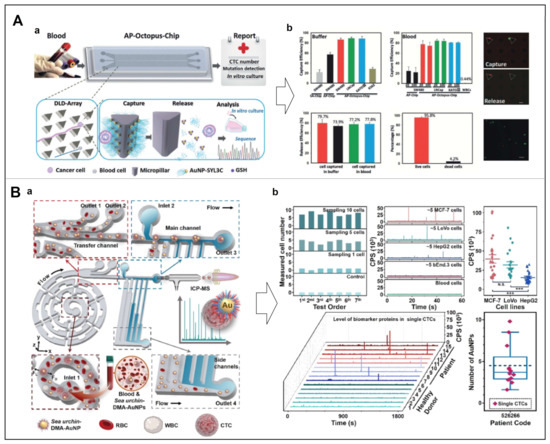
Figure 9.
(A) Design of the NP-HBCTC-Chip for the capture and release of CTCs. (a) Based on the DLD principle, CTCs could continuously interact with aptamer-functionalized AuNPs in the channel; afterward, the captured CTCs could be released by the Au-S bond disrupted by excess GSH. (b) Capture performance in buffer and blood samples. Reproduced with permission from [82]. (B) Working principle of the AP-octopus-chip for CTCs recognition. (a) The CTCs captured by sea urchin-DMA-AuNPs were aligned and sorted in the microchip, then the captured CTCs were introduced to process efficient single-cell analysis. (b) Detection performance of the AP-octopus-chip, using different number of MCF-7 cells, various cell types from healthy and patients, respectively (N.S. for p > 0.05, *** for p < 0.00). Reproduced with permission from [85].
Prospects analysis. In addition to the aforementioned nanoparticles, an important polymer-coated microfluidic device shows great potential to enable CTCs capture as well. Yu et al. used an electrostatic spinning process to prepare poly(lactidylethanolic acid) (PLGA) nanofiber arrays on glass slides in random or aligned orientations, after which these PLGA nanofibers were embedded in poly(methyl methacrylate) (PMMA)-based microfluidic chips to achieve efficient CTCs capture (Figure 10A) [86]. Moreover, a transparent microfluidic device based on graphene oxide (GO) was proposed by Kim’s team for the isolation of CTCs within the in vivo retention vessel (Figure 10B) [87]. Specifically, they assembled anti-EpCAM antibodies-functionalized GO on a gold layer, so as to maximize the frequency of cell-substrate contact. In this work, they successfully achieved sensitive trapping and high throughput via the herringbone structure. Unfortunately, the MIPs have not yet been well integrated with microfluidic devices for CTCs detection, nor have they achieved ideal application results. Because MIPs with small size are difficult to adapt to the mechanical environment in microfluidics, their recognition performance is easily deformed in such environment. Furthermore, there is no particularly good microchannel imprinting method, so as to not interfere with the final package, other than photolithographic etching techniques. To note, given that polymer materials have already played an excellent role in microfluidic devices and are conducive to the era of wearable devices, we believe that there is still much work to be done in this blank area, and it is worth exploring.
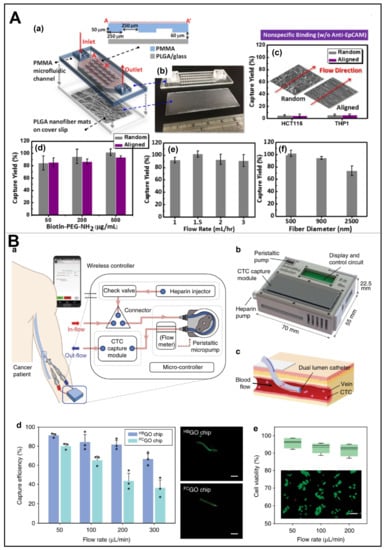
Figure 10.
(A) Schematic representation of the PLGA nanofibers-embedded microfluidic chip. (a,b) Design and photograph of the microfluidic chip. (c) Performance for preventing nonspecific binding on random and aligned arrays. (d–f) Cell-capture performance under different conditions. Reproduced with permission from [86]. (B) The in vivo aphaeretic CTCs isolation system. (a,b) Schematic representation of the system by functional components and manifold. (c) Application for in vivo CTCs isolation. (d) CTCs isolation performance of two chips, as well as fluorescent MCF7 cell within the two different channels. (e) Measure of cell viability as a function of flow rate after capture, as well as fluorescent image of MCF7 cells (green cells were alive, scale bar: 100 µm). Reproduced with permission from [87].
3.3. Wearable Sensors
Wearable sensors are integrated analytical devices that enable real-time and continuous noninvasive measurements by using physicochemical or biological principles [88]. The history of wearable devices dates back to the 1960s [89], and in the last decade, the first generation of wearable devices have received great attention due to their advantages in quality, manufacturing cost, high flexibility, and adaptability. Nowadays, the wearable sensors are mainly used for tracking physical activity and vital signs, showing great potential in predictive analytics, and personalized medicine therapy [90]. With the advent of the smart era, wearable sensors have also become the future trend. The core of the next generation of wearable sensors has shifted to the development of biometric recognition for biofluids [91]. Since the unique context provided by neoplastic lesions not only contains variegated forms but also varies in individual conditions, the wearable devices may meet the needs of personalized diagnosis and the purpose of monitoring tumor development progress in real time. Combining advanced cell capture technology with flexible materials, wearable sensors can be expected to help predict, screen, diagnose, and treat relevant diseases in the human body, thus providing more in-depth personalized medical diagnosis.
