1. Introduction
Individualized tumor spheroid models can help patients identify appropriate treatment plans (including the use of different drugs, different drug concentrations, different rays, different dose segmentation methods, etc.), and develop personalized treatment plans for patients. Among these models, the simple handling of two-dimensional (2D) cultures and standardized analysis methods make them the initial model of choice for many biological studies [
1]. In 2D culture, cell-to-cell interactions are confined to the narrow, common shared area between cells, whereas direct contact exists between each cell and both the culture medium and the underlying culture substrate [
2]. Direct cell culture medium contact results in more significant exposure of cells to nutrients, oxygen, and drugs [
3]. This can lead to implementation of the wrong treatment plan and poor treatment results. Moreover, 2D cultures are insufficient in recapitulating the heterogeneous features of the tumor microenvironment (TME) [
4]. Compared to 2D culture, 3D culture simulates the real environment in which the tumor is located, and drug/irradiation test results are more accurate. In vivo patient-derived xenografts (PDXs) have been utilized to determine patients’ drug responsiveness but have had limited success due to their low success rates, long turnaround times and high costs [
5]. Cancer-cell-line-derived models fail to fully capture the histopathological features exhibited in a clinical setting, although, compared to patients in clinical trials, PDX models largely preserve the genetic and epigenetic abnormalities of the original tumors [
6]. Compared with PDX, 3D culture has the advantages of having a high throughput, low cost and a short detection cycle, which makes it very suitable for clinical application.
Spheroids are now re-emerging three-dimensional (3D) cell culture technologies that are progressively used for studies on developmental biology, regenerative medicine, tissue engineering and tumor biology [
7,
8,
9]. Among these technologies, the cellular spheroid is one of the simplest but most effective 3D cell culture models and is formed by the aggregation of a wide range of cell types [
10]. 3D culture can also generate a more realistic concentration gradient of oxygen, nutrients, metabolic wastes and signaling molecules, as well as more adequate cell-to-cell interactions, intercellular adhesion, signal transduction and cell differentiation [
7]. In vitro 3D models cannot fully reflect the complex physiological functions, metabolic pathways and mechanisms of cellular interactions in vivo. However, they do mimic physiological conditions in vivo to a certain extent, thus making up for some of the deficiencies of 2D culture and PDX. Therefore, tumor spheroids can be used as cancer models to identify patients with sensitive treatment options at an early stage, avoiding the adverse consequences of drug resistance.
Several methods are currently used for 3D culture, including hanging drops, low-adherence substrates, bioreactors, magnetic manipulation, microwells, microfluidics and so on [
11]. These technologies have various advantages and disadvantages. For instance, hanging drop cultures are very laborious and have a low throughput. This approach requires the disruption of culture conditions [
12] and transfer of the formed spheroids into a secondary plate for end point analysis [
13]. Hanging drop cultures also suffer from elevated osmolarity caused by the evaporation of media from the droplets, which limits the time for growth and analysis [
13]. Tumor spheroids formed from low-adherence substrates and spinner flasks are often heterogeneous in size [
9]. There are two main types of microfluidic chips used for cell spheroid generation [
13]. The first type produces droplets that encapsulate cells with an encapsulation medium such as alginate [
14] and polyethylene glycol [
15]. The exchange of nutrients between tumor spheroids prepared in this way and the external environment is hindered, and the proliferation and survival of cells are affected. The second type of microwell directly aggregates cells without the use of any encapsulation medium [
13]. However, the resulting tumor spheroids are difficult to collect for subsequent analysis. In general, the above methods have the disadvantages of being laborious and producing cell spheroids with an inhomogeneous size and low throughput; these factors affect cell proliferation and survival and are inconvenient for subsequent analysis. These shortcomings limit the general applicability of these three-dimensional culture methods in clinical practice.
At present, there is still a lack of a culture methods for the fast and efficient preparation of three-dimensional tumor spheroids with simple operation and high throughput. Here, we developed a 384-well plate/96-well plate microwell array microdevice for the simple and rapid high-throughput culture of tumor spheroids. The 3D cell culture microdevice can rapidly generate spheroids within 24 h, and repeatedly form spheroids of a relatively uniform size, which is also suitable for commercial sample loading instruments or detection instruments, such as a microplate reader. To test the concept, breast cancer tumor spheroids were generated in the 3D microdevice, and then drug and irradiation sensitivity tests were performed. The results demonstrated that the microdevice has the potential to generate uniformly multicellular tumor spheroids highly efficiently and support drug and radiosensitivity testing, suggesting that the microdevice may provide an efficient model for personalized drug screening. It can help patients find the appropriate treatment plan as soon as possible and undergo effective early treatment.
