Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers
Abstract
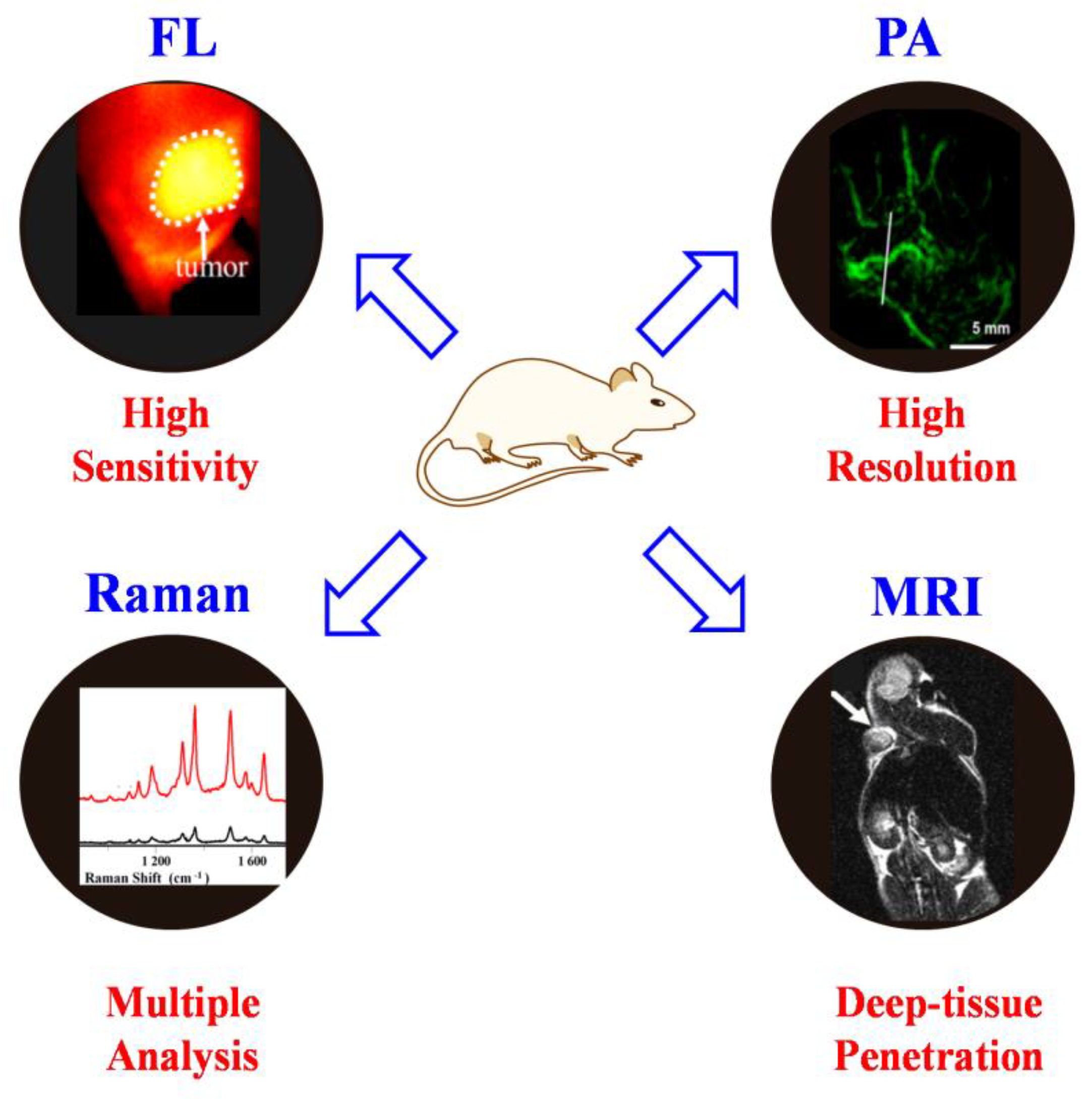
1. Introduction
2. The Design Principle of SERS Nanotags
3. Morphology-Tuned Plasmonic Nanostructures in SERS Nanotags for Multi-Biomarker Detection
3.1. Isotropic Nanostructures
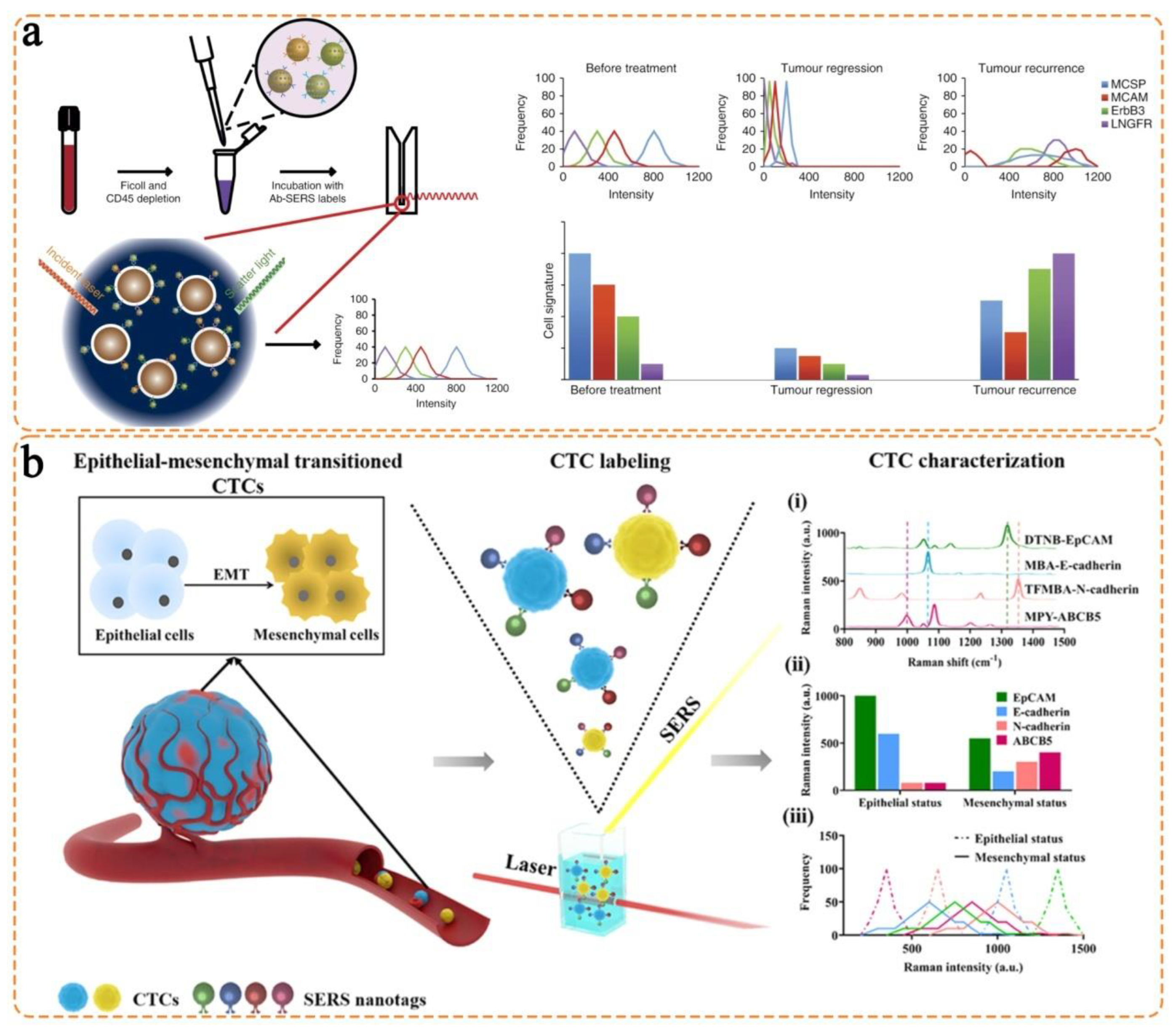
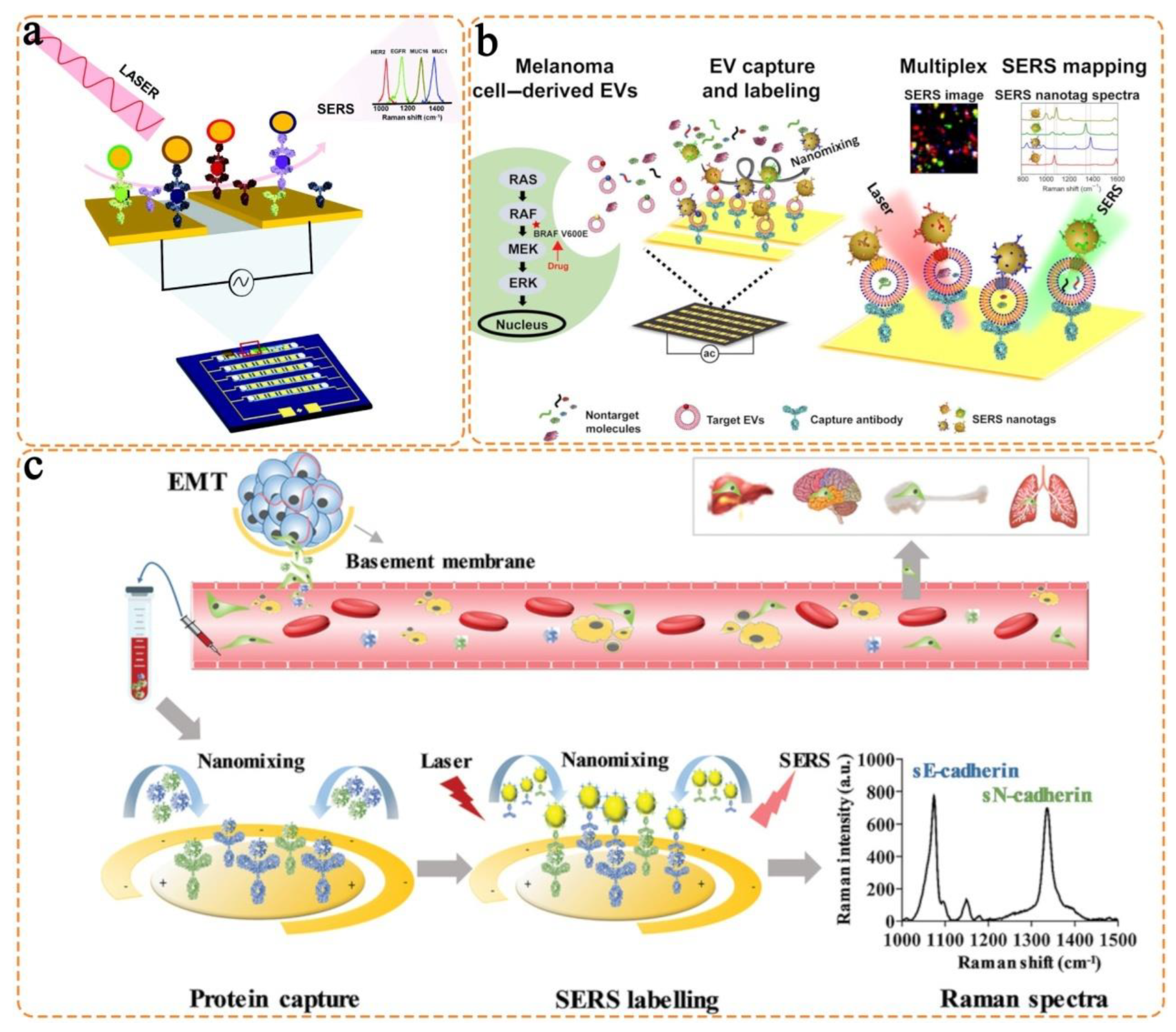
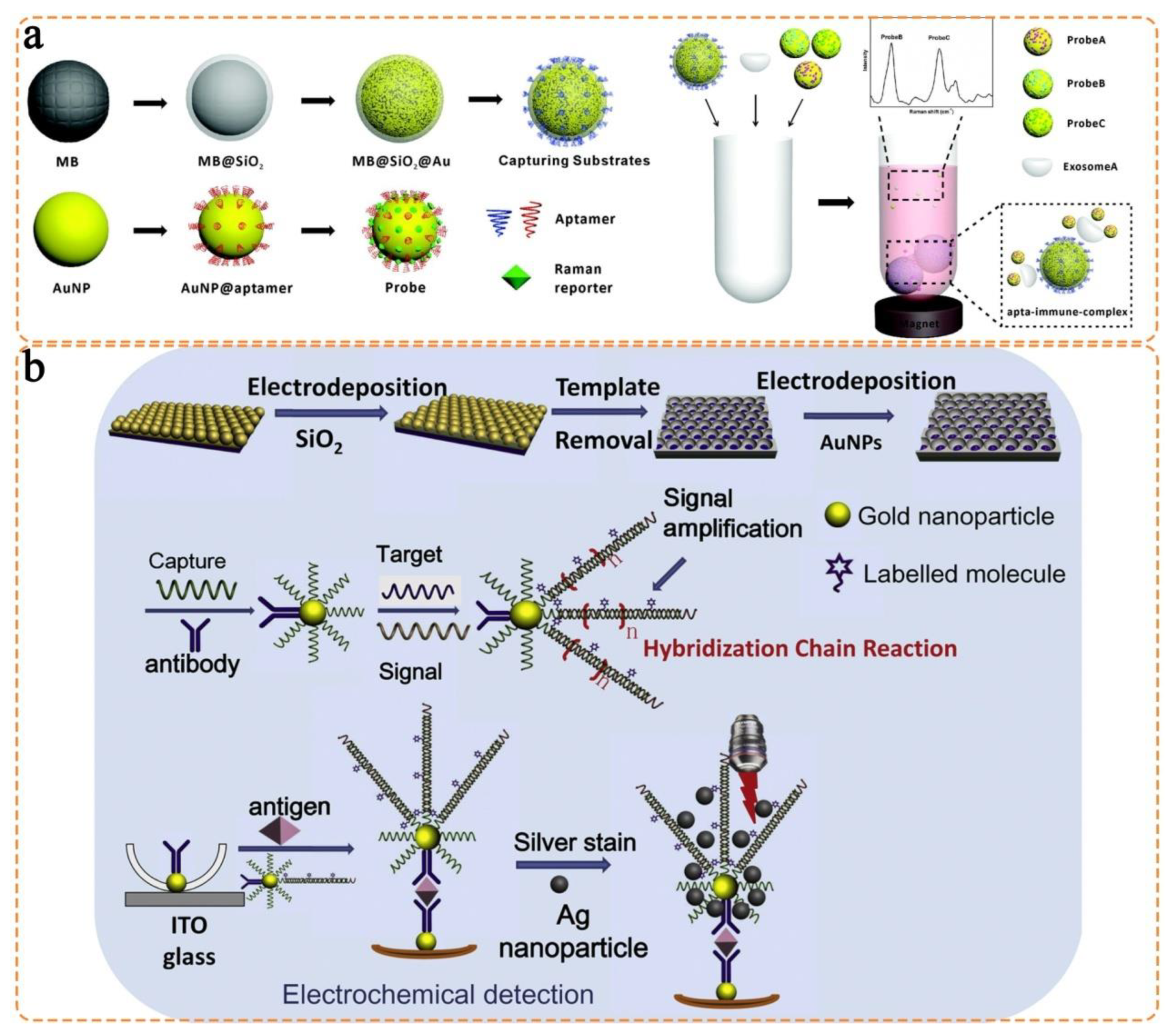
3.2. Anisotropic Nanostructures
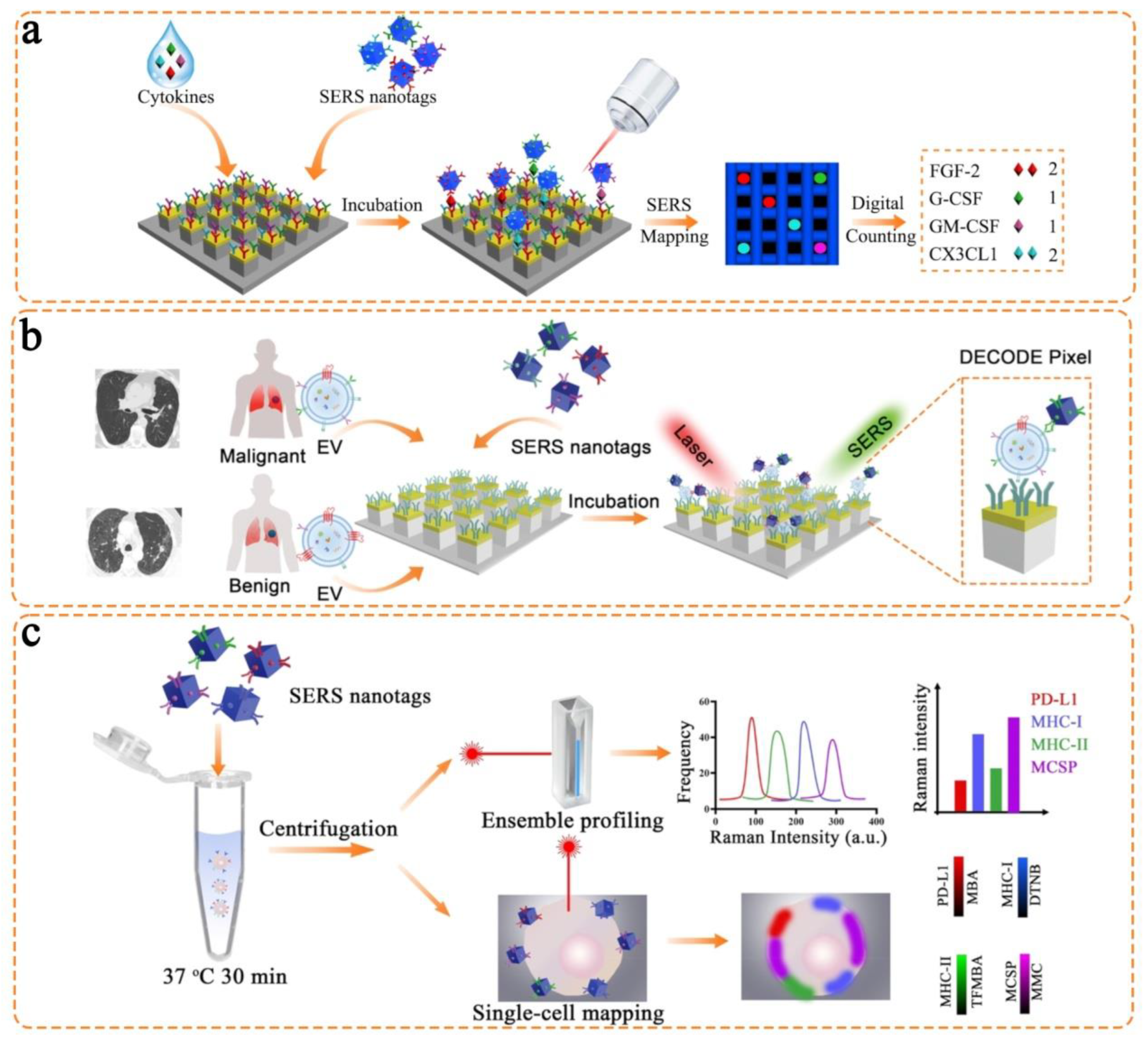
4. Triple Bond-Modulated Raman Reporters in SERS Nanotags for Multi-Biomarker Detection
5. Challenges and Outlooks
5.1. Improve the Nanostructure Reproducibility and Stability
5.2. Explore New Strategies to Tune the Raman Shifts of Triple Bond-Modulated Raman Reporters
5.3. Promote Broad Collaborations
5.4. Realizing SERS with Multifunctional Abilities for Multimodal Imaging and Theranostics
5.5. Integration of SERS with Other Imaging or Therapy Modalities toward Precision Medicine
5.6. Test the Assay Performance on a Large Cohort of Clinical Samples
5.7. Pushing SERS Nanoprobesfrom In Vitro to In Vivo Applications
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SERS | surface-enhanced Raman scattering | CTCs | circulating tumor cells |
| NAs | nucleic acids | EVs | extracellular vesicles |
| PSA | prostate-specific antigen | BSA | bovine serum albumin |
| LSPR | localized surface plasma resonance | MCSP | melanoma–chondroitin sulfate proteoglycan |
| MCAM | melanoma cell adhesion molecule | EpCAM | epithelial cell adhesion molecule |
| EMT | mesenchymal transition | EHD | electrohydrodynamic |
| CEA | carcinoembryonic antigen | HER2 | human epidermal growth factor receptor 2 |
| AFP | alpha-fetoprotein | GMA | microelectrode array |
| HCR | hybridization chain reaction | CA | cancer antigen |
| SEM | scanning electron microscope | TEM | transmission electron microscope |
| NSE | neuron-specific enolase | IGF | insulin-like growth factor |
| THBS2 | thrombospondin 2 | VCAN | versican |
| TNC | tenascin C | PD-L1 | programmed cell death-ligand 1 |
| MHC | major histocompatibility | MARS | Manhattan Raman scattering |
| SRS | stimulated Raman scattering | MBA | 4-mercaptobenzoic acid |
| TFMBA | 2,3,5,6-tetrafluoro-4-mercaptobenzoic acid | MNBA | 4-mercapto-3-nitro benzoic acid |
| MPY | 4-mercaptopyridine | DTNB | 5,5-dithiobis(2-nitrobenzoic acid) |
| PATP | p-aminothiophenol | PNTP | p-nitrothiophenol |
| 4MSTP | 4-(methylsulfanyl)thiophenol | FGF-2 | fibroblast growth factor 2 |
| G-CSF | granulocyte colony-stimulating factor | AS1411 | oligonucleotide aptamer |
| cRGD | cyclic arginine–glycine–aspartic acid | FL | fluorescence imaging |
| PA | photoacoustic imaging |
References
- World Health Organization (WHO). Cancer. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 3 February 2022).
- Borrebaeck, C.A.K. Precision siagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.M.; Wee, E.J.H.; Mainwaring, P.N.; Wang, Y.; Trau, M. Toward precision medicine: A cancer molecular subtyping nano-strategy for RNA biomarkers in tumor and urine. Small 2016, 12, 6233–6242. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, C.; Ma, X.; Tuo, W.; Tu, L.; Li, X.; Sun, Y.; Stang, P.J.; Sun, Y. Long wavelength-emissive Ru(II) metallacycle-based photosensitizer assisting in vivo bacterial diagnosis and antibacterial treatment. Proc. Natl. Acad. Sci. 2022, 119, e2209904119. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Zhong, M.; Ye, F.; Zhang, X. Low-frequency HIFU induced cancer immunotherapy: Tempting challenges and potential opportunities. Cancer Bio. Med. 2019, 16, 714. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472. [Google Scholar] [CrossRef]
