Abstract
Convective PCR (CPCR) can perform rapid nucleic acid amplification by inducing thermal convection to continuously, cyclically driving reagent between different zones of the reactor for spatially separate melting, annealing, and extending in a capillary tube with constant heating temperatures at different locations. CPCR is promoted by incorporating an FTA membrane filter into the capillary tube, which constructs a single convective PCR reactor for both sample preparation and amplification. To simplify fluid control in sample preparation, lysed sample or wash buffer is driven through the membrane filter through centrifugation. A movable resistance heater is used to heat the capillary tube for amplification, and meanwhile, a smartphone camera is adopted to monitor in situ fluorescence signal from the reaction. Different from other existing CPCR systems with the described simple, easy-to-use, integrated, real-time microfluidic CPCR system, rapid nucleic acid analysis can be performed from sample to answer. A couple of critical issues, including wash scheme and reaction temperature, are analyzed for optimized system performance. It is demonstrated that influenza A virus with the reasonable concentration down to 1.0 TCID50/mL can be successfully detected by the integrated microfluidic system within 45 min.
1. Introduction
Nucleic acid-based diagnostics provide ultrasensitive, specific, accurate DNA or RNA assays for detecting pathogen- or pathogen-related nucleic acids (NAs) in different resources, such as medical samples from human bodies or other biological samples from food or environment [1]. Nucleic acid-based diagnostics or nucleic acid amplification and tests (NAATs) gain their high performance with desired sensitivity and specificity by utilizing enzymatic nucleic acid amplification to replicate the target NAs by more than a million-fold, facilitating detection by optical (e.g., fluorescence) or electrochemical means [2].
Typically—and for best results with respect to LOD (limit of detection), specificity, and reliability—NAATs incorporate sample processing in order to extract NAs from the crude samples and concentrate the nucleic acid by 100-fold to remove substances that interfere or inhibit the amplification process [3]. Further, the most common amplification technique, PCR (polymerase chain reaction) is implemented generally in a thermal cycler that provides precise time–temperature control. For these reasons, NAATs have been largely restricted to benchtop laboratory use, requiring comparatively sophisticated instruments and trained technicians [4]. Nevertheless, the availability of portable, easy-to-use systems for NAATs, especially for testing outside the laboratory in resource-poor settings, would prove to be a highly useful diagnostic tool, enabling a new paradigm in rapid and on-site detection [5]. Such tests would enable rapid diagnosis of infectious diseases, wider screening for cancer and genetic mutations, detection for food contamination throughout the supply chain from farm to consumer, and would eliminate the need to transport and store clinical specimens [6].
Consequently, there have been extensive efforts over the last several decades to develop simplified NAATs appropriate for nontraditional venues, such as schools, pharmacies, and clinics, and especially for developing appropriate diagnostics technology for resource-limited settings in less developed regions [7]. As a promising technology, microfluidic-based NAATs demonstrate their potentiality to perform nucleic acid diagnosis in an easy-to-use, rapid, efficient, and affordable way at point-of-care (POC) diagnosis [8]. The microfluidic system enables automatic, integrated diagnosis with the functional fluid network consisting of micro-channel, micro-valve, micro-pump, and micro-reactor [9]. Especially, microfluidic-based PCR reactors in tiny size allow ultra-fast nucleic acid amplification [10]. Fully integrated microfluidic NAAT systems combining sample preparation, amplification, and detection together have been extensively studied [11]. Existing microfluidic NAAT systems still have a couple of remarkable limitations [12]. First, most of them rely on time-consuming conventional PCR to perform nucleic acid amplification, which requires repeated and precise temperature control with sophisticated and costly thermal cyclers [13]. Second, because of the trivial nucleic acid extraction procedure, an existing fully integrated microfluidic NAAT system with sample preparation is normally quite complicated with high cost. Therefore, many works still must be performed to allow microfluidic NAAT system to perform rapid, simple, affordable nucleic acid analysis with high convenience.
Compared to continuous-flow PCR, which always relies on active pumping [14], convective PCR (CPCR) can be implemented with much simpler supporting instrumentation without any external pumps in short time [15,16]. Different from conventional PCR, CPCR relies on spontaneous thermal convection to perform nucleic acid amplification [17,18]. For a properly designed CPCR reactor, a consistent temperature field on the reactor must be established to achieve the stable and repeatable thermal convection for robust amplification [19,20]. Compared to conventional PCR, which normally takes 1–2 h, CPCR can be performed within 30 min, which is competitive in POC diagnostics [21,22,23,24]. Recently, few microfluidic CPCR NAAT systems with complicated sample preparation have been reported [25], which nevertheless demonstrate their potentiality in POC diagnostics with simple heating and ultra-fast amplification.