Compared with natural antibodies or receptors, the non-invasive handling mode of MIPs at flexible interfaces exhibits considerable capture efficiency, good cell selectivity, and more durable molecular recognition sites, promising to be a useful and versatile tool for CTCs detection and collection in wearable sensors. Usually, the efficiency of the cell sheet system is limited by the increasing cell adhesion and needs to become repulsive to cells after forming a fused cell monolayer, so as to avoid damaging the cells. As a pioneer in this field, Pan et al. first used imprinting technology to introduce the cell adhesion peptide Arg-Gly-Asp-Ser (RGDS) into a heat-responsive cell culture substrate, resulting in an efficient cell sheet harvesting system (Figure 11A) [92]. Cell sheeting technology mainly utilized poly(n-isopropylacrylamide) (PNIPAAm) as a thermoresponsive cell culture substrate to achieve cell harvest at 37 °C [93,94,95]. In Pan’s work, the imprinted hydrogel recognized and bound RGDS molecules at cell culture temperature (37 °C), and then rapidly released RGDS when the temperature decreased (e.g., 20 °C). In the field of MIP, it is Zhang et al. that really fabricated a wearable sensor, which was later successfully applied to monitor lactate level in human sweat (Figure 11B) [96]. Specifically, they performed electropolymerization of 3-aminobenzenesulfonic acid (3-APBA) on silver nanowires (AgNWs)-coated electrodes in the presence of lactic acid. The obtained sensor could detect lactate in the range of 1 μM to 0.1 M with a LOD of 0.22 μM in human sweat, and could be reused 200 times.
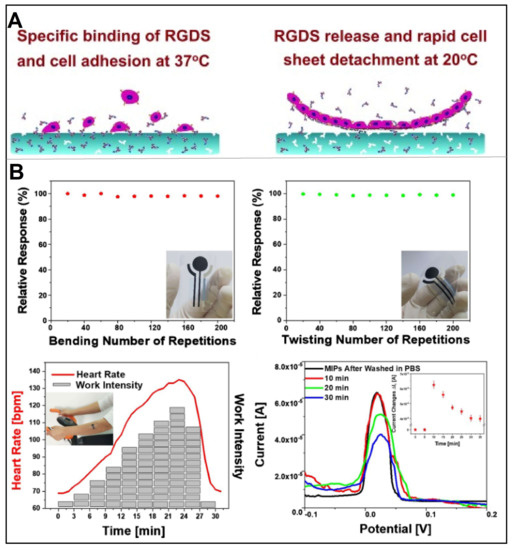
Figure 11.
(A) Schematic diagram of a heat-responsive hydrogel layer for reversible cell adhesion and cell sheet harvesting using RGDS peptide blotting. Reproduced with permission from [92]. (B) Results of the wearable electrochemical sensor for lactate detection in human sweat: The DPV relative response of MIPs-AgNWs biosensor undergoing repeated inward bending performed in PBS from −0.2 V to 0.3 V, as well as the stability results (* represents negligible data). Reproduced with permission from [96].
Prospects analysis. It is a pity that experts in the field of MIP have not given a case of wearable sensors for CTC detection. Combining the two strategies, we believe that there must be peers with cutting-edge ideas who are trying to synthesize some imprinted hydrogel-based wearable sensors that may not only regulate reversible cell adhesion behavior, but also monitor more complicated cell behavior on the flexible interfaces.
4. Artificial Intelligence-Enabled Sensors for CTCs Detection
Nowadays, artificial intelligence (AI) has come into the spotlight for medical imaging and minimally invasive biomarkers, drawing significant interests in machine learning and deep learning. Along with the popular machine learning algorithm in predictive models, the accurate quantification of targets and the hidden-space feature extraction have made huge progress in recent years [97]. Meanwhile, a great deal of machine-learning algorithms, such as support vector machines (SVMs), random forests (RFs), and artificial neural networks (ANNs), have been developed in the biomedical field [98,99,100]. Based on machine learning, a deep learning algorithm dubbed “convolutional neural networks” (CNNs) has emerged and shown great potential in processing and interpreting radiological and pathological images. Since then, pathology researchers have made a great effort in developing AI-enabled sensors for tumor detection, tumor classification and grading, pathological property identification, and genomic mutation location.
At the cellular level, CTCs identification is usually performed by physicians according to the morphological characteristics, such as size, texture, and shape. Therefore, the output pathological results are always affected by physicians’ experience and subjective judgments, leading to tremendous, misdiagnosed cases. To address this issue, deep learning algorithms have been widely used to empower machines, bringing high efficiency and accuracy. Regarding CTCs analysis, state-of-the-art algorithms have achieved high-throughput, automated quantification and accurate identification via integrated feature extraction (including biomarker level, morphological features, etc.) [101]. On this basis, Guo et al. developed a deep migration learning-based algorithm for CTCs recognition, lesion tracking, and gene marker identification (Figure 12A) [102]. Specifically, their algorithm enabled knowledge migration from primary tumor cells to CTCs via deep transfer learning. To note, the deep transfer learning was established on the large amount of primary tumor cell sequencing data (scRNA-seq), which could reveal the location of cancer lesions in CTCs. Their algorithm displayed excellent efficiency and accuracy while analyzing scRNA-seq data of various tumors from different platforms, thus emphasizing its great potential for analyzing a broad range of CTC data sets.
Owning to the software advancement, the performance of hardware has also improved a lot. For instance, the CellSearch® apparatus may apply anti-EpCAM antibodies-coupled ferromagnetic fluid to capture CTCs, meanwhile providing accurate classification information of CTCs. Based on the CellSearch® apparatus, Sanne et al. analyzed all 4′6-diamidino-2-phenylindole (DAPI)-stained nuclei in EpCAM rich blood samples from 192 patients with metastatic non-small cell lung cancer (NSCLC), with respect to 162 control samples from healthy people, so as to develop an open-source imaging program that could automatically identify, count, and classify the NSCLC [103]. Using their program, a significant increase in the number of nuclei was detected in 300 patient samples with a mean and standard deviation of 73,570 ± 74,948 compared to 4191 ± 4463 in 359 control samples (p < 0.001). Moreover, they innovatively added CD16-PerCP for granulocyte staining, used an LED as the light source for CD45-APC excitation, and applied wheat germ agglutinin to stain plasma membrane, leading to significantly improved EpCAM-cells enrichment, resulting in the identification of 94% ± 5%. Later, Zeune et al. hit on a novel idea to identify CTCs in fluorescent images that were automatically obtained by the CellSearch® system (Figure 12B) [104]. Specifically, they developed a dimensionality reduction CNN to decipher the information in hidden space of the fluorescent raw images containing CTCs, after which the well trained CNN network might classify individual cell into five categories and perform CTC counts. Their CNN network exhibited a significant accuracy and sensitivity, with specificity of ~96%, while allowing monitoring and evaluation of overall therapeutic effects in patients.