- Li, J.; Wuethrich, A.; Dey, S.; Lane, R.E.; Sina, A.A.I.; Wang, J.; Wang, Y.; Puttick, S.; Koo, K.M.; Trau, M. The growing impact of micro/nanomaterial-based systems in precision oncology: Translating “multiomics” technologies. Adv. Funct. Mater. 2020, 30, 1909306. [Google Scholar] [CrossRef]
- Wang, T.; Li, J.; Yu, G.; Yu, K. Effects of siRNA interference and over-expression of HMGA2 on proliferation and apoptosis of colorectal cancer cells. Int. J. Clin. Exp. Pathol. 2017, 10, 4611. [Google Scholar]
- Li, W.; Wang, H.; Zhao, Z.; Gao, H.; Liu, C.; Zhu, L.; Wang, C.; Yang, Y. Emerging nanotechnologies for liquid biopsy: The detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 2018, 31, 1805344. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zhang, Z.; Zheng, F.; Wang, L.; Guo, J.; Zhang, T.; Dai, X.; Zhang, S.; Yang, D.; Kuang, R.; et al. Combining protein and miRNA quantification for bladder cancer analysis. ACS Appl. Mater. Interfaces 2017, 9, 23420–23427. [Google Scholar] [CrossRef]
- Liu, C.; Xu, X.; Li, B.; Situ, B.; Pan, W.; Hu, Y.; An, T.; Yao, S.; Zheng, L. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018, 18, 4226–4232. [Google Scholar] [CrossRef]
- Ning, Z.; Gan, J.; Chen, C.; Zhang, D.; Zhang, H. Molecular functions and significance of the MTA family in hormone-independent cancer. Cancer Metast. Rev. 2014, 33, 901. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Chandran, B.K.; Lim, C.T.; Chen, X. Rational design of materials interface for efficient capture of circulating tumor cells. Adv. Sci. 2015, 2, 1500118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Wang, Y.; Trau, M. Simple and rapid colorimetric detection of melanoma circulating tumor cells using bifunctional magnetic nanoparticles. Analyst 2017, 142, 4788–4793. [Google Scholar] [CrossRef]
- Bostwick, D.G. Prostate-specific antigen. Current role in diagnostic pathology of prostate cancer. Am. J. Clin. Pathol. 1994, 102, S31–S37. [Google Scholar] [PubMed]
- Ankerst, D.P.; Thompson, I.M. Sensitivity and specificity of prostate-specific antigen for prostate cancer detection with high rates of biopsy verification. Arch. Ital. Urol. Androl. 2006, 78, 125–129. [Google Scholar]
- Ferrari, M. Cancer nanotechnology: Opportunities and challenges. Nat. Rev. Cancer 2005, 5, 161–171. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and future of surface-enhanced Raman scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, B.; Chen, L. SERS tags: Novel optical nanoprobes for bioanalysis. Chem. Rev. 2013, 113, 1391–1428. [Google Scholar]
- Mir-Simon, B.; Reche-Perez, I.; Guerrini, L.; Pazos-Perez, N.; Alvarez-Puebla, R.A. Universal one-pot and scalable synthesis of SERS encoded nanoparticles. Chem. Mater. 2015, 27, 950–958. [Google Scholar] [CrossRef]
- Tan, T.; Tian, C.; Ren, Z.; Yang, J.; Chen, Y.; Sun, L.; Li, Z.; Wu, A.; Yin, J.; Fu, H. LSPR-dependent SERS performance of silver nanoplates with highly stable and broad tunable LSPR prepared through an improved seed-mediated strategy. Phys. Chem. Chem. Phys. 2013, 15, 21034–21042. [Google Scholar] [CrossRef]