2. Materials and Methods
2.1. Device Design and Fabrication
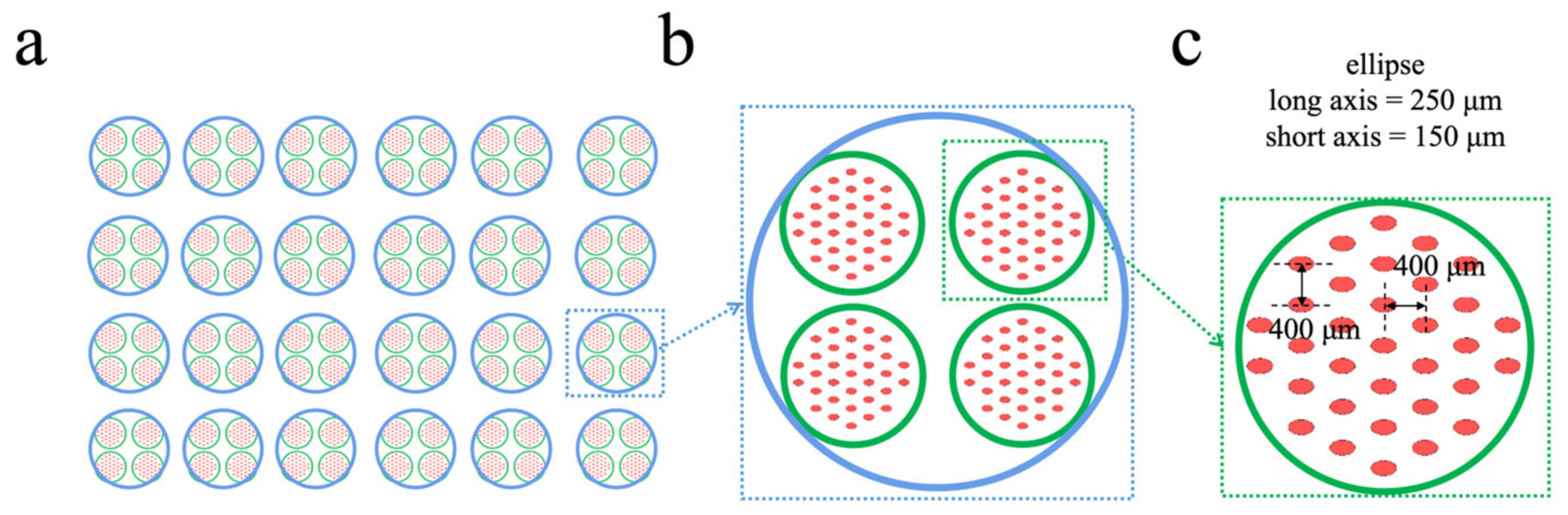
L-Edit (Tanner) was used to draw a mold design for a 384-well/96-well microwell array (
Figure 1). A well of a 96-well plate contains 4 wells of a 384-well plate, whereas a well of a 384-well plate contains 33 microwells, and the flux is 33 times that of a 384-well plate. The cross-section of the micropore is elliptical, and its major and minor axes are 250 µm and 150 µm, respectively. The bottom of the microwell is designed to be arc-shaped, with a height of 220 µm at the deepest part of the bottom. This makes it easier for cells to aggregate into clusters [
16].