Since cancer is a self-sustaining and adaptive disease that associates dynamically with the environment, it haunts patients like a devil and hinders the progress of researchers. In the context of cancer screening, Hashemzadeh et al. developed a reliable computer-aided diagnosis (CAD) system for automated cancer screening via the combined microfluidic deep learning approach (Figure 12C) [105]. In this study, microfluidic devices were used to culture prevalent lung cancer cells and normal cells, so as to establish the baseline accuracy of lung cancer cell line classification desired by modern deep learning models. The images of lung cancer cells were divided into five different cancer cell lines and one normal cell line. As result, their CAD system have achieved 98.37% accuracy and a 97.29% f1 score, demonstrating that the system could not only achieve high-throughput data processing with high precision and robustness, but also completely eliminate the need for user intervention.
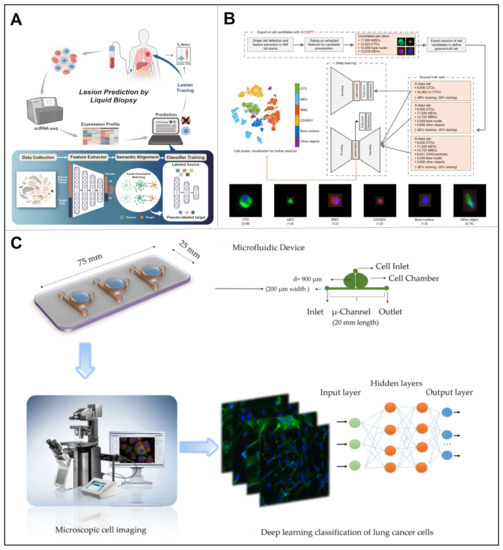
Figure 12.
(A) Overview of the main function of CTC-Tracer and its application. Reproduced with permission from [102]. (B) Overview of the analysis workflow used [104]. (C) Overview of the combined microfluidic deep learning approach. Reproduced with permission from [105].
Prospects analysis. It is clear that AI has the potential to outperform most pathologists in detecting CTCs, and is not affected by inherent inter observer variability. However, the current challenges include training pathologists and physicians to use AI methods and helping them to accept the results, as well as the fact that AI algorithms rely heavily on the quantity and quality of data used to do training. Due to intra- and inter-tumor heterogeneity, it becomes difficult to select cohorts that represent the entire tumor cell plasticity, which may lead to a non-negligible risk of over fitting [106]. Therefore, the algorithms and computational techniques must be validated with large-scale clinical studies.
Unlike identification and classification tasks, manually labeling pixel annotations for segmentation tasks is an expensive and time-consuming task. In the field of medical images, collecting a large number of pixel-level semantic labels requires extremely high costs. For example, CT images themselves are grayscale images, and their foreground boundary areas are very prone to ambiguity, which requires physicians to be extremely professional and spend a lot of time to be accurate. Therefore, we believe that model training based on a semi-supervised approach is the mainstream direction in the future. It can use a small amount of labeled data and a large amount of unlabeled data for network learning at the same time, and can also reduce the burden and cost of obtaining a large amount of human-labeled data for medical images. Additionally, we believe that AI will open an exciting avenue for the identification of new biological phenomena. Moreover, the combination of fluorescent MIP-based detection and AI technology can only be a long-term goal.
5. Conclusions
In summary, the efficient and dynamic capture of CTCs has been addressed with the development of 3D aptamers and MIPs. Currently, detection based on the whole cell and related biomarkers is primarily important in the field of precise medicine and clinical therapy [107], hence the importance of liquid biopsy technology. However, the commercialization of MIP-based sensors is restricted by the lack of programmability, lack of order in functional monomers, and inability to adapt to fluid environments while being small enough, etc. [108,109,110]. Hopefully, with the development of new sensing technology, new materials and artificial intelligence, new varieties, new structures, and new applications of chemosensors are constantly emerging.
Nowadays, the “five modernizations” have become an important trend of development, namely intelligence, motorization, miniaturization, integration, and diversification. Thus, an interesting part concerning computer-aided diagnosis and intelligent image processing was briefly introduced. Although various medical image data, including radiological images (CT, MRI, X-ray), stained H&E histopathological images, fluorescently labeled confocal images, endoscopic, and fully digital images containing both low-level and high-level semantic information about cancer can be extracted to meet the requirements of cancer diagnosis. Moreover, with the advancement of detection methods, the treatment of cancer is gradually becoming patterned and professional. Currently, precision medicine and treatment are highly dependent on the most accurate data of cancer patients. Diagnostic information is derived from different diagnostic manners and data. Thus, multimodal fusion may achieve the effect of complementary information, which can improve the accuracy of prediction to a certain extent. We believe that the fusion of cancer modalities can eliminate the physical limitations of sensing techniques and achieve the target of high resolution and low noise. Other modal information, mainly DNA sequences, biomarker content, clinical data, and physician’s diagnostic text, are commonly fused at the input level, feature level, and the decision level. Furthermore, the fusion of multimodal information is promising for survival analysis and postoperative recurrence prediction in cancer patients. Additionally, the topic of mobile deployment represents a new future direction, where have been initial attempts in skin cancer diagnosis. With the 5G technology and high demand for future application scenarios, along with client application memory and processing limitations, mobile computer-aided diagnostic devices based on 5G, and cloud storage technologies are bound to be hotspots in future research.