- Im, H.; Bantz, K.C.; Lee, S.H.; Johnson, T.W.; Haynes, C.L.; Oh, S.-H. Self-assembled plasmonic nanoring cavity arrays for SERS and LSPR biosensing. Adv. Mater. 2013, 25, 2678–2685. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koo, K.M.; Wang, Y.; Trau, M. Native microRNA targets trigger self-assembly of nanozyme-patterned hollowed nanocuboids with optimal interparticle gaps for plasmonic-activated cancer detection. Small 2019, 15, 1904689. [Google Scholar] [CrossRef] [PubMed]
- Jeon, T.Y.; Park, S.-G.; Lee, S.Y.; Jeon, H.C.; Yang, S.-M. Shape control of Ag nanostructures for practical sers substrates. ACS Appl. Mater. Interfaces 2013, 5, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Chikkaraddy, R.; Salmon, A.; Ohadi, H.; de Nijs, B.; Mertens, J.; Carnegie, C.; Bowman, R.W.; Baumberg, J.J. SERS of individual nanoparticles on a mirror: Size does matter, but so does shape. J. Phys. Chem. Lett. 2016, 7, 2264–2269. [Google Scholar] [CrossRef]
- Shen, X.S.; Wang, G.Z.; Hong, X.; Zhu, W. Nanospheres of silver nanoparticles: Agglomeration, surface morphology control and application as SERS substrates. Phys. Chem. Chem. Phys. 2009, 11, 7450–7454. [Google Scholar] [CrossRef]
- Wei, L.; Chen, Z.; Shi, L.; Long, R.; Anzalone, A.V.; Zhang, L.; Hu, F.; Yuste, R.; Cornish, V.W.; Min, W. Super-multiplex vibrational imaging. Nature 2017, 544, 465–470. [Google Scholar] [CrossRef]
- Zheng, X.-S.; Hu, P.; Cui, Y.; Zong, C.; Feng, J.-M.; Wang, X.; Ren, B. Bsa-coated nanoparticles for improved SERS-based intracellular ph sensing. Anal. Chem. 2014, 86, 12250–12257. [Google Scholar] [CrossRef]
- Indrasekara, A.S.D.S.; Paladini, B.J.; Naczynski, D.J.; Starovoytov, V.; Moghe, P.V.; Fabris, L. Dimeric gold nanoparticle assemblies as tags for SERS-based cancer detection. Adv. Healthc. Mater. 2013, 2, 1370–1376. [Google Scholar] [CrossRef]
- Fales, A.M.; Yuan, H.; Vo-Dinh, T. Silica-coated gold nanostars for combined surface-enhanced Raman scattering (SERS) detection and singlet-oxygen generation: A potential nanoplatform for theranostics. Langmuir 2011, 27, 12186–12190. [Google Scholar] [CrossRef]
- Farahavar, G.; Abolmaali, S.S.; Nejatollahi, F.; Safaie, A.; Javanmardi, S.; Khajeh Zadeh, H.; Yousefi, R.; Nadgaran, H.; Mohammadi-Samani, S.; Tamaddon, A.M.; et al. Single-chain antibody-decorated Au nanocages@liposomal layer nanoprobes for targeted SERS imaging and remote-controlled photothermal therapy of melanoma cancer cells. Mater. Sci. Eng. C 2021, 124, 112086. [Google Scholar] [CrossRef]
- Kumar, A.R.; Shanmugasundaram, K.B.; Li, J.; Zhang, Z.; Ibn Sina, A.A.; Wuethrich, A.; Trau, M. Ultrasensitive melanoma biomarker detection using a microchip sers immunoassay with anisotropic au–ag alloy nanoboxes. RSC Adv. 2020, 10, 28778–28785. [Google Scholar] [CrossRef] [PubMed]
- Farokhinejad, F.; Li, J.; Hugo, L.E.; Howard, C.B.; Wuethrich, A.; Trau, M. Detection of dengue virus 2 with single infected mosquito resolution using yeast affinity bionanofragments and plasmonic sers nanoboxes. Anal. Chem. 2022, 94, 14177–14184. [Google Scholar] [CrossRef] [PubMed]
- Berciaud, S.; Cognet, L.; Tamarat, P.; Lounis, B. Observation of intrinsic size effects in the optical response of individual gold nanoparticles. Nano Lett. 2005, 5, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Walkenfort, B.; Yoon, J.H.; Schlucker, S.; Xie, W. Gold and silver nanoparticle monomers are non-sers-active: A negative experimental study with silica-encapsulated raman-reporter-coated metal colloids. Phys. Chem. Chem. Phys. 2015, 17, 21120–21126. [Google Scholar] [CrossRef]