So far, portable microfluidic NAAT systems have not become widespread, and improvements are needed in performance, cost, and ease-of-use. Based on our experience on CPCR [21,22,23,24], by incorporating sample preparation, this work aims to demonstrate an integrated, simple, low-cost, portable, microfluidic CPCR NAAT system with simplified instrumentation. For simplified sample preparation, the CPCR reactor is integrated with a PCR compatible solid phase membrane on the bottom for nucleic acid extraction, which provides the nucleic acid templates without elution. The capillary tube works as a connection channel in nucleic acid extraction as well as an amplification reactor in thermal convection. To simplify fluid control, centrifugation is applied to drive different fluid reagent by mounting the capillary tube on a spinning disk. With centrifugal force, the lysed sample or wash buffer flows through the solid phase membrane until it reaches the downstream waste chamber, while nucleic acids are captured by the solid phase membrane. After nucleic acid extraction, PCR mix is loaded into the capillary tube reactor, which allows CPCR amplification with the nucleic acid templates from the bottom solid phase membrane. Since there are no venting holes in downstream flow network, CPCR can be performed without closing any valves once the PCR mix is covered with mineral oil on top. To perform real-time CPCR, a portable and compact companion instrument is developed to rotate the disk for sample preparation, heat the capillary tube for amplification, and collect the real-time fluorescent signal by a smartphone camera. As a proof of concept, the critical performance of the portable, integrated real-time CPCR NAAT microfluidic system is demonstrated with successful detection to influenza A virus from sample to answer within 45 min.
2. Material and Methods
2.1. Single CPCR Reactor with Sample Preparation
Previously, we successfully developed a couple of CPCR systems for rapid nucleic acid amplification within 30 min, which all relied on a capillary tube reactor (inner and outside diameter: 1.6 mm and 3.3 mm) to introduce consistent thermal convection with pseudo-isothermally heating [21,22,23,24]. The relationship of the sample/reaction volume vs. thermal characteristics of the CPCR reactor was found in our previously published study [26]. Once a consistent temperature field was established along the capillary tube with properly heating, CPCR amplification was achieved by inducing thermal convection to continuously and cyclically drive reagent between different zones of the reactor for spatially separate denaturation, annealing, and extension. These disposable and low-cost capillary tubes were massively manufactured with injection molding. In this study, based on our previous work, the capillary tube was equipped with a solid phase membrane for sample preparation, which constructs a single CPCR reactor combining nucleic acid extraction and CPCR amplification. By doing this, the capillary tube worked not only as the introduction channel in nucleic acid extraction but also as the reactor in CPCR amplification, which reduced the system complexity. As shown in Figure 1, for the single CPCR reactor, a solid phase membrane disk was integrated into the capillary tube for sample preparation.

Figure 1.
Single CPCR reactor equipped with a solid phase membrane for sample preparation. (A) Schematic depiction of real-time CPCR with an FTA membrane; (B) a photograph of capillary tube equipped with an FTA membrane.
FTA membrane was incorporated into microfluidic chips for nucleic acid extraction without elution since it did not inhibit PCR with a limited size [27,28,29]. In this study, as shown in Figure 1, an FTA membrane disk (Whatman) with a diameter of ~2.0 mm was deposited into the bottom of the capillary tube for sample preparation. Following the flow path provided by capillary tube, lysed sample or wash buffer was driven through the FTA membrane disk, while the purified nucleic acids were captured by the FTA membrane. Without elution, the captured nucleic acids on the FTA membrane worked as the templates for CPCR amplification after the capillary tube was filled with PCR reaction mix. In principle, the CPCR tube and its heating system in this study were basically identical to those developed in our previously published study. In this study, the size ~1 mm in thickness and ~2 mm in diameter of the FTA nucleic acid extraction membrane in the CPCR reactor was quite small compared to the whole reactor (~18 mm in height), so its influence on physical modeling was ignored. The thermal management model inside the CPCR reactor was found in our previously published studies, including physical modeling and simulation analysis [22,26]. Since the FTA membrane disk was located at the bottom, thermal convection inside the capillary tube, which was essential to CPCR amplification, was deteriorated. Therefore, based on the concept of single CPCR reactor, both sample preparation and CPCR amplification were reasonably joint with each other by integrating an FTA membrane disk into the capillary tube, which was helpful to simplify the microfluidic CPCR NAAT system.
2.2. Integrated Real-Time Microfluidic CPCR NAAT System
2.2.1. CPCR Reactor Module with FTA Membrane
To facilitate fluid reagent handling, an integrated CPCR reactor module was developed properly. As shown in Figure 2, the CPCR reactor module included three major parts: a reagent storage chamber, a capillary tube with an embedded FTA membrane disk, and a waste storage chamber without any venting holes.
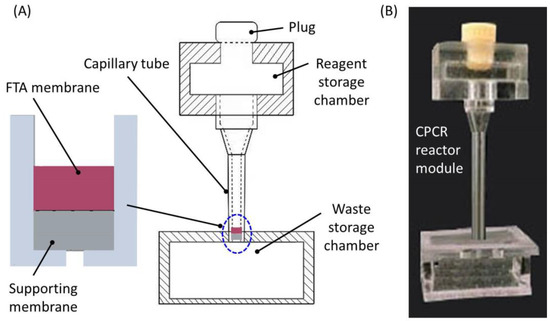
Figure 2.
Integrated PCR reactor module. (A) Schematic depiction of integrated PCR reactor; (B) a photograph of the CPCR reactor module made from PMMA.