In the near future, with the deepening of technology, the reduction of cost, the improvement of performance and reliability, and driven by the rapid development of the Internet of Things, mobile Internet, and high-end equipment manufacturing, chemosensors based on MIPs will shine in the application market.
Author Contributions
Conceptualization, Writing—Original draft: R.D. and M.Y.; Assistant for writing: Y.Z. (Yijie Zhu), Y.Z. (Yingyan Zhao) and Q.L.; Conceptualization, Writing—Review and editing, Supervision, Funding acquisition: Y.C. and J.X. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the financial support from the National Natural Science Foundation of China (22001162) as well as the Shanghai Sailing Program (20YF1414200).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Pe’er, D.; Ogawa, S.; Elhanani, O.; Keren, L.; Oliver, T.G.; Wedge, D. Tumor heterogeneity. Cancer Cell 2021, 39, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, S.S. Insights on CTC biology and clinical impact emerging from advances in capture technology. Cells 2019, 8, 553. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Koopman, C.D.; Werb, Z. Cancer Circulating tumor cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Liu, H.; Li, G.; Cui, H.; Jiang, L.; Wang, S. Photo and thermo dual-responsive copolymer surfaces for efficient cell capture and release. ChemPhysChem 2018, 19, 2107–2112. [Google Scholar] [CrossRef]
- Ruan, H.; Wu, X.; Yang, C.; Li, Z.; Xia, Y.; Xue, T.; Shen, Z.; Wu, A. A supersensitive CTC analysis system based on triangular silver nanoprisms and SPION with function of capture, enrichment, detection, and release. ACS Biomater. Sci. Eng. 2018, 4, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Hu, Z.; Tian, C.; Xiao, H.; Liu, L.; Yang, X.; Li, J.; Wu, H.; Lu, J.; Gao, J.; et al. In vivo detection of CTC and CTC plakoglobin status helps predict prognosis in patients with metastatic breast cancer. Pathol. Oncol. Res. 2020, 26, 2435–2442. [Google Scholar] [CrossRef]
- Gascoyne, P.R.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Deng, Y.; Sun, Z.; Wang, L.; Wang, M.; Yang, J.; Li, G. Biosensor-based assay of exosome biomarker for early diagnosis of cancer. Front. Med. 2022, 16, 157–175. [Google Scholar] [CrossRef]
- Polyakov, M.V. Adsorption properties and structure of silica gel. Zh. Fiz. Khim. 1931, 2, 799–805. [Google Scholar]
- Vlatakis, G.; Andersson, L.I.; Müller, R.; Mosbach, K. Drug assay using antibody mimics made by molecular imprinting. Nature 1993, 361, 645–647. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Trzpis, M.; McLaughlin, P.M.; de Leij, L.M.; Harmsen, M.C. Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007, 171, 386–395. [Google Scholar] [CrossRef]
- Pantel, K.; Brakenhoff, R.H.; Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 2008, 8, 329–340. [Google Scholar] [CrossRef]
- Huang, C.; Yang, G.; Ha, Q.; Meng, J.; Wang, S. Multifunctional “smart” particles engineered from live immunocytes: Toward capture and release of cancer cells. Adv. Mater. 2015, 27, 310–313. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, Q.; Zheng, W.; Li, W.; Li, P.; Zhu, L.; Liu, X.; Shao, B.; Li, H.; Wang, C.; et al. Peptide-functionalized nanomaterials for the efficient isolation of HER2-positive circulating tumor cells. ACS Appl. Mater. Interfaces 2017, 9, 18423–18428. [Google Scholar] [CrossRef]
- Tian, H.; Xu, Y.; Liu, S.; Jin, D.; Zhang, J.; Duan, L.; Tan, W. Synthesis of gibberellic acid derivatives and their effects on plant growth. Molecules 2017, 22, 694. [Google Scholar] [CrossRef]
- Pang, X.; Cui, C.; Wan, S.; Jiang, Y.; Zhang, L.; Xia, L.; Li, L.; Li, X.; Tan, W. Bioapplications of cell-SELEX-generated aptamers in cancer diagnostics, therapeutics, theranostics and biomarker discovery: A comprehensive review. Cancers 2018, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Y.; Zhu, L.; Liu, Y.; Wang, T.; Liu, D.; Song, Y.; Yang, C. Aptamer-based liquid biopsy. ACS Appl. Bio Mater. 2020, 3, 2743–2764. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.S.; Dabrowski, M.; D’Souza, F.; Kutner, W. Surface development of molecularly imprinted polymer films to enhance sensing signals. TrAC Trends Anal. Chem. 2013, 51, 146–157. [Google Scholar] [CrossRef]