- Kleinman, S.L.; Frontiera, R.R.; Henry, A.-I.; Dieringer, J.A.; Van Duyne, R.P. Creating, characterizing, and controlling chemistry with sers hot spots. Phys. Chem. Chem. Phys. 2013, 15, 21–36. [Google Scholar] [CrossRef]
- Willets, K.A. Super-resolution imaging of sers hot spots. Chem. Soc. Rev. 2014, 43, 3854–3864. [Google Scholar] [CrossRef]
- Pearce, A.K.; Wilks, T.R.; Arno, M.C.; O’Reilly, R.K. Synthesis and applications of anisotropic nanoparticles with precisely defined dimensions. Nat. Rev. Chem. 2021, 5, 21–45. [Google Scholar] [CrossRef]
- Sajanlal, P.R.; Sreeprasad, T.S.; Samal, A.K.; Pradeep, T. Anisotropic nanomaterials: Structure, growth, assembly, and functions. Nano Rev. 2011, 2, 5883. [Google Scholar] [CrossRef]
- Mulvihill, M.J.; Ling, X.Y.; Henzie, J.; Yang, P. Anisotropic etching of silver nanoparticles for plasmonic structures capable of single-particlesers. J. Am. Chem. Soc. 2010, 132, 268–274. [Google Scholar] [CrossRef]
- Ye, S.; Benz, F.; Wheeler, M.C.; Oram, J.; Baumberg, J.J.; Cespedes, O.; Christenson, H.K.; Coletta, P.L.; Jeuken, L.J.; Markham, A.F. One-step fabrication of hollow-channel gold nanoflowers with excellent catalytic performance and large single-particle sers activity. Nanoscale 2016, 8, 14932–14942. [Google Scholar] [CrossRef]
- Tran, V.; Thiel, C.; Svejda, J.T.; Jalali, M.; Walkenfort, B.; Erni, D.; Schlücker, S. Probing the sers brightness of individual au nanoparticles, hollow au/ag nanoshells, au nanostars and au core/au satellite particles: Single-particle experiments and computer simulations. Nanoscale 2018, 10, 21721–21731. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Kang, H.; Yang, J.-K.; Jo, A.; Lee, H.-Y.; Lee, Y.-S.; Jeong, D.H. Ag shell–au satellite hetero-nanostructure for ultra-sensitive, reproducible, and homogeneous nirsers activity. ACS Appl. Mater. Interfaces 2014, 6, 11859–11863. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.C.-H.; Wang, J.; Wang, Y.; Behren, A.; Cebon, J.; Trau, M. Characterising the phenotypic evolution of circulating tumour cells during treatment. Nat. Commun. 2018, 9, 1482. [Google Scholar] [CrossRef]
- Zhang, Z.; Wuethrich, A.; Wang, J.; Korbie, D.; Lin, L.L.; Trau, M. Dynamic monitoring of emt in ctcs as an indicator of cancer metastasis. Anal. Chem. 2021, 93, 16787–16795. [Google Scholar] [CrossRef] [PubMed]
- Kamil Reza, K.; Wang, J.; Vaidyanathan, R.; Dey, S.; Wang, Y.; Trau, M. Electrohydrodynamic-induced SERS immunoassay for extensive multiplexed biomarker sensing. Small 2017, 13, 1602902. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Shanmugasundaram, K.B.; Yeo, B.; Möller, A.; Wuethrich, A.; Lin, L.L.; Trau, M. Tracking drug-induced epithelial–mesenchymal transition in breast cancer by a microfluidic surface-enhanced Raman spectroscopy immunoassay. Small 2020, 16, 1905614. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wuethrich, A.; Sina, A.A.I.; Lane, R.E.; Lin, L.L.; Wang, Y.; Cebon, J.; Behren, A.; Trau, M. Tracking extracellular vesicle phenotypic changes enables treatment monitoring in melanoma. Sci. Adv. 2020, 6, eaax3223. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef]
- Gu, X.; Wang, K.; Qiu, J.; Wang, Y.; Tian, S.; He, Z.; Zong, R.; Kraatz, H.-B. Enhanced electrochemical and SERS signals by self-assembled gold microelectrode arrays: A dual readout platform for multiplex immumoassay of tumor biomarkers. Sens. Actuators B Chem. 2021, 334, 129674. [Google Scholar] [CrossRef]