As shown in Figure 2A, on the top, a reagent storage chamber was incorporated to temporarily hold lysed sample, purification buffer, or wash buffer, respectively, at different times. A soft plug was used to seal the reagent storage chamber from the top when the fluid was flown through the FTA membrane. As shown in Figure 2A, under the FTA membrane disk (2 mm in diameter), a supporting membrane layer (2 mm in diameter) was used to properly hold the FTA membrane in position, and meanwhile, it was helpful to prevent leakage from the capillary tube in CPCR amplification. As shown in Figure 2A, a waste storage chamber on downstream was used to collect waste sample and buffer in sample preparation. Since all the fluid reagent was driven through the FTA membrane with centrifugation, venting holes were not needed in the waste storage chamber. Different from existing microfluidic PCR reactors, which always needed to be sealed with valves in thermal cycling for the described CPCR reactor module, CPCR amplification was performed without any valves once the PCR mix was covered with mineral oil from the top. Therefore, with significantly simplified setup for both fluid and thermal control, rapid nucleic acid analysis was achieved with the described microfluidic CPCR reactor module. As shown in Figure 2B, both the reagent and waste storage chambers were made from PMMA by CNC machining (JDEM_V, Beijing Jingdiao Group Co., Ltd, Beijing, China). All three parts were assembled together with solvent (acetonitrile) binding.
2.2.2. Centrifugation-Based Fluid Control
Similar to other existing microfluidic disk chips [30,31], centrifugation was used to perform fluid control in sample preparation, for example, to drive lysed sample or wash buffer from the reagent storage chamber to the waste storage chamber by flowing through the FTA membrane inside the capillary tube. With centrifugation, fluid was smoothly driven through the FTA membrane with low risk of trapping any air bubbles. In principle, the binding efficiency of nucleic acids to FTA membrane as well as the washing efficiency of purification buffer or TE buffer was sensitive to the flowing speed of different fluid. With centrifugation, the fluid flow speed was conveniently regulated according to the centrifugation speed, which was helpful to ensure the efficiency of nucleic acid extraction. As shown in Figure 3A, once the CPCR reactor module was mounted on a disk rotated by a motor, centrifugation-based fluid control was conveniently implemented. The centrifugation speed was 1500–2000 revolutions per minute (RPM), and the radius of the centrifugal disk was 45 mm.
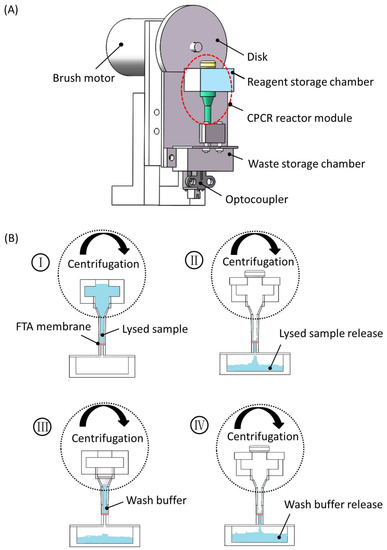
Figure 3.
Centrifugation-based fluid control to CPCR reactor module. (A) Schematic depiction of CPCR reactor module with centrifugation; (B) schematic depiction of sample preparation process based on centrifugation.
As shown in Figure 3A, when the disk was rotated, the mounted CPCR reactor module rotated accordingly to generate centrifugation force for draining fluid into the waste chamber through the FTA membrane filter. An optocoupler was used to indicate the initial position of the disk. A brush motor was adopted to rotate the disk to generate centrifugation force, and the centrifugation speed was adjusted conveniently by controlling the applied voltage of the brush motor. As shown in Figure 3B, with centrifugation, lysed sample, or wash buffer was introduced into the reagent storage chamber, and it was flown smoothly through the FTA membrane until it reached the waste storage chamber. Therefore, with centrifugation, sample preparation with simplified fluid control was conveniently achieved.
2.2.3. Portable Real-Time Microfluidic CPCR NAAT System
An integrated system was developed to perform integrated nucleic acid analysis with the described microfluidic CPCR module. The portable device performed sample preparation, CPCR amplification, and real-time fluorescence collection. As shown in Figure 4A, all the components were enclosed in a sealed box, which was equipped with a smartphone for fluorescence detection from outside. The component cost of our device was less than $400 (excluding smartphone), which was significantly lower than that of commercially available PCR devices.
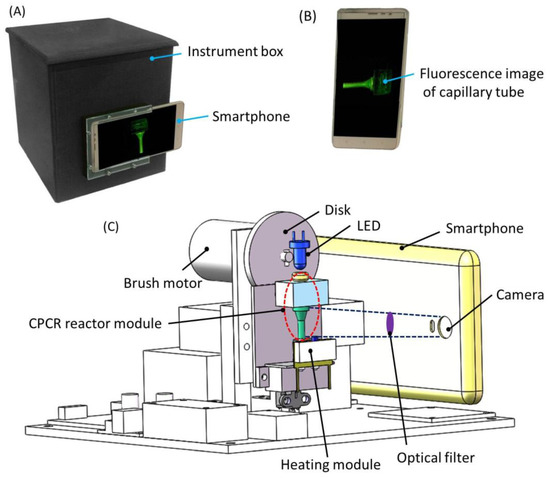
Figure 4.