- Bai, W.; Spivak, D.A. A double-imprinted diffraction-grating sensor based on a virus-responsive super-aptamer hydrogel derived from an impure extract. Angew. Chem. Int. Ed. 2014, 53, 2095–2098. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Wang, Z.; Wu, Z.; Zhu, W.; Liu, L.; Sun, N.; Pei, R. Aptamer-based nanostructured interfaces for the detection and release of circulating tumor cells. J. Mater. Chem. B 2020, 8, 3408–3422. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Phillips, J.A.; Yan, J.; Li, Q.; Fan, Z.H.; Tan, W. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal. Chem. 2009, 81, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.H.; Cha, A.; Tam, K.A.; Poellmann, M.; Borgeat, A.; Sharifi, R.; Molokie, R.E.; Votta-Velis, G.; Hong, S. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew. Chem. 2011, 123, 11973–11976. [Google Scholar] [CrossRef]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, Y.; Allen, P.B.; Asghar, W.; Mahmood, M.A.; Tan, J.; Duhon, H.; Kim, Y.T.; Ellington, A.D.; Iqbal, S.M. Capture, isolation and release of cancer cells with aptamer-functionalized glass bead array. Lab Chip 2012, 12, 4693–4701. [Google Scholar] [CrossRef]
- Li, Z.; Wang, G.; Shen, Y.; Guo, N.; Ma, N. DNA-templated magnetic nanoparticle-quantum dot polymers for ultrasensitive capture and detection of circulating tumor cells. Adv. Funct. Mater. 2018, 28, 1707152. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef]
- Zheng, F.; Cheng, Y.; Wang, J.; Lu, J.; Zhang, B.; Zhao, Y.; Gu, Z. Aptamer-functionalized barcode particles for the capture and detection of multiple types of circulating tumor cells. Adv. Mater. 2014, 26, 7333–7338. [Google Scholar] [CrossRef]
- Pei, H.; Li, F.; Wan, Y.; Wei, M.; Liu, H.; Su, Y.; Chen, N.; Huang, Q.; Fan, C. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA–gold nanoparticle nanoconjugates. J. Am. Chem. Soc. 2012, 134, 11876–11879. [Google Scholar] [CrossRef]
- Li, M.; Ding, H.; Lin, M.; Yin, F.; Song, L.; Mao, X.; Li, F.; Ge, Z.; Wang, L.; Zuo, X.; et al. DNA framework-programmed cell capture via topology-engineered receptor–ligand interactions. J. Am. Chem. Soc. 2019, 141, 18910–18915. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef] [PubMed]
- Prokup, A.; Hemphill, J.; Deiters, A. DNA computation: A photochemically controlled AND gate. J. Am. Chem. Soc. 2012, 134, 3810–3815. [Google Scholar] [CrossRef]
- Fan, D.; Wang, J.; Wang, E.; Dong, S. Propelling DNA computing with materials’ power: Recent advancements in innovative DNA logic computing systems and smart bio-applications. Adv. Sci. 2020, 7, 2001766. [Google Scholar] [CrossRef] [PubMed]
- Andrianova, M.; Kuznetsov, A. Logic gates based on DNA aptamers. Pharmaceuticals 2020, 13, 417. [Google Scholar] [CrossRef]
- Yoshida, W.; Yokobayashi, Y. Photonic Boolean logic gates based on DNA aptamers. Chem. Commun. 2007, 2, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Fu, X.; Zhang, Q.; Mao, D.; Song, Y.; Feng, C.; Zhu, X. A DNA logic gate with dual-anchored proximity aptamers for the accurate identification of circulating tumor cells. Chem. Commun. 2020, 56, 6961–6964. [Google Scholar] [CrossRef]
- You, M.; Zhu, G.; Chen, T.; Donovan, M.J.; Tan, W. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc. 2015, 137, 667–674. [Google Scholar] [CrossRef]
- Peng, R.; Zheng, X.; Lyu, Y.; Xu, L.; Zhang, X.; Ke, G.; Liu, Q.; You, C.; Huan, S.; Tan, W. Engineering a 3D DNA-logic gate nanomachine for bispecific recognition and computing on target cell surfaces. J. Am. Chem. Soc. 2018, 140, 9793–9796. [Google Scholar] [CrossRef]
- Hoshino, Y.; Koide, H.; Urakami, T.; Kanazawa, H.; Kodama, T.; Oku, N.; Shea, K.J. Recognition, neutralization, and clearance of target peptides in the bloodstream of living mice by molecularly imprinted polymer nanoparticles: A plastic antibody. J. Am. Chem. Soc. 2010, 132, 6644–6645. [Google Scholar] [CrossRef]
- Chianella, I.; Guerreiro, A.; Moczko, E.; Caygill, J.S.; Piletska, E.V.; De Vargas Sansalvador, I.M.; Whitcombe, M.J.; Piletsky, S.A. Direct replacement of antibodies with molecularly imprinted polymer nanoparticles in ELISA development of a novel assay for vancomycin. Anal. Chem. 2013, 85, 8462–8468. [Google Scholar] [CrossRef]
- Wang, S.; Yin, D.; Wang, W.; Shen, X.; Zhu, J.J.; Chen, H.Y.; Liu, Z. Targeting and imaging of cancer cells via monosaccharide-imprinted fluorescent nanoparticles. Sci. Rep. 2016, 6, 22757. [Google Scholar] [CrossRef] [PubMed]
- Mashinchian, O.; Bonakdar, S.; Taghinejad, H.; Satarifard, V.; Heidari, M.; Majidi, M.; Sharifi, S.; Peirovi, A.; Saffar, S.; Taghinejad, M.; et al. Cell-imprinted substrates act as an artificial niche for skin regeneration. ACS Appl. Mater. Interfaces 2014, 6, 13280–13292. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, K.; Gao, H.; Li, X.; Chen, Y.; Zhang, L.; Peng, X.; Zhang, Y. Artificial antibody with site-enhanced multivalent aptamers for specific capture of circulating tumor cells. Anal. Chem. 2019, 91, 2591–2594. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Chen, S.; Lu, Q. Cell-imprinted biomimetic interface for intelligent recognition and efficient capture of CTCs. Biomater. Sci. 2019, 7, 4027–4035. [Google Scholar] [CrossRef]
- Armutcu, C.; Bereli, N.; Bayram, E.; Uzun, L.; Say, R.; Denizli, A. Aspartic acid incorporated monolithic columns for affinity glycoprotein purification. Colloids Surf. B Biointerfaces 2014, 114, 67–74. [Google Scholar] [CrossRef]