- Li, M.; Kang, J.W.; Sukumar, S.; Dasari, R.R.; Barman, I. Multiplexed detection of serological cancer markers with plasmon-enhanced Raman spectro-immunoassay. Chem. Sci. 2015, 6, 3906–3914. [Google Scholar] [CrossRef]
- Song, C.; Yang, Y.; Yang, B.; Min, L.; Wang, L. Combination assay of lung cancer associated serum markers using surface-enhanced Raman spectroscopy. J. Mater. Chem. B 2016, 4, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Nima, Z.A.; Mahmood, M.; Xu, Y.; Mustafa, T.; Watanabe, F.; Nedosekin, D.A.; Juratli, M.A.; Fahmi, T.; Galanzha, E.I.; Nolan, J.P.; et al. Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci. Rep. 2014, 4, 4752. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, G.; Wang, J.; Maksymov, I.S.; Greentree, A.D.; Hu, J.; Shen, A.; Wang, Y.; Trau, M. Facile one-pot synthesis of nanodot-decorated gold–silver alloy nanoboxes for single-particle surface-enhanced Raman scattering activity. ACS Appl. Mater. Interfaces 2018, 10, 32526–32535. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wuethrich, A.; Sina, A.A.I.; Cheng, H.-H.; Wang, Y.; Behren, A.; Mainwaring, P.N.; Trau, M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat. Commun. 2021, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sina, A.A.I.; Antaw, F.; Fielding, D.; Möller, A.; Lobb, R.; Wuethrich, A.; Trau, M. Digital decoding of single extracellular vesicle phenotype differentiates early malignant and benign lung lesions. Adv. Sci. 2022, 10, 2204207. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wuethrich, A.; Zhang, Z.; Wang, J.; Lin, L.L.; Behren, A.; Wang, Y.; Trau, M. SERS multiplex profiling of melanoma circulating tumor cells for predicting the response to immune checkpoint blockade therapy. Anal. Chem. 2022, 94, 14573–14582. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Grewal, Y.S.; Howard, C.B.; Raftery, L.J.; Mahler, S.; Wang, Y.; Trau, M. Multiplexed SERS detection of soluble cancer protein biomarkers with gold–silver alloy nanoboxes and nanoyeast single-chain variable fragments. Anal. Chem. 2018, 90, 10377–10384. [Google Scholar] [CrossRef]
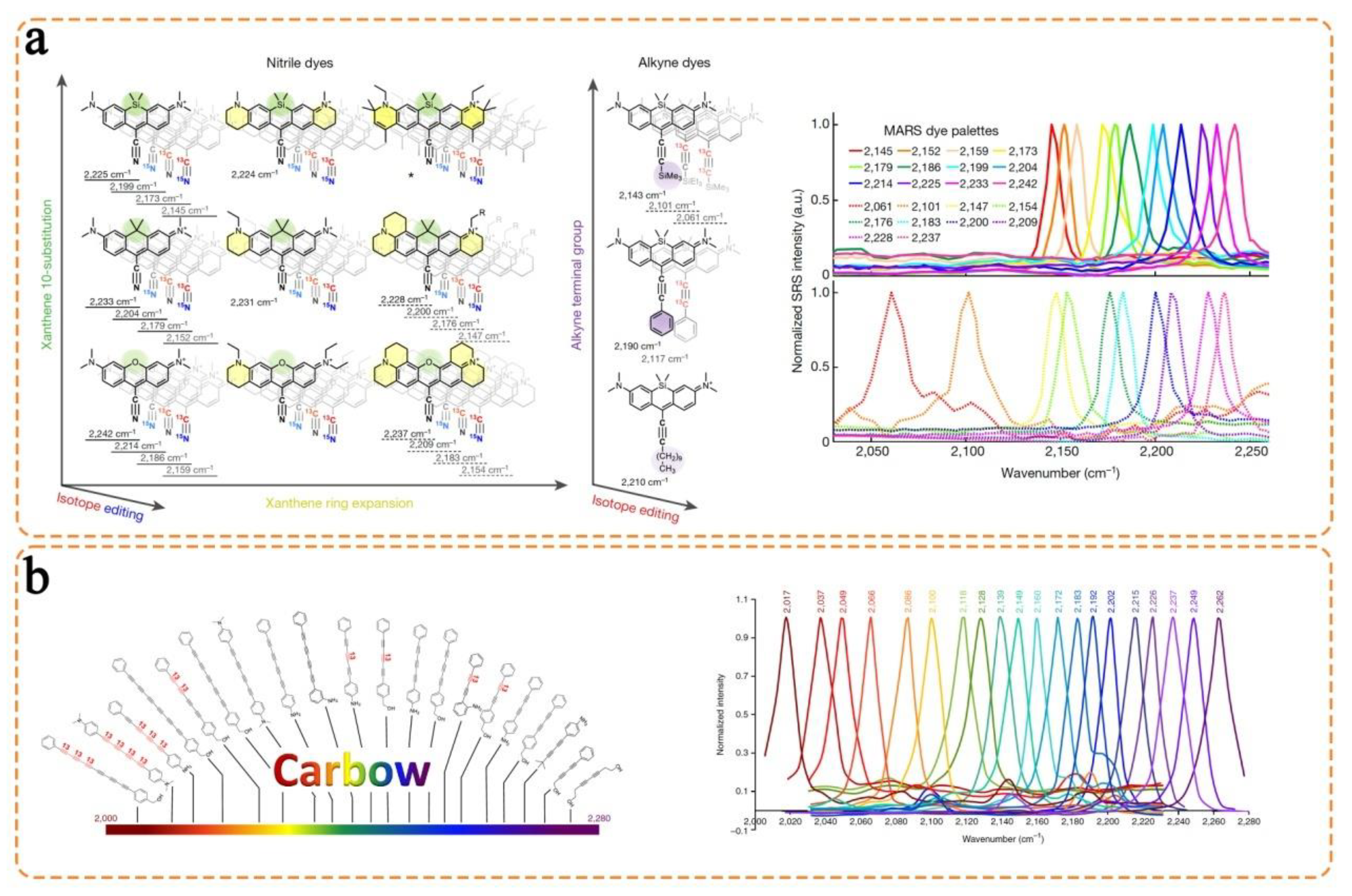
- Miao, Y.; Qian, N.; Shi, L.; Hu, F.; Min, W. 9-Cyanopyronin probe palette for super-multiplexed vibrational imaging. Nat. Commun. 2021, 12, 4518. [Google Scholar] [CrossRef]
- Hu, F.; Zeng, C.; Long, R.; Miao, Y.; Wei, L.; Xu, Q.; Min, W. Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat. Methods 2018, 15, 194–200. [Google Scholar] [CrossRef]
- Hu, F.; Shi, L.; Min, W. Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nat. Methods 2019, 16, 830–842. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, Z.; Qian, N.; Wei, S.; Hu, F.; Min, W. Multiplexed live-cell profiling with Raman probes. Nat. Commun. 2021, 12, 3405. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wei, M.; Miao, Y.; Qian, N.; Shi, L.; Singer, R.A.; Benninger, R.K.P.; Min, W. Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nat. Biotechnol. 2022, 40, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ren, J.-Q.; Zhang, X.-G.; Wu, D.-Y.; Shen, A.-G.; Hu, J.-M. Alkyne-modulated surface-enhanced Raman scattering-palette for optical interference-free and multiplex cellular imaging. Anal. Chem. 2016, 88, 6115–6119. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.-R.; Wang, L.-H.; Ren, J.-Q.; Bai, X.-W.; Zeng, L.-W.; Shen, A.-G.; Hu, J.-M. Accurate clinical diagnosis of liver cancer based on simultaneous detection of ternary specific antigens by magnetic induced mixing surface-enhanced Raman scattering emissions. Anal. Chem. 2019, 91, 2955–2963. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, D.; Feng, J.; Tang, X. Multicolor cocktail for breast cancer multiplex phenotype targeting and diagnosis using bioorthogonal surface-enhanced Raman scattering nanoprobes. Anal. Chem. 2019, 91, 11045–11054. [Google Scholar] [CrossRef]