Integrated Microfluidic CPCR NAAT System. (A) A photograph of CPCR device in this paper; (B) a smartphone and in-situ fluorescent signal collected; (C) schematic depiction of CPCR device with centrifugation, heating and fluorescence detection modules.
In situ fluorescent signal was collected by a smartphone camera (as shown in Figure 4B). As shown in Figure 4C, in addition to the centrifugation module, two other modules, heating and fluorescence detection modules, were integrated into the portable instrument. To simplify system design, single-end heating from the bottom was adopted. A movable heating module was adopted to allow it to contact with or separate from the CPCR reactor module for heating, if necessary. A thermistor was used to monitor the heating temperature for feedback temperature control. As shown in Figure 4C, for fluorescence detection, a LED (light-emitting diode) with a proper wavelength coupled with a short-pass optical filter (λcut-off = 495 nm) was used to illuminate the capillary tube from the top. Meanwhile, a smartphone camera behind an optical filter (λcut-off = 515 nm) was used to collect the fluorescent image from the capillary tube. Two short-pass optical filters, λcut-off = 495 nm and λcut-off = 515 nm, were chosen for excitation and emission in fluorescence detection based on the property of the Taqman probes (FAM at the 5′ end and Eclipse at the 3′ end), respectively. Based on experimental results, it was found that satisfied fluorescence images could be obtained with the properly configured optical filters. In amplification, multiple fluorescent images were collected by the smartphone camera with a predefined time interval. Fluorescent signal at different time was extracted from multiple images with custom image processing software running on the smartphone. Meanwhile, a friendly interface was provided by the smartphone to allow people to operate the system conveniently.
Previously, real-time CPCR based on Taqman probes was successfully implemented on our previously developed real-time CPCR device similar to conventional PCR device [22,24]. The performance of fluorescence detection of our previously developed real-time CPCR device was evaluated and confirmed with conventional PCR. In the developed system, the same Taqman probes were used for real-time CPCR with smartphone-based detection. Therefore, the performance of fluorescence detection of the developed system should be comparable to that achieved with our previously developed real-time CPCR device based on photodiode-based florescence detection.
3. Results and Discussion
3.1. Evaluation of Temperature Control
To evaluate the performance of the heating module, a temperature calibration capillary tube was constructed by depositing a thermistor (diameter: 0.76 mm) into the bottom of the capillary tube. The thermistor connected an existing temperature reader (USB-TEMP, Measurement Computing Corporation, Norton, MA 02766, USA) to monitor the inside temperature of the capillary tube. Figure 5 depicts the thermal time-response of the capillary tube when it is heated from the bottom side. After ~10 min, the inner temperature of the capillary tube reaches and stays at 95 °C. Similar results were achieved with repeated experiments.
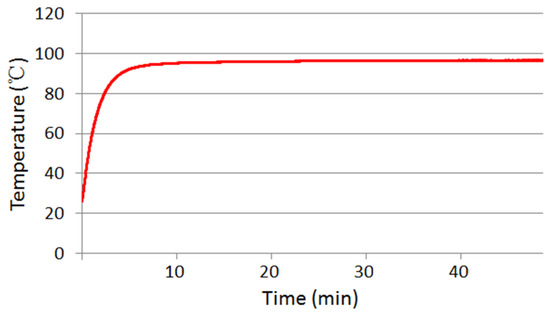
Figure 5.
Thermal time-response of the temperature calibration capillary tube.
Before CPCR amplification, the heater was moved by a linear motor to tightly contact with the CPCR reactor module for desirable thermal conduction. As shown in Figure 5, the movable heater can heat and maintain the CPCR reaction tube at 95 ± 0.1 °C after ~10 min. In principle, the ramping rate can be significantly improved by increasing the power of the heating module. In the future, the device will be optimized to further improve the thermal response of the system.