- Jing, T.; Du, H.; Dai, Q.; Xia, H.; Niu, J.; Hao, Q.; Mei, S.; Zhou, Y. Magnetic molecularly imprinted nanoparticles for recognition of lysozyme. Biosens. Bioelectron. 2010, 26, 301–306. [Google Scholar] [CrossRef]
- Chen, L.; Liu, J.; Zeng, Q.; Wang, H.; Yu, A.; Zhang, H.; Ding, L. Preparation of magnetic molecularly imprinted polymer for the separation of tetracycline antibiotics from egg and tissue samples. J. Chromatogr. A 2009, 1216, 3710–3719. [Google Scholar] [CrossRef]
- Turan, E.; Şahin, F. Molecularly imprinted biocompatible magnetic nanoparticles for specific recognition of Ochratoxin A. Sens. Actuators B Chem. 2016, 227, 668–676. [Google Scholar] [CrossRef]
- Sun, B.; Ni, X.; Cao, Y.; Cao, G. Electrochemical sensor based on magnetic molecularly imprinted nanoparticles modified magnetic electrode for determination of Hb. Biosens. Bioelectron. 2017, 91, 354–358. [Google Scholar] [CrossRef]
- Ansell, R.J.; Mosbach, K. Magnetic molecularly imprinted polymer beads for drug radioligand binding assay. Analyst 1998, 123, 1611–1616. [Google Scholar] [CrossRef]
- Kuhn, J.; Aylaz, G.; Sari, E.; Marco, M.; Yiu, H.H.P.; Duman, M. Selective binding of antibiotics using magnetic molecular imprint polymer (MMIP) networks prepared from vinyl-functionalized magnetic nanoparticles. J. Hazard. Mater. 2020, 387, 121709. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Guo, Z.; Ding, Q.; Zhao, C.; Yu, H.; Zhu, X.; Fu, M.; Liu, Q. Magnetic-graphene oxide based molecular imprinted polymers for selective extraction of glycoprotein at physiological pH. Polymer 2021, 215, 123384. [Google Scholar] [CrossRef]
- An, K.; Kang, H.; Tian, D. Fabrication and evaluation of controllable core/shell magnetic molecular imprinted polymers based on konjac glucomannan for trichlorfon. J. Appl. Polym. Sci. 2020, 137, 48910. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Du, X.; Liu, M. Application of electrochemical biosensors in tumor cell detection. Thorac. Cancer 2020, 11, 840–850. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.; Yan, C.; Chen, W. A polyamidoamine dendrimer-based electrochemical immunosensor for label-free determination of epithelial cell adhesion molecule-expressing cancer cells. Sensors 2019, 19, 1879. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef]
- Gao, T.; Li, L.; Wang, B.; Zhi, J.; Xiang, Y.; Li, G. Dynamic electrochemical control of cell capture-and-release based on redox-controlled host–guest interactions. Anal. Chem. 2016, 88, 9996–10001. [Google Scholar] [CrossRef]
- Shen, H.; Yang, J.; Chen, Z.; Chen, X.; Wang, L.; Hu, J.; Ji, F.; Xie, G.; Feng, W. A novel label-free and reusable electrochemical cytosensor for highly sensitive detection and specific collection of CTCs. Biosens. Bioelectron. 2016, 81, 495–502. [Google Scholar] [CrossRef]
- Kordasht, H.K.; Hasanzadeh, M. Aptamer based recognition of cancer cells: Recent progress and challenges in bioanalysis. Talanta 2020, 220, 121436. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Y.; Xu, X.; Liu, Y.; Lin, B.; Zhang, M.; Zhang, J.; Wan, S.; Yang, C.; Tan, W. Aptamer-based detection of circulating targets for precision medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef]
- Ou, D.; Sun, D.; Liang, Z.; Chen, B.; Lin, X.; Chen, Z. A novel cytosensor for capture, detection and release of breast cancer cells based on metal organic framework PCN-224 and DNA tetrahedron linked dual-aptamer. Sens. Actuators B Chem. 2019, 285, 398–404. [Google Scholar] [CrossRef]
- Luo, J.; Liang, D.; Zhao, D.; Yang, M. Photoelectrochemical detection of circulating tumor cells based on aptamer conjugated Cu2O as signal probe. Biosens. Bioelectron. 2020, 151, 111976. [Google Scholar] [CrossRef] [PubMed]
- Hassan, E.M.; DeRosa, M.C. Recent advances in cancer early detection and diagnosis: Role of nucleic acid based aptasensors. TrAC Trends Anal. Chem. 2020, 124, 115806. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, J.; Xu, J.J.; Zhang, S.S.; Chen, H.Y. Optical nano-biosensing interface via nucleic acid amplification strategy: Construction and application. Chem. Soc. Rev. 2018, 47, 1996–2019. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pan, G.; Zhang, Y.; Guo, X.; Zhang, H. Narrowly dispersed hydrophilic molecularly imprinted polymer nanoparticles for efficient molecular recognition in real aqueous samples including river water, milk, and bovine serum. Angew. Chem. 2013, 125, 1551–1554. [Google Scholar] [CrossRef]
- Ye, L. Molecularly imprinted polymers with multi-functionality. Anal. Bioanal. Chem. 2016, 408, 1727–1733. [Google Scholar] [CrossRef]
- Guan, G.; Liu, B.; Wang, Z.; Zhang, Z. Imprinting of molecular recognition sites on nanostructures and its applications in chemosensors. Sensors 2008, 8, 8291–8320. [Google Scholar] [CrossRef] [PubMed]