A uPG-501 (Heidelberg) laser direct writer was used to convert and accurately expose the pattern to the quartz chrome plate mask. Then, the photoresist developer and the chrome etchant were used for sequential development, and finally the chrome plate mask presented the pattern. The fabrication of the microwell array mold was similar to the fabrication of other chip molds, but the silicon wafer was replaced by a chrome plate because the microfluidic chip that was fabricated in this way had higher precision. After the chrome mask was made, the exposure machine was used to expose the photoresist on the chrome mask, and then the photoresist developer was used to wash off all the photoresist, leaving only a layer of chrome. This occurred due to the firmer bond between the SU-8 photoresist and the chrome substrate. SU-83002 was poured onto a chrome plate mask, air bubbles were removed with a pipette and the plate was spun at 2000 rpm for 30 s in order to form a thin layer of SU-8 substrate. The chrome plate was placed on a 95 °C electric hot plate for 5 min, cooled, exposed to UV light for 40 s and then placed on a 95 °C electric hot plate for 5 min. The SU-83050 photoresist was placed upside down on the chrome plate, the air bubbles were removed with a pipette, the plate was spun at 500 rpm for 10 s and then spun at 800 rpm for 30 s. After this, the chrome was placed on a 95 °C electric hot plate for 30 min. Then, SU-83025 was poured onto a chrome plate mask, air bubbles were removed with a pipette, the plate was spun at 500 rpm for 10 s and was then spun at 1200 rpm for 30 s. The chrome was placed on a 95 °C electric hot plate for 40 min. Then, the chrome plate mold was directly inverted and a light diffuser (Opal diffuser, Edmund Optics Inc., Barrington, NJ, USA) [
17] was placed on the back of the chrome plate mask for UV exposure for 40 s. After the ultraviolet light was scattered, the end of the SU-8 photoresist formed an arc-shaped surface and the top of the formed column was arc-shaped when viewed from the mold. The exposed chrome plate was baked on a 95 °C hot plate for 15 min. By using PDMS to make chips, microwells with curved bottoms can be obtained.
After the mold was made, PDMS chips were produced from the mold using a Sylgard 184 elastomer kit (Momentive Corporation, Huntersville, NC, USA) with a crosslinker-to-polymer ratio of 1:8. Then, the mold was put into a vacuum pump for defoaming for 30 min and was cured at 75 °C for 1 h in order to obtain a microwell array chip. The microwell array chip was used as the first layer of substrate and two layers of reservoirs were added to it. The reservoirs were also made of PDMS and the corresponding holes were prepared using a punch. The macroporous reservoirs were made with an 8 mm hole punch (
Figure 1b) and the small hole reservoirs were made with a 4 mm hole punch (
Figure 1c). After removing the surface dust with tape, the microwell array chip, the small pore reservoir layer and the macroporous reservoir layer were placed in sequence. The three layers of PDMS were bonded using a plasma cleaning machine (Harrick Plasma, Ithaca, NY, USA) for 5 min. After the bonding was complete, the microdevice was placed in a petri dish which was then placed in a 75 °C oven overnight.
2.2. Cell Culture
Human breast cancer MCF-7 cells and their cancer stem-like cells (CSCs) were used in this study. The MCF-7 CSC subtype was sorted by flow cytometry, as reported previously [
18]. Briefly, the cell suspension was concentrated at 1 × 10
6 cells/100 µL. Ten microliters of anti-CD44-PE antibody (BD Pharmingen, San Diego, CA, USA) and ten microliters of anti-CD24-FITC antibody (BD Pharmingen) were added and incubated at 4 °C for 20 min (for the single staining group and control group, optionally with/without the corresponding antibody). After staining, the cells were washed twice with PBS. The expression of cell surface markers and cells were analyzed and sorted by flow cytometry (BD AriaIII) [
19].
The cells were cultured in a standard incubator at 37 °C with 5% CO2. DMEM high-glucose medium (HyClone, Logan, UT, USA) for MCF-7 cells was supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (HyClone, Logan, UT, USA). DMEM/F12 1:1 medium (HyClone, Logan, UT, USA) for CSCs was supplemented with N2 (Gibco, Grand Island, NY, USA), B27 (Gibco, Grand Island, NY, USA), EGF (10 ng/mL, PeproTech, Rocky Hill, USA), bFGF (20 ng/mL, PeproTech, Rocky Hill, CT, USA) and 1% penicillin-streptomycin (HyClone, Logan, UT, USA). CSC culture required the addition of 1% Geltrex (Gibco, Grand Island, NY, USA) to the dish. Geltrex was removed from a 4 °C refrigerator and diluted 100-fold with DMEM/F-12 (1:1) medium. The entire procedure was carried out on ice in order to avoid overheating, which would otherwise cause the Geltrex to solidify. The diluted Geltrex was added to a 100 mm petri dish (5 mL/dish), and then the petri dish was gently shaken to ensure that the Geltrex covered the bottom of the dish. The dishes were kept in the incubator at 37 °C for 1–2 h and were then sealed with a sealing film. The dishes were stored at 4 °C for later use. The remaining Geltrex in the dish was aspirated before use.