3.2. Sample Preparation with the Integrated Microfluidic CPCR NAAT System
The utility and performance of sample preparation with the integrated microfluidic CPCR NAAT system was evaluated with detection to one of popular infectious viral pathogens, H1N1 influenza A virus. Throat swabs were used in the experiments. Throat swabs were collected by the Children’s Hospital, Capital Institute of Pediatrics, Bejing, China, and transferred to Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beiijng, China. Independent ethics committee approval was obtained from the Ethics Committee of the Children’s Hospital, Capital Institute of Pediatrics, Bejing, China. All experiments were performed in accordance with relevant guidelines and regulations approved by the Ethics and Safety Committee of Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beiijng, China. A simplified sample preparation protocol with CPCR reactor module is defined as the following. First, 200 μL of culture stock with H1N1 viruses is mixed with 400 μL of lysis buffer (Beijing Wantai Biological Pharmacy Enterprise, Ltd., Beijing, China) outside the chip for 8 min. Secondly, a total of 600 μL of lysed sample is loaded into the reagent storage chamber of the CPCR reactor module, and then it is driven through the FTA membrane filter with centrifugation within 2 min. Thirdly, purification buffer (Global Life Sciences Solutions Operations, Ltd., Sheffield, UK) or TE buffer (PH 8.0, Beijing Wantai Biological Pharmacy Enterprise, Ltd., Beijing, China) is loaded into the reagent storage chamber to wash the FTA membrane filter with centrifugation within 1–2 min. Wash can be repeated to achieve efficient sample preparation. Finally, PCR reaction mix is loaded into the capillary tube with a bottom FTA membrane filter, and the reaction is sealed from the top with mineral oil. Since there are no venting holes in the waste storage chamber, the FTA membrane with the supporting membrane can prevent PCR reagent draining from the capillary tube into the waste storage chamber. Purified viral RNA captured on the FTA membrane filter is amplified with RT-PCR in a single capillary tube. The total volume of PCR reaction mix is 40 μL. It includes 0.4 U of AMV reverse transcriptase (Promega, Madison, WI, USA), 3 μL primers (Beijing Wantai Biological Pharmacy Enterprise, Ltd., Beijing, China) with Taqman probes, 4 mM dNTP and 4 μL of Fast Buffer I (Mg ion buffer) (all Takara Bio Inc., Shiga, Japan), 1 U of SpeedSTAR HS DNA polymerase (Takara), and molecular-free water. To ensure the detection specificity, Taqman probes (FAM at the 5′ end and Eclipse at the 3′ end) were used to label the DNA amplicons. The amplicons of H1N1 templates are 105-bp. In principle, the CPCR tube and its heating system in this study are basically identical to those developed in our previously published study. The amplification quality of DNA amplicons has been verified by the gel electrophoresis in our previously published studies [32,33]. A smartphone camera was used to continually collect the image of the capillary tube according to a timing cycle. The images were further processed to extract the intensity of fluorescence signal with custom image processing application software developed by Java. Finally, a fluorescence curve was plotted with multiple data points of intensity of fluorescence signal at different times. Because CPCR relies on spontaneous thermal convection to perform nucleic acid amplification, its thermal cycling cannot be strictly controlled cycle by cycle just like traditional PCR. Therefore, CPCR intends to perform qualitative detection, and even for the repeated tests to the samples with the same concentration, their fluorescence curves may be different from each other. Therefore, to demonstrate the performance of the developed system, typical amplification curves of CPCR are adopted for discussion.
Based on our or others’ previous work [29,34], a simplified wash strategy, which included one wash step with purification buffer and two repeated wash steps with TE buffer, was adopted to ensure the efficiency of nucleic acid extraction. To find out optimal size of wash buffer, different experiments with various wash volume were performed for comparison. In details, three different wash options with different volume of purification buffer/TE buffer (20/40 μL, 40/80 μL, and 80/160 μL) were evaluated and compared. Especially, the volume for TE buffer (40 μL, 80 μL, and 160 μL) corresponded to two repeated wash steps. A total of 200 μL of H1N1 virus culture stock (10 TCID50/mL) was used as the test sample. To ensure the efficiency of sample preparation, a proper rotation speed of centrifugation was chosen to allow lysed sample or reagent to flow through the FTA membrane filter with a proper speed. In fact, the centrifugation time for each step was intentionally lengthened to further dry the FTA membrane filter after sample or reagent pass through. Accordingly, it took ~2 min, 1–2 min, respectively, for lysed sample, purification buffer, or TE buffer to be driven through the FTA membrane filter, which contributed to a total sample preparation time of ~15 min, including another 8 min for lysis.
Based on our or others’ experience [29,34], comparing to a membrane with a standard size in a spin column, the FTA membrane filter with a small size around 2 mm incorporated in a microfluidic chip can be effectively washed using significantly lower volume of wash buffer. As shown in Figure 6A, end-point fluorescent images of amplicons were obtained after washing with 20/40 μL, 40/80 μL, and 80/160 μL purified buffer/TE buffer, respectively. It can be found that the fluorescence signal intensity is different from each other with three wash options. As shown in Figure 6B, when the volume of purification buffer or TE buffer is not less than 40 μL or 80 μL, satisfied efficiency for sample preparation can be reasonably achieved. Otherwise, when the volume of wash buffer is not enough, for example 20 μL and 40 μL, respectively, for purification buffer and TE buffer, the efficiency of sample preparation will be deteriorated because the lysis detergent, adsorbed proteins or other cellular debris in FTA membrane cannot be removed sufficiently after wash. Consequently, it also has been proved that it is unnecessary to wash the FTA membrane with larger volume of wash buffer, for example, 80 μL for purification buffer and 160 μL for TE buffer, because of the limited size of the adopted FTA membrane filter. Similar results were achieved with multiple repeated experiments. Therefore, an optimized wash scheme, including single wash with 40 μL of purification buffer combined with two duplicated washes with 80 μL of TE buffer, was adopted for efficient sample preparation.