- Eersels, K.; van Grinsven, B.; Ethirajan, A.; Timmermans, S.; Jiménez Monroy, K.L.; Bogie, J.F.; Punniyakoti, S.; Vandenryt, T.; Hendriks, J.J.; Cleij, T.J.; et al. Selective identification of macrophages and cancer cells based on thermal transport through surface-imprinted polymer layers. ACS Appl. Mater. Interfaces 2013, 5, 7258–7267. [Google Scholar] [CrossRef]
- Lv, S.W.; Liu, Y.; Xie, M.; Wang, J.; Yan, X.W.; Li, Z.; Dong, W.G.; Huang, W.H. Near-infrared light-responsive hydrogel for specific recognition and photothermal site-release of circulating tumor cells. ACS Nano 2016, 10, 6201–6210. [Google Scholar] [CrossRef]
- Welch, E.C.; Powell, J.M.; Clevinger, T.B.; Fairman, A.E.; Shukla, A. Advances in biosensors and diagnostic technologies using nanostructures and nanomaterials. Adv. Funct. Mater. 2021, 31, 2104126. [Google Scholar] [CrossRef]
- Chen, J.F.; Zhu, Y.; Lu, Y.T.; Hodara, E.; Hou, S.; Agopian, V.G.; Tomlinson, J.S.; Posadas, E.M.; Tseng, H.R. Clinical applications of NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Theranostics 2016, 6, 1425. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xue, P.; Kang, Y.; Hui, K.M. Highly specific and ultrasensitive graphene-enhanced electrochemical detection of low-abundance tumor cells using silica nanoparticles coated with antibody-conjugated quantum dots. Anal. Chem. 2013, 85, 3166–3173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chu, M.; Wang, X.; Niu, Q.; Ma, G.; Fang, Y.; Liu, J.; Lv, W.; Zhao, W. Electrochemical cytosensor for detection of cell surface sialic acids based on 3D biointerface. Electrochim. Acta 2018, 282, 923–930. [Google Scholar] [CrossRef]
- Zhang, J.; Misra, R.D.K. Nanomaterials in microfluidics for disease diagnosis and therapy development. Mater. Technol. 2019, 34, 92–116. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, Y.; Zhao, Y.; Zhang, L.; Zhang, L.; Mao, H.; Huang, C. Nanotechnology-assisted isolation and analysis of circulating tumor cells on microfluidic devices. Micromachines 2020, 11, 774. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, Y.; Wang, F.; Ding, J.; Meng, S.; Li, C.; Tang, D.; Yin, X. Functional and biocompatible polymeric ionic liquid (PIL)-decorated immunomagnetic nanospheres for the efficient capture of rare number CTCs. Anal. Chim. Acta 2018, 1044, 162–173. [Google Scholar] [CrossRef]
- Lin, Z.; Lin, S.Y.; Xie, P.; Lin, C.Y.; Rather, G.M.; Bertino, J.R.; Javanmard, M. Rapid Assessment of Surface Markers on cancer cells Using immuno-Magnetic Separation and Multi-frequency impedance cytometry for targeted therapy. Sci. Rep. 2020, 10, 3015. [Google Scholar] [CrossRef]
- Lee, T.Y.; Hyun, K.A.; Kim, S.I.; Jung, H.I. An integrated microfluidic chip for one-step isolation of circulating tumor cells. Sens. Actuators B Chem. 2017, 238, 1144–1150. [Google Scholar] [CrossRef]
- Shi, W.; Wang, S.; Maarouf, A.; Uhl, C.G.; He, R.; Yunus, D.; Liu, Y. Magnetic particles assisted capture and release of rare circulating tumor cells using wavy-herringbone structured microfluidic devices. Lab Chip 2017, 17, 3291–3299. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, L.; Huang, M.; Song, J.; Zhang, H.; Song, Y.; Wang, W.; Yang, C. Aptamer-based microfluidics for isolation, release and analysis of circulating tumor cells. TrAC Trends Anal. Chem. 2019, 117, 69–77. [Google Scholar] [CrossRef]
- Park, M.H.; Reátegui, E.; Li, W.; Tessier, S.N.; Wong, K.H.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L.; et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J. Am. Chem. Soc. 2017, 139, 2741–2749. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem. Int. Ed. 2019, 58, 2236–2240. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.; Zandberg, W.; Hsiao, D.; Erno, Z.; Sen, D.; Gates, B.D.; Branda, N.R. Photothermal release of single-stranded DNA from the surface of gold nanoparticles through controlled denaturating and Au- S bond breaking. ACS Nano 2010, 4, 6395–6403. [Google Scholar] [CrossRef]
- Yerramilli, D.; Johnsen, S. Spatial vision in the purple sea urchin Strongylocentrotus purpuratus (Echinoidea). J. Exp. Biol. 2010, 213, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, X.; Men, X.; Wu, C.X.; Bai, J.J.; Li, W.T.; Yang, T.; Chen, M.L.; Wang, J.H. Dual-multivalent-aptamer-conjugated nanoprobes for superefficient discerning of single circulating tumor cells in a microfluidic chip with inductively coupled plasma mass spectrometry detection. ACS Appl. Mater. Interfaces 2021, 13, 43668–43675. [Google Scholar] [CrossRef]
- Yu, C.C.; Chen, Y.W.; Yeh, P.Y.; Hsiao, Y.S.; Lin, W.T.; Kuo, C.W.; Chueh, D.Y.; You, Y.W.; Shyue, J.J.; Chang, Y.C.; et al. Random and aligned electrospun PLGA nanofibers embedded in microfluidic chips for cancer cell isolation and integration with air foam technology for cell release. J. Nanobiotechnol. 2019, 17, 31. [Google Scholar] [CrossRef]