- Li, M.; Wu, J.; Ma, M.; Feng, Z.; Mi, Z.; Rong, P.; Liu, D. Alkyne- and nitrile-anchored gold nanoparticles for multiplex SERS imaging of biomarkers in cancer cells and tissues. Nanotheranostics 2019, 3, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sun, Y.; Cao, Y.; Kang, S.H. Plasmonic nanostructure-based bioimaging and detection techniques at the single-cell level. Trend Anal. Chem. 2019, 117, 58–68. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, Z.; Li, Q.; Wang, R.; Yu, F. Toward Sensitive and Reliable Surface-Enhanced Raman Scattering Imaging: From Rational Design to Biomedical Applications. ACS Sens. 2012, 6, 3912–3932. [Google Scholar] [CrossRef] [PubMed]
- Eremina, O.E.; Czaja, A.T.; Fernando, A.; Aron, A.; Eremin, D.B.; Zavaleta, C. Expanding the Multiplexing Capabilities of Raman Imaging to Reveal Highly Specific Molecular Expression and Enable Spatial Profiling. ACS Nano 2022, 16, 10341–10353. [Google Scholar] [CrossRef]
- Zhong, Q.; Zhang, R.; Yang, B.; Tian, T.; Zhang, K.; Liu, B. A Rational Designed Bioorthogonal Surface-Enhanced Raman Scattering Nanoprobe for Quantitatively Visualizing Endogenous Hydrogen Sulfide in Single Living Cells. ACS Sens. 2022, 7, 893–899. [Google Scholar] [CrossRef]
- Dodo, K.; Fujita, K.; Sodeoka, M. Raman Spectroscopy for Chemical Biology Research. J. Am. Chem. Soc. 2022, 144, 19651–19667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, X.; Wu, G.; Cheng, L. Controlled Self-Assembly of Metallacycle -Bridged Gold Nanoparticles for Surface-Enhanced Raman Scattering. Chem. Eur. J. 2020, 26, 11695–11700. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Gong, K.; Liu, Y.; Zhang, L. Strategies and Challenges of Identifying Nanoplastics in Environment by Surface-Enhanced Raman Spectroscopy. Environ. Sci. Technol. 2023, 57, 25–43. [Google Scholar] [CrossRef]
- Wu, G.; Zheng, W.; Yang, X.; Liu, Q.; Cheng, L. Supramolecular metallacycle-assisted interfacial self-assembly: A promising method of fabricating gold nanoparticle monolayers with precise interparticle spacing for tunable SERS activity. Tetrahedron Lett. 2022, 94, 153716. [Google Scholar] [CrossRef]
- Morsby, J.J.; Thimes, R.L.; Olson, J.E.; McGarraugh, H.H.; Payne, J.N.; Camden, J.P.; Smith, B.D. Enzyme Sensing Using 2-Mercaptopyridine-Carbonitrile Reporters and Surface-Enhanced Raman Scattering. ACS Omega 2022, 7, 6419–6426. [Google Scholar] [CrossRef]
- Li, M.; Tian, S.; Meng, F.; Yin, M.; Yue, Q.; Wang, S.; Bu, W.; Luo, L. Continuously Multiplexed Ultrastrong Raman Probes by Precise Isotopic Polymer Backbone Doping for Multidimensional Information Storage and Encryption. Nano Lett. 2022, 22, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Plou, J.; Valera, P.S.; Garcia, I.; de Albuquerque, C.D.L.; Carracedo, A.; Liz-Marzan, L.M. Prospects of Surface-Enhanced Raman Spectroscopy for Biomarker Monitoring toward Precision Medicine. ACS Photonics 2022, 9, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, F.; Ye, J. Boosting the Brightness of Thiolated Surface-Enhanced Raman Scattering Nanoprobes by Maximal Utilization of the Three-Dimensional Volume of Electromagnetic Fields. J. Phy. Chem. Lett. 2022, 13, 6496–6502. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yang, H.; Hu, Z.; Fan, Y.; Guan, X.; Feng, W.; Liu, Z.; Sun, Y. Rigidity Bridging Flexibility to Harmonize Three Excited-State Deactivation Pathways for NIR-II-Fluorescent-Imaging-Guided Phototherapy. Adv. Healthc. Mater. 2021, 10, 2101003. [Google Scholar] [CrossRef]