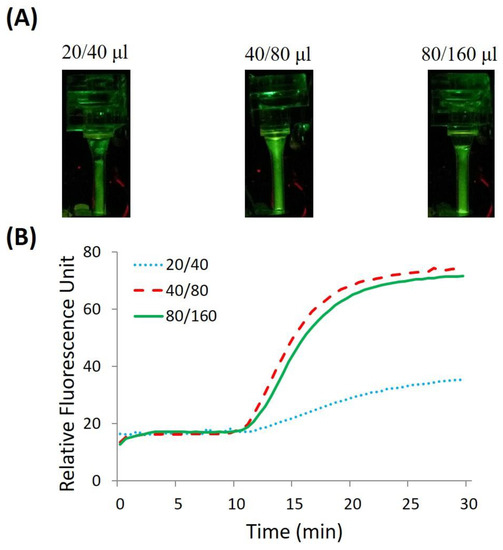
Figure 6.
CPCR with H1N1 templates. In details: (A) end-point fluorescent images of amplicons with different wash options; (B) analyzed real-time fluorescence curves with different wash options, the solid line, dashed line, and dotted line, respectively, correspond to different wash options of 80/160 μL, 40/80 μL, and 20/40 μL of purification buffer/TE buffer.
3.3. Amplification with the Integrated Microfluidic CPCR NAAT System
As described before, it is necessary to establish stable and desired temperature gradient along the capillary tube to achieve consistent and robust thermal convection for effective thermal cycling of PCR amplification. To determine the appropriate heating temperature for efficient amplification, multiple experiments with different heating temperatures were performed. A total of 200 μL of H1N1 virus culture stock (10 TCID50/mL) was used as the test sample. Sample preparation with the integrated microfluidic CPCR NAAT system was implemented as described above, and the proper heating temperature for effective amplification was discovered with experiments. Figure 7 shows that 95 °C was found to be the optimal temperature for consistent and sensitive amplification. Comparable results were achieved with multiple repeated experiments.
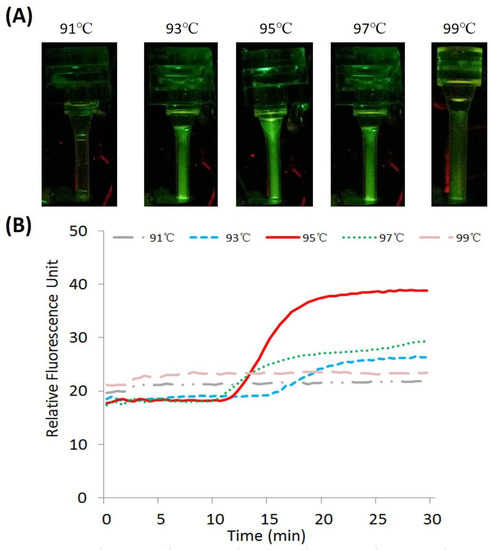
Figure 7.
CPCR with H1N1 virus templates. In details: (A) end-point fluorescent images of amplicons with different heating temperatures; (B) analyzed real-time fluorescence curves with different heating temperatures, the long-dashed line, dotted line, solid line, short-dashed line, and dashed and dotted line, respectively, correspond to experiments with different setpoint temperatures, 99 °C, 97 °C, 95 °C, 93 °C, and 91 °C.
As shown in Figure 7A, the end-point fluorescent images of amplicons were obtained when the reactor was heated to different temperatures, 91 °C, 93 °C, 95 °C, 97 °C, and 99 °C, respectively. The fluorescence signal intensity was found to be different from each other with five heating temperatures. As shown in Figure 7B, when the reaction temperature is too high or too low, for example, 99 °C or 91 °C, failed CPCR amplification occurs because they cannot establish desired thermal convection for effective thermal cycling. Furthermore, when the temperature is close to 95 °C, for example, 97 °C or 93 °C, CPCR amplification can be performed with low efficiency. Since CPCR amplification relies on space-domain thermal cycling introduced by thermal convection, the heating temperature will directly affect not only CPCR amplification, just like conventional PCR, but also decide the temperature gradient on the capillary tube, which is essential to consistent thermal convection. Therefore, to achieve efficient amplification, the optimal setpoint temperature for bottom-end heating should be set to 95 °C.
3.4. Detection of Influenza Virus with the Integrated Microfluidic CPCR NAAT System
The performance of the integrated microfluidic CPCR NAAT system is demonstrated with detection to H1N1 influenza viruses. A total of 200 μL of H1N1 virus culture stock with different concentrations (1000 to 1.0 TCID50/mL) was used in the experiments. A total of 1.0 TCID50/mL or better (101.29 TCID50/mL) is regarded as an acceptable limit for H1N1 detection of existing diagnosis kits. As described above, lysed sample, purification buffer, and TE buffer were consecutively loaded into the reagent storage chamber and then driven through the FTA membrane filter with centrifugation. Then, PCR reaction mix was loaded into the capillary tube and stayed above the bottom FTA membrane filter. Once the PCR mix was covered with mineral oil on top, CPCR amplification was ready. As proved by our previous work [21,22,23,24], it takes at most 30 min for CPCR amplification to be completed. With the developed system, rapid and simple nucleic acid test from sample to answer can be conveniently performed within 45 min. As shown in Figure 8B, at different times, real-time curves of all the positive tests start to increase, except that the negative control stays on the bottom all the time.