- Kim, T.H.; Wang, Y.; Oliver, C.R.; Thamm, D.H.; Cooling, L.; Paoletti, C.; Smith, K.J.; Nagrath, S.; Hayes, D.F. A temporary indwelling intravascular aphaeretic system for in vivo enrichment of circulating tumor cells. Nat. Commun. 2019, 10, 1478. [Google Scholar] [CrossRef]
- Ates, H.C.; Nguyen, P.Q.; Gonzalez-Macia, L.; Morales-Narváez, E.; Güder, F.; Collins, J.J.; Dincer, C. End-to-end design of wearable sensors. Nat. Rev. Mater. 2022, 7, 887–907. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, W. Wearable and flexible electronics for continuous molecular monitoring. Chem. Soc. Rev. 2019, 48, 1465–1491. [Google Scholar] [CrossRef]
- Iqbal, S.M.; Mahgoub, I.; Du, E.; Leavitt, M.A.; Asghar, W. Advances in healthcare wearable devices. NPJ Flex. Electron. 2021, 5, 9. [Google Scholar] [CrossRef]
- Jameson, J.L.; Longo, D.L. Precision medicine—Personalized, problematic, and promising. Obstet. Gynecol. Surv. 2015, 70, 612–614. [Google Scholar] [CrossRef]
- Pan, G.; Guo, Q.; Ma, Y.; Yang, H.; Li, B. Thermo-responsive hydrogel layers imprinted with RGDS peptide: A system for harvesting cell sheets. Angew. Chem. Int. Ed. 2013, 52, 6907–6911. [Google Scholar] [CrossRef]
- Brun-Graeppi, A.K.A.S.; Richard, C.; Bessodes, M.; Scherman, D.; Merten, O.W. Thermoresponsive surfaces for cell culture and enzyme-free cell detachment. Prog. Polym. Sci. 2010, 35, 1311–1324. [Google Scholar] [CrossRef]
- Ma, Y.; Yin, Y.; Ni, L.; Miao, H.; Wang, Y.; Pan, C.; Tian, X.; Pan, J.; You, T.; Li, B.; et al. Thermo-responsive imprinted hydrogel with switchable sialic acid recognition for selective cancer cell isolation from blood. Bioact. Mater. 2020, 6, 1308–1317. [Google Scholar] [CrossRef]
- Pan, G.; Shinde, S.; Yeung, S.Y.; Jakštaitė, M.; Li, Q.; Wingren, A.G.; Sellergren, B. An epitope-imprinted biointerface with dynamic bioactivity for modulating cell–biomaterial interactions. Angew. Chem. Int. Ed. 2017, 56, 15959–15963. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, D.; Xu, C.; Ge, Y.; Liu, X.; Wei, Q.; Huang, L.; Ren, X.; Wang, C.; Wang, Y. Wearable electrochemical biosensor based on molecularly imprinted Ag nanowires for noninvasive monitoring lactate in human sweat. Sens. Actuators B Chem. 2020, 320, 128325. [Google Scholar] [CrossRef]
- Yang, X.; Mu, D.; Peng, H.; Li, H.; Wang, Y.; Wang, P.; Wang, Y.; Han, S. Research and application of artificial intelligence based on electronic health records of patients with cancer: Systematic review. JMIR Med. Inform. 2022, 10, e33799. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. New Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Roshan, A.; Byrne, M.F. Artificial intelligence in colorectal cancer screening. CMAJ 2022, 194, E1481–E1484. [Google Scholar] [CrossRef]
- Suzuki, H.; Yoshitaka, T.; Yoshio, T.; Tada, T. Artificial intelligence for cancer detection of the upper gastrointestinal tract. Dig. Endosc. 2021, 33, 254–262. [Google Scholar] [CrossRef]
- Guo, X.; Lin, F.; Yi, C.; Song, J.; Sun, D.; Lin, L.; Zhong, Z.; Wu, Z.; Wang, X.; Zhang, Y.; et al. Deep transfer learning enables lesion tracing of circulating tumor cells. Nat. Commun. 2022, 13, 7687. [Google Scholar] [CrossRef]
- de Wit, S.; Zeune, L.L.; Hiltermann, T.J.N.; Groen, H.J.M.; Dalum, G.V.; Terstappen, L.W.M.M. Classification of cells in CTC-enriched samples by advanced image analysis. Cancers 2018, 10, 377. [Google Scholar] [CrossRef]
- Zeune, L.L.; Boink, Y.E.; van Dalum, G.; Nanou, A.; de Wit, S.; Andree, K.C.; Swennenhuis, J.F.; van Gils, S.A.; Terstappen, L.W.M.M.; Brune, C. Deep learning of circulating tumour cells. Nat. Mach. Intell. 2020, 2, 124–133. [Google Scholar] [CrossRef]
- Hashemzadeh, H.; Shojaeilangari, S.; Allahverdi, A.; Rothbauer, M.; Ertl, P.; Naderi-Manesh, H. A combined microfluidic deep learning approach for lung cancer cell high throughput screening toward automatic cancer screening applications. Sci. Rep. 2021, 11, 9804. [Google Scholar] [CrossRef]
- Ma, J.; Yang, J.; Jin, Y.; Cheng, S.; Huang, S.; Zhang, N.; Wang, Y. Artificial intelligence based on blood biomarkers including CTCs predicts outcomes in epithelial ovarian cancer: A prospective study. OncoTargets Ther. 2021, 14, 3267. [Google Scholar] [CrossRef]
- Chen, M.; Xie, S. Therapeutic targeting of cellular stress responses in cancer. Thorac. Cancer 2018, 9, 1575–1582. [Google Scholar] [CrossRef]
- Arabi, M.; Ostovan, A.; Wang, Y.; Mei, R.; Fu, L.; Li, J.; Wang, X.; Chen, L. Chiral molecular imprinting-based SERS detection strategy for absolute enantiomeric discrimination. Nat. Commun. 2022, 13, 5757. [Google Scholar] [CrossRef]
- Rebelo, P.; Costa-Rama, E.; Seguro, I.; Pacheco, J.G.; Nouws, H.P.A.; Cordeiro, M.N.D.S.; Delerue-Matos, C. Molecularly imprinted polymer-based electrochemical sensors for environmental analysis. Biosens. Bioelectron. 2021, 172, 112719. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).