- Yang, H.; Tu, L.; Li, J.; Bai, S.; Hu, Z.; Yin, P.; Lin, H.; Yu, Q.; Zhu, H.; Sun, Y. Deep and precise lighting-up/combat diseases through sonodynamic agents integratiing molecular imaging and therapy modalities. Corrdin. Chem. Rev. 2022, 453, 214333. [Google Scholar]
- Sun, Y.; Ding, F.; Chen, Z.; Zhang, R.; Li, C.; Xu, Y.; Zhang, Y.; Ni, R.; Li, X.; Yang, G.; et al. Melanin-dot-mediat4ed delivery of metallacycle for NIR-II/photoacoustic dual-modal imaging-guided chemo-photothermal synergistic therapy. Proc. Nat. Acad. Sci. USA 2019, 116, 16729–16735. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; Xu, Y.; Fan, Y.; Li, J.; Qiu, M.; Xiong, X.; Li, X.; Sun, Y. Biomedical applications of Pt(II) metallacycle/metallacage-based agents From mono-chemotherapy to versatile imaging contrasts and theranostic platforms. Coordin. Chem. Rev. 2021, 443, 214017. [Google Scholar] [CrossRef]
- Li, Q.; Ge, X.; Ye, J.; Li, Z.; Su, L.; Wu, Y.; Yang, H.; Song, J. Dual Ratiometric SERS and Photoacoustic Core-Satelite Nanoprobe for Quantitatively Visualizing Hydrogen Peroxide in Inflammation and Cancer. Angew. Chem. Int. Ed. 2021, 60, 7323–7332. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, Y.; Tu, L.; Choi, M.; Fan, Y.; Chen, X.; Sessler, J.L.; Kim, J.S.; Sun, Y. Rationally designed Ru(II)-metallacycle chemo-phototheranostic that emits beyond 1000 nm. Chem. Sci. 2022, 13, 6541–6549. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, C.; An, J.; Ma, X.; Yang, J.; Luo, L.; Deng, Y.; Kim, J.S.; Sun, Y. Construction of a 980 nm laser-activated Pt(II) metallacycle nanosystem for efficient and safe photo-induced bacteria sterilization. Sci. China Chem. 2023, 66, 155–163. [Google Scholar] [CrossRef]
- Xu, Y.; Tuo, W.; Yang, L.; Sun, Y.; Li, C.; Chen, X.; Yang, W.; Yang, G.; Stang, P.J.; Sun, Y. Design of a Metallacycle-Based Supramolecular Photosensitizer for In Vivo Image-Guided Photodynamic Inactivation of Bacteria. Angew. Chem. Int. Ed. 2022, 61, e202110048. [Google Scholar]
- Tu, L.; Li, C.; Liu, C.; Bai, S.; Yang, J.; Zhang, X.; Xu, L.; Xiong, X.; Sun, Y. Rationally designed Ru(II) metallacycles with tunable imidazole ligands for synergistical chemo-phototherapy of cancer. Chem. Commun. 2022, 58, 9068–9071. [Google Scholar] [CrossRef]
- Henry, A.; Sharma, B.; Cardinal, M.F.; Kurouski, D.; van Duyne, R.P. Surface-Enhanced Raman Spectroscopy Biosensing: In Vivo Diagnosics and Multimodal Imaging. Anal. Chem. 2016, 88, 6638–6647. [Google Scholar] [CrossRef]
- Chen, Z.; Mas, J.; Forbes, L.H.; Andrews, M.R.; Dholakia, K. Depth-resolved multimodal imaging: Wavelength modulated spatially offset Raman spectroscopy with optical coherence tomography. J. Biophotonics 2018, 11, e201700129. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Zhang, Y.; Guan, X.; Mei, L.; Feng, H.; Li, J.; Tu, L.; Feng, G.; Deng, G.; et al. Construction of Long-Wavelength Emissive Organic Nanosonosensitizer Targeting Mitochondria for Precise and Efficient In Vivo Sonotherapy. Adv. Funct. Mater. 2022, 32, 2207259. [Google Scholar] [CrossRef]