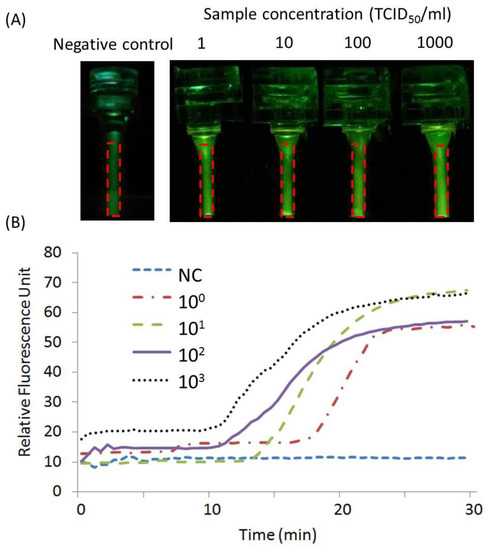
Figure 8.
Real-time CPCR with H1N1 templates. Real-time curves of intensity of fluorescence signal with diluted samples. In details: (A) end-point fluorescent images of amplicons with different concentrations and a negative control (NC); (B) analyzed real-time fluorescence curves, the short-dashed line, dashed and dotted line, long dashed line, solid line, and dotted line, respectively, correspond to negative control (NC), and samples with different concentrations from 1.0 to 1000 TCID50/mL.
As shown in Figure 8A, from the end-point fluorescent images of amplicons, the fluorescence signal intensity for all positive samples was found to be larger than the one with the negative control. From multiple real-time fluorescence curves (as shown in Figure 8B), all positive samples with different concentrations from 1.0 to 1000 TCID50/mL are found to be successfully amplified and detected within 30 min. Comparable results were achieved with multiple repeated experiments.
For the negative control, due to the property of Taqman probes, the test sample has background fluorescence before amplification. After amplification, based on the real-time fluorescence curve, the signal intensity of the negative control remains unchanged. In contrast, the positive test can be identified from the intensity change in the real-time fluorescence signal. Therefore, the test result is decided based on the change in the fluorescence signal intensity, instead of the end-point fluorescence signal intensity itself.
In principle, for isothermal amplification, the required reaction time for one positive test to reach the threshold value can be used to indicate the concentration of templates for quantitative or semi-quantitative detection. As proved by our previous work [21,22,23,24], nucleic acid amplification is achieved not only without periodic cycling, but also the dynamics of thermal convection from run to run may be slightly different at some degree; quantification detection cannot be achieved with the described CPCR system. Compared to conventional PCR (normally between 1–2 h), the amplification time can be significantly shortened with CPCR (within 30 min). Furthermore, the total detection time from sample to answer is shortened by combining a simplified, efficient sample preparation method with existing CPCR amplification, which finally becomes a highly efficient qualitative detection within just 45 min, which is beneficial to POC diagnostics.
Different from silicon membrane or magnetic beads [35], FTA membrane is not intentionally developed for standard sample preparation, for example, nucleic acid extraction with a large allowable size range for the processed sample. The unique functions of FTA membrane include cell lysis, nucleic acid extraction, storage, and protection. Especially, nucleic acid templates captured by the FTA membrane can be directly used for amplification without elution because of its PCR compatibility. For the described microfluidic system, the critical performance can be significantly improved by incorporating an FTA membrane filter into a CPCR capillary tube. However, because of the limited size of FTA membrane (~2 mm), which can be put into amplification without inhabitation, it is more reasonable for the FTA membrane to handle detection samples with small size or relatively high concentration. For test samples with a relatively low concentration in a relatively large size, the developed microfluidic system is incapable of detecting targets successfully. To handle large size of detection samples, one solution is to pre-concentrate the original samples first. Especially, when the original concentration of the detection sample is not an issue, the described microfluidic CPCR NAAT system can perform rapid, simple, and convenient nucleic acid analysis from sample to answer with significantly less time comparing to other existing systems. In the future, the performance of the described microfluidic system can be further improved with optimized sample preparation. This can be achieved by incorporating a pre-concentration module to further improve the limit of detection, especially for large-size samples with low concentration, or alternatively, by constructing a large CPCR reactor equipped with a PCR-compatible matrix with a large ratio of surface to volume for highly efficient nucleic acid extraction.
In terms of LOD (1.0 TCID50/mL), the reaction time (30 min), and the heating system, the CPCR device developed in this study is basically identical to the CPCR device in our previously published studies [24,36]. The previously reported CPCR systems developed by our group focus more on amplification and detection based on different system design. However, in this work, a partly integrated nucleic acid analysis system, including nucleic acid extraction and amplification, is developed based on CPCR. For nucleic acid extraction, an FTA membrane filter is integrated into the capillary tube reactor. Meanwhile, the capillary tube reactor embedded with an FTA filter also works as the CPCR amplification reactor, which is helpful to reduce the complexity of the nucleic acid analysis system. To assist partly automatic nucleic acid analysis, a portable companion device is developed to drive reagent for nucleic acid extraction based on centrifugal force, heat the capillary tube reactor for amplification, and collect the fluorescence signal with a smartphone camera as well. Different from our previous work, by combining CPCR with a simplified method for nucleic acid extraction, an integrated, real-time convective PCR system for isolation, amplification, and detection of nucleic acids is developed.