During tumor spheroid culture, the medium was changed twice a day. Two-thirds of the medium was slowly removed from the macroporous reservoir, and the new medium was carefully and slowly added from above the macroporous reservoir using a smaller pipette tip head in order to avoid flushing the tumor spheroids out of the microwells.
2.3. Cell Seeding
To prevent cell adhesion, the microdevice was coated with 4% Pluronic F-127 prior to seeding the cells. Then, the microdevice was washed twice with PBS and was UV-sterilized for 30 min. After disinfection, the microdevice was vacuumized for 10 min, and 120μL of tumor cell suspension was added into the 96-well large-hole storage tank at the top of every culture device. Under the force of gravity, the cell suspension traveled from the macroporous reservoir to the small pore reservoir and settled in the microwells. This design made the operation simple and convenient. After being allowed to stand for 15 min, the cell suspension that was not precipitated in the microwells was aspirated, the medium was refreshed and the cells were placed in a standard incubator for culture. Aggregation was completed within 24 h and 3D tumor spheroids were obtained. The medium was changed every 2–3 days.
2.4. Growth of Tumor Spheroids in the Microdevice
The growth and viability of MCF-7 tumor spheroids was monitored in the microdevice. Then, 120 μL of MCF-7 cell suspensions at different cell densities (1 × 105/mL, 3 × 105/mL, 5 × 105/mL and 10 × 105/mL) were added to the macroporous reservoir of the microdevice (where indicated, the cell seeding density was not the final cell density that was seeded in the wells). Aggregation was completed within 24 h and 3D tumor spheroids were obtained. The tumor spheroids were photographed 1, 3, 5, 7 and 8 days after inoculation.
2.5. Use of the Microdevice for Drug Screening
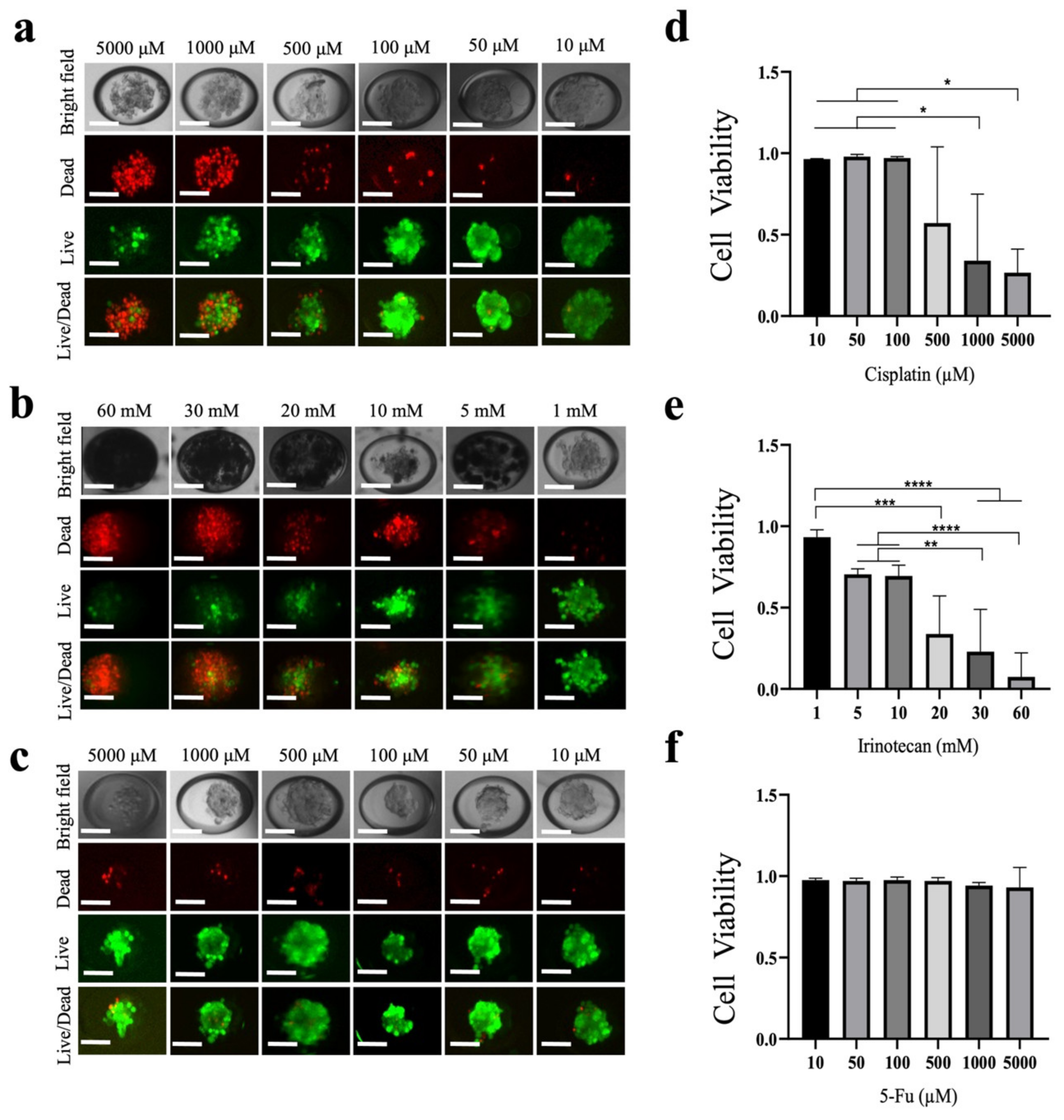
Then, 120 µL of MCF-7 cell suspension at a density of 3 × 105/mL was added to the microdevice for culture, and 3D tumor spheroids with a good size, good roundness and good firmness were obtained after 48 h. Six concentrations of the three different drugs to be tested (cisplatin, irinotecan and 5-fluorouracil (5-FU) were added to the microdevice to act on the tumor spheroids. The final concentrations of 5-FU and cisplatin were 5000, 1000, 500, 100, 50 and 10 µM. The final concentrations of irinotecan were 60, 30, 20, 10, 5 and 1 mM. After the tumor spheroids were treated with the drugs in the incubator for 48 h, live/dead cell staining (Calcein-AM/PI double staining, Bestbio) was performed on the tumor spheroids in the microdevice containing different concentrations of the drugs.
2.6. Use of the Microdevice for Irradiation Testing
2.6.1. Irradiation Testing of 2D Cells and 3D Tumor Spheroids
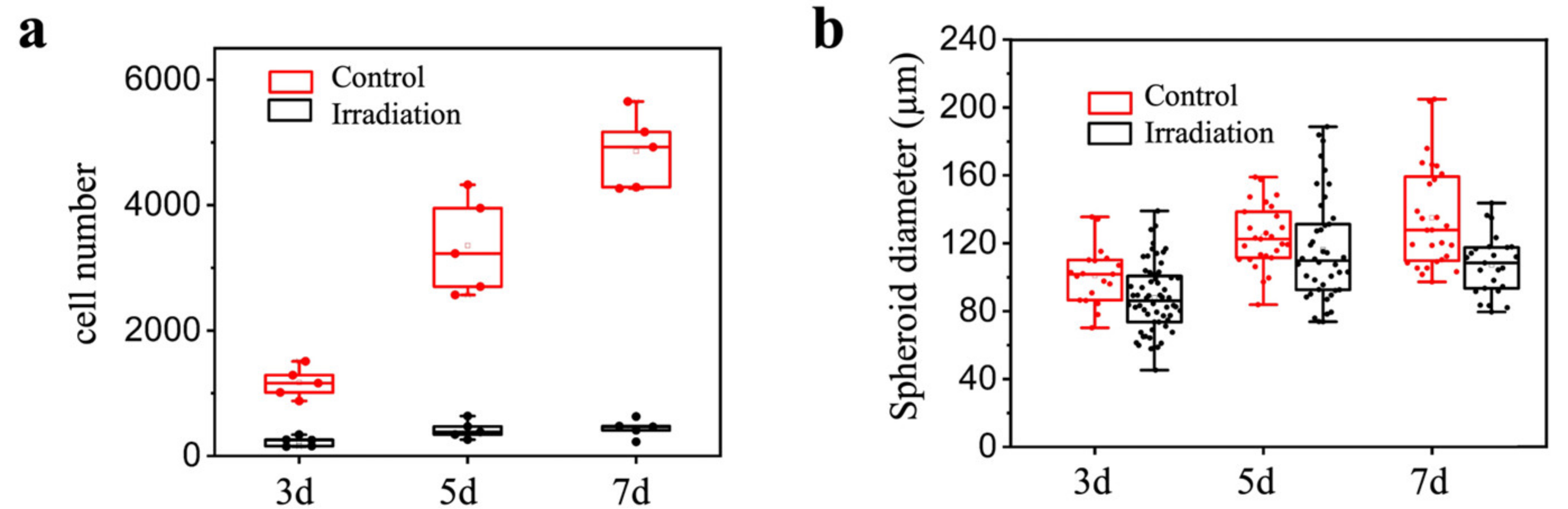