- Fan, Y.; Li, C.; Bai, S.; Ma, X.; Yang, J.; Guan, X.; Sun, Y. NIR-II Emissive Ru(II) Metallacycle Aaaisting Fluorescence Imaging and Cancer Therapy. Small 2022, 18, 2201625. [Google Scholar] [CrossRef]
- Li, C.; Guan, X.; Zhang, X.; Zhou, D.; Son, S.; Xu, Y.; Deng, M.; Guo, Z.; Sun, Y.; Kim, J.S. NIR-II bioimaging of small molecule fluorophores: From basic research to clinical applications. Biosen. Bioelec. 2022, 216, 114620. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhang, Y.; Zhu, S.; Ye, J.; Lin, L. Resonant Strategy in Designing NIR-II SERS Nanotages: A Quantitative Study. J. Phys. Chem. C. 2022, 126, 12575–12581. [Google Scholar] [CrossRef]
- Merone, G.M.; Tartaglia, A.; Locatelli, M.; D’Ovidio, C.; Rosato, E.; de Grazia, U.; Santavenere, F.; Rossi, S.; Savini, F. Analytical chemistry in the 21st century: Challenges, solutions, and future perspectives of complex matrices quantitative analyses in biological/clinical field. Analytica 2020, 1, 44–59. [Google Scholar] [CrossRef]
- Wen, C.; Wang, L.; Liu, L.; Shen, X.; Chen, H. Surface-enhanced Raman Probes Based on Gold Nanomaterials for in vivo Diagnosis and Imaging. Chem. Asian J. 2022, 17, e202200014. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Pan, Y.; Wang, S.; Wang, X.; Zhao, X.; Ren, W.; Lu, G.; Wu, A. Raman Reporter-Coupled Agcore@Aushell Nanostars for in Vivo Improved Surface Enhanced Raman Scattering Imaging and Near-infrared-Triggered Photothermal Therapy in Breast Cancers. ACS Appl. Mater. Interfaces 2015, 7, 16781–16791. [Google Scholar] [CrossRef] [PubMed]
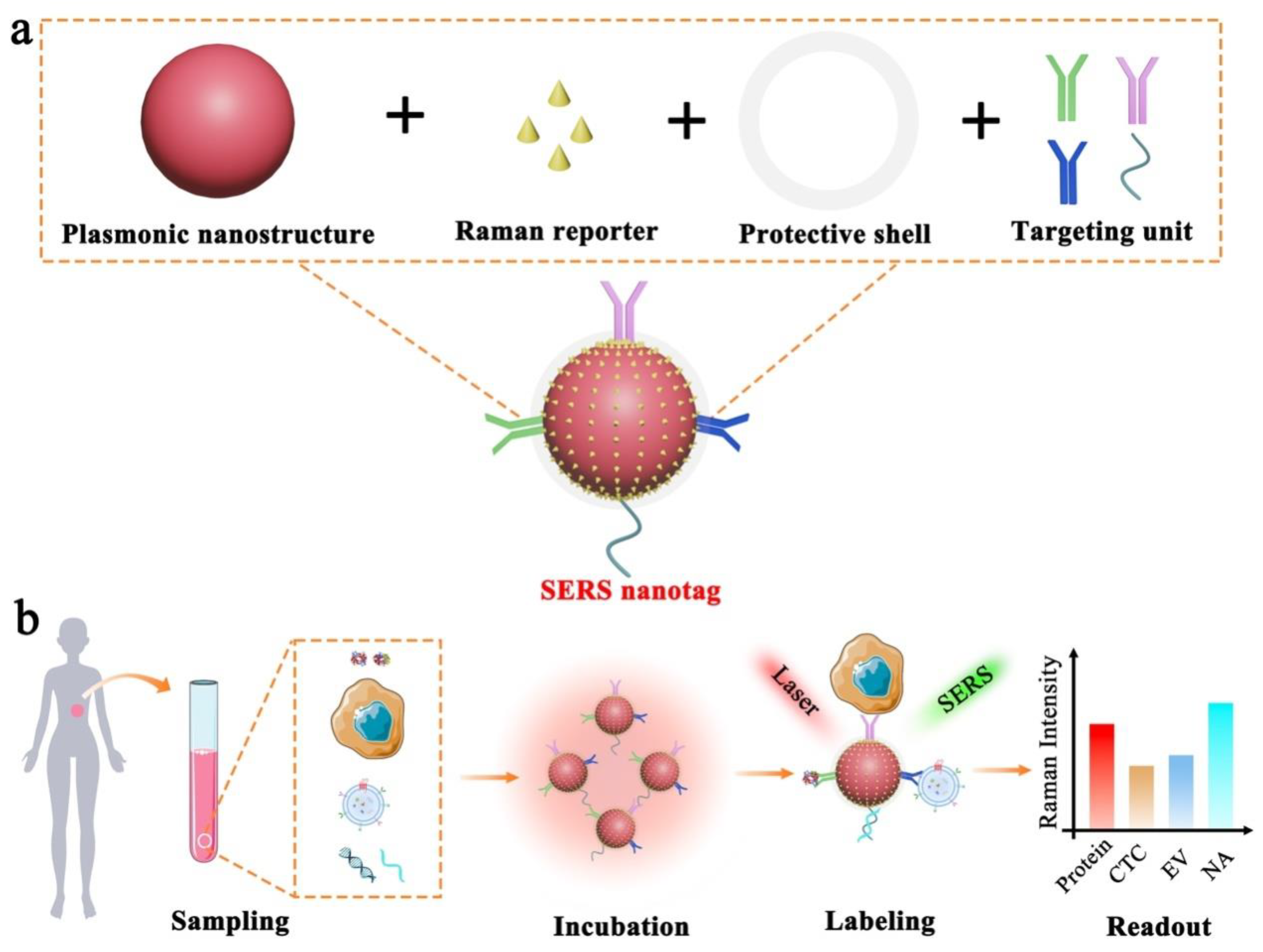
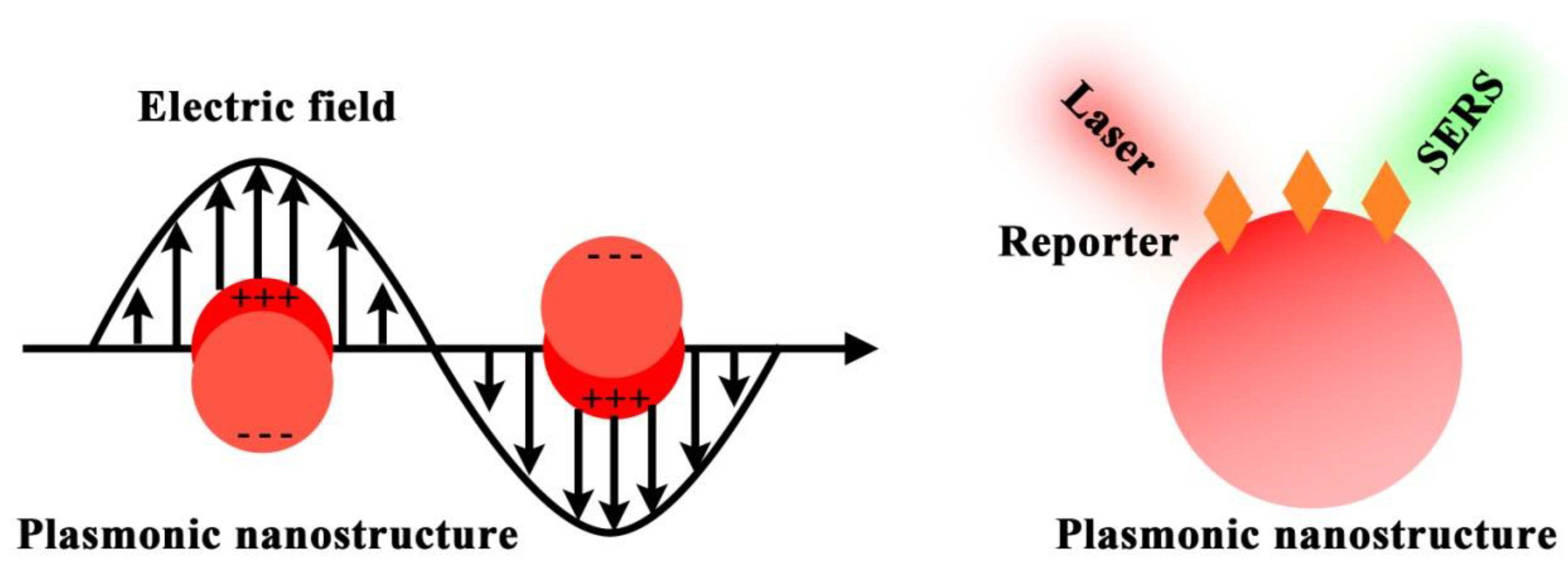

| Component | Role | Example | Working Principle |
|---|---|---|---|
| Plasmonic nanostructure | Enhancing Raman signals |
| Localized surface plasmon resonance (LSPR) |
| Raman reporter | Generating Raman signals |
| Intrinsic molecular vibration and rotation |
| Protective shell |
|
|
|
| Targeting unit | Providing required specificity |
| Specific molecular interactions |
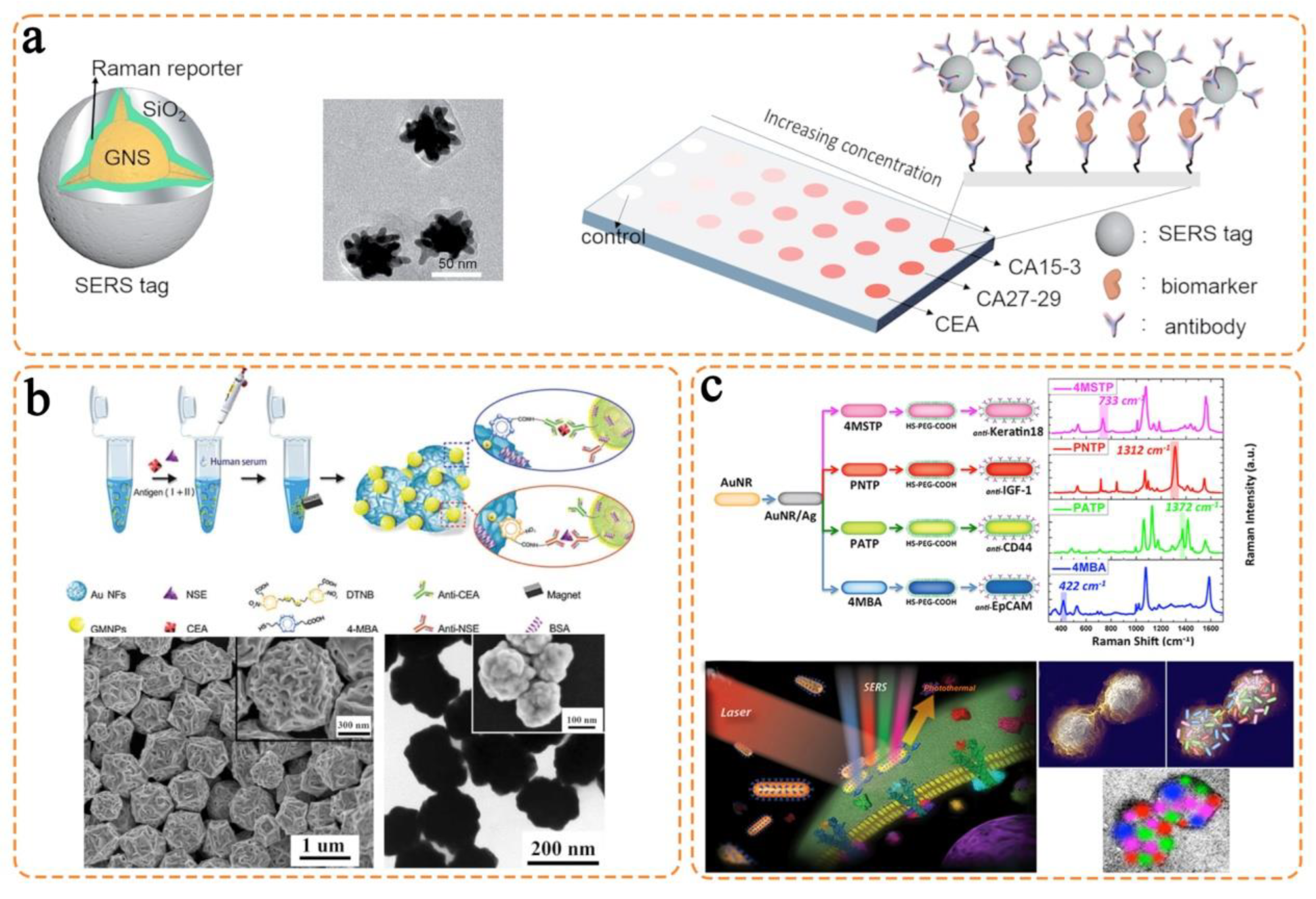
| Plasmonic Nanostructure | Reporter | Detected Biomarker | Samples | Application | Ref. |
|---|---|---|---|---|---|
| Gold nanospheres |
|
| Human serum | Melanoma cancer detection and treatment monitoring | [44] |
| Gold nanospheres |
|
| Human serum | Breast cancer metastasis monitoring | [45] |
| Silver–gold nanorods |
|
| Human blood | Breast cancer detection | [53] |
| Gold–silver alloy nanoboxes |
|
| Human serum | Melanoma cancer treatment monitoring | [55] |
| Gold nanospheres |
|
| Human serum | Liver cancer detection | [65] |
| Gold nanoflowers |
|
| Human cell lines and mice | Breast cancer detection | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Guan, R.; Li, J.; Sun, Y. Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers. Chemosensors 2023, 11, 110. https://doi.org/10.3390/chemosensors11020110
Zhang Z, Guan R, Li J, Sun Y. Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers. Chemosensors. 2023; 11(2):110. https://doi.org/10.3390/chemosensors11020110
Chicago/Turabian StyleZhang, Zhipeng, Rui Guan, Junrong Li, and Yao Sun. 2023. "Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers" Chemosensors 11, no. 2: 110. https://doi.org/10.3390/chemosensors11020110
APA StyleZhang, Z., Guan, R., Li, J., & Sun, Y. (2023). Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers. Chemosensors, 11(2), 110. https://doi.org/10.3390/chemosensors11020110