Then, 200 µL of MCF-7 cell suspension with a density of 2.5 × 104/mL was added to each well in a 96-well plate for 2D culture, and 120 µL of MCF-7 cell suspension with a density of 3 × 105/mL was added to the microdevice for 48 h to form tumor spheroids. The 3D tumor spheroids and 2D cells were irradiated with γ-rays for two consecutive days, with a dose of 15 Gy per irradiation at a dose rate of 1 Gy/min each day. The number of cells/size of tumor spheroids under 2D and 3D conditions was determined 3, 5 and 7 days after the first irradiation in order to detect the killing effect of irradiation on the cells.
2.6.2. Irradiation Testing of Tumor Spheroids Cocultured with MCF-7 Cells and CSCs at Different Ratios
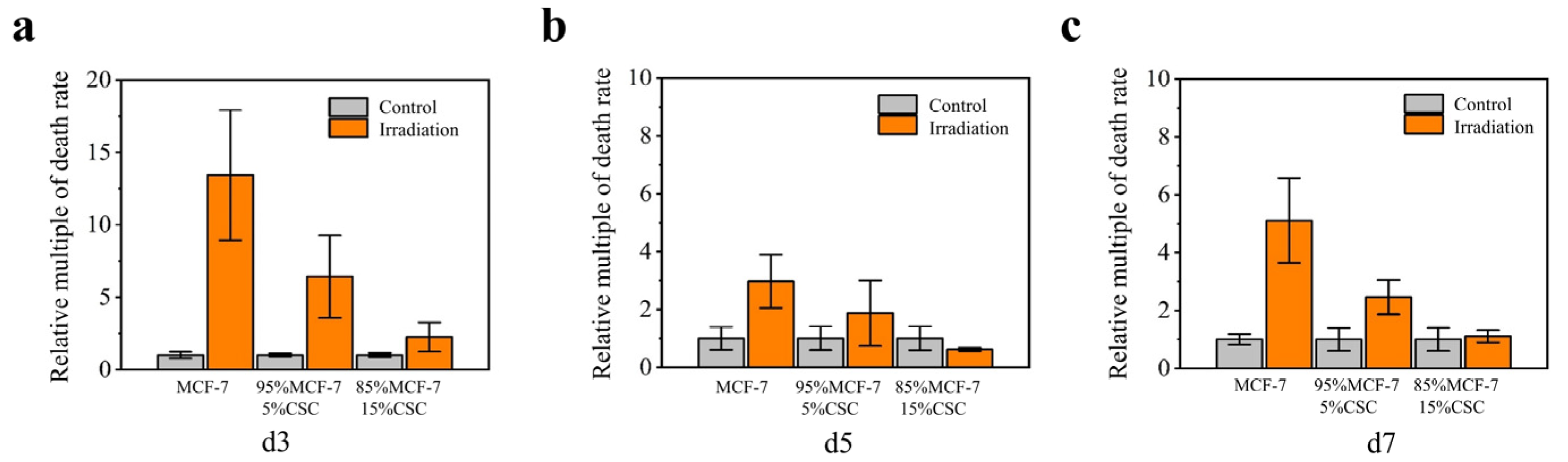
After digestion, the MCF-7 cells and CSCs were adjusted to the same density and were then mixed by volume. For example, 50 µL of CSC suspension and 950 µL of MCF-7 cell suspension were mixed well and added to the microdevice in order to obtain tumor spheroids containing 5% CSCs and 95% McF-7 cells. Three ratios were set as follows: 0% CSCs + 100% MCF-7, 5% CSCs + 95% MCF-7 cells and 15% CSCs + 85% MCF-7 cells. After 48 h, the spheroids were irradiated with 8 Gy γ-ray at a dose rate of 1 Gy/min. The tumor spheroid size was measured and live/dead staining of tumor spheroids was performed 3, 5 and 7 days post-irradiation. Irradiation with γ-rays was performed in the 60Co source room at the School of Chemistry and Molecular Engineering, Peking University. The radioactivity of 60Co was 2.6 × 105 Ci and the dose rate was fixed at 1 Gy/min.
2.7. Microscopy and Image Processing
An Olympus IX71 inverted fluorescence microscope/Nikon Ti-E inverted fluorescence microscope was used to obtain images of the tumor spheroids. To measure tumour spheroid diameter and determine the proliferative status of tumor spheroids, ImageJ was used. The green area (live cells) and the red area (dead cells) were circled. The following formula was used: cell viability = area of all green areas/(area of all green areas + area of all red areas) ×100%.
2.8. Statistical Analyses
The data from this experiment were statistically analyzed using ImageJ, Origin and GraphPad Prism software. Comparisons between multiple groups of data were performed using one-way ANOVA, with a statistical difference at p < 0.05 and a statistically significant difference at p < 0.01.
4. Conclusions
In this study, we developed a novel microdevice that aggregates cells to form three-dimensional cell spheroids. We used a design based on the bottom of the microwell (using the force and gravity of the wall on the cells can quickly make the cells aggregate and shortens the spheroidization time) and a standard 384 commercial well plate/96 commercial well plate microarray design. Each large well (96-well plate well) consisted of 33 × 4 microwells and used gravity to transfer cells or reagents into the microwells. This feature reduces the operation time and number of steps in the process, while achieving rapid (~24 h) and high throughput (the throughput is 33 times that of a 384-well plate). The formed cell spheroids have good roundness and high viability, with significant advantages compared to existing methods. These microdevices provide efficient and rapid in situ sensitivity testing, avoiding damage and loss of cell spheroids. The microdevice is simple to manufacture, easy to operate and compatible with commercial instruments. Collectively, these advantages greatly facilitate the use of this microdevice as an effective tool for 3D culture and various sensitivity analyses.
We prepared MCF-7 spheroids using microdevices. The results showed that the cells completed spontaneous aggregation within 24 h and a single tumor spheroid was formed in each microwell. The survival rate of cells in spheroids was > 95%. In addition to the culture of cell spheroids, our microdevices were used for sensitivity analyses. Since the 3D structure of cell spheroids better mimics the diffusion limitations of in vivo tumors, it is suitable for the study of drug penetration. We performed toxicity studies on MCF-7 tumor spheroids using cisplatin, irinotecan and 5-FU as model drugs to demonstrate the feasibility of the microdevices for drug research. Compared with 2D cultures, 3D tumor cultures, such as spherical tumors, have more accurate and diverse characteristics (such as spatial arrangement, cellular interactions and phenotypic gradients related to proliferation and metabolism) in tumor tissue in vivo [
30].
Next, we compared the response of conventional monolayer MCF-7 cell lines and three-dimensional MCF-7 cell line-derived spheroids to radiotherapy. Consistent with previous publications, such as that by Jana Koch et al. [
31], we found that the tumor spheroids were more resistant to irradiation. Subsequently, we further compared the radiosensitivity of tumor spheroids with different CSC doping ratios. The results showed that the radiation resistance of tumor spheroids varied according to the proportion of CSCs, that is, the higher the proportion of CSCs in the tumor spheroids, the stronger the radiation resistance of the tumor spheroids.
Future research is still needed, including research investigating whether tumor cells from patients can be successfully cultured, whether successfully cultured tumor spheroids truly reflect the physiological status of patients’ tumors in vivo and whether results from tumor spheroid sensitivity analyses can predict patient disease. Our microdevices might provide models and references for precise treatment.