4. Conclusions and Outlook
Compared to conventional PCR, CPCR can substantially shorten the amplification time (by at least 50%, from about 1 h to less than 30 min) with efficient thermal cycling introduced by thermal convection. Previously, we developed different CPCR systems for rapid nucleic acid amplification [21,22,23,24]. To further take advantage of the characteristics of CPCR, it is highly desired to integrate CPCR amplification with sample preparation to perform rapid and simple nucleic acid test from sample to answer, which is beneficial to POC test. A simplified sample preparation method is developed by incorporating an FTA membrane filter into a CPCR capillary tube, which constructs a concise single CPCR reactor combining nucleic acid extraction, purification, amplification, and detection. Based on the single CPCR reactor, an integrated concise microfluidic CPCR NAAT system is developed for nucleic acid detection from sample to answer. To simplify fluid control in sample preparation, centrifugation is applied to smoothly drive lysed sample or wash buffer through the FTA membrane filter consecutively within a reasonable time. For CPCR amplification, the capillary tube is heated with a movable resistance heater. Meanwhile, a smartphone camera is applied to monitor the in situ fluorescence signal by taking pictures from the capillary tube. The wash scheme implemented on the microfluidic system is optimized to ensure the efficiency of nucleic acid extraction. An appropriate heating temperature range for efficient nucleic acid amplification is discovered for sensitive detection. Experimental results demonstrate that the integrated microfluidic CPCR NAAT system can perform H1N1 virus detection from sample to answer with reasonable sensitivity within just 45 min. Rapid, integrated, POC nucleic acid analysis from sample to answer has been proved to be conveniently achieved with the promoted CPCR system. In the next step, efforts can be made to further improve the performance of sample preparation for a more powerful microfluidic CPCR NAAT system with high sensitivity, repeatability, and consistency.
Author Contributions
Data curation, G.M. and M.G.; Formal analysis, M.G.M.; Funding acquisition, X.Q.; Investigation, K.L. and X.Y.; Methodology, S.G., N.X., D.Y. and X.Q.; Project administration, M.G.M.; Resources, X.Y.; Software, M.G.; Validation, G.M. and K.L.; Writing—original draft, G.M. and M.G.; Writing—review and editing, D.Y. and X.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 81871505, 81371711), National Science and Technology Major Project (Grant No. 2018ZX10732101-001-009), the Fundamental Research Funds for the Central Universities (Grant No. XK1802-4, PYBZ1830, PT1908) and the research fund to the top scientific and technological innovation team from Beijing University of Chemical Technology (Grant No. buctylkjcx06).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Dahl, F.; Banér, J.; Gullberg, M.; Mendel-Hartvig, M.; Landegren, U.; Nilsson, M. Circle-to-circle amplification for precise and sensitive DNA analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 4548–4553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, M.; Johnson, B.; Brahmasandra, S.; Handique, K.; Webster, J.; Krishnan, M.; Sammarco, T.; Man, P.; Jones, D.; Heldsinger, D. An integrated nanoliter DNA analysis device. Science 1998, 282, 484–487. [Google Scholar] [CrossRef] [Green Version]
- Dineva, M.A.; Mahilum-Tapay, L.; Lee, H. Sample preparation: A challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst 2007, 132, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.S.; Marshall, T.M. Challenges in implenting HIV load testing in South Africa. J. Infect. Dis. 2010, 201, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Technol. 2011, 29, 240–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, G.; Song, D.; Shrestha, S.; Miao, J.; Cui, L.; Guan, W. A field-deployable mobile molecular diagnostic system for malaria at the point of need. Lab Chip 2016, 16, 4341–4349. [Google Scholar] [CrossRef]
- Norian, H.; Field, R.M.; Kymissis, I.; Shepard, K.L. An integrated CMOS quantitative-polymerase-chain-reaction lab-on-chip for point-of-care diagnostics. Lab Chip 2014, 14, 4076–4084. [Google Scholar] [CrossRef]
- Jung, J.H.; Park, B.H.; Oh, S.J.; Choi, G.; Seo, T.S. Integrated centrifugal reverse transcriptase loop-mediated isothermal amplification microdevice for influenza A virus detection. Biosens. Bioelectron. 2015, 68, 218–224. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, S.; Yang, S.; Wang, T.H. An all-in-one microfluidic device for parallel DNA extraction and gene analysis. Biomed. Microdevices 2010, 12, 1043–1049. [Google Scholar] [CrossRef]
- Son, J.H.; Cho, B.; Hong, S.; Lee, S.H.; Hoxha, O.; Haack, A.J.; Lee, L.P. Ultrafast photonic PCR. Light Sci. Appl. 2015, 4, e280. [Google Scholar] [CrossRef] [Green Version]
- Oblath, E.A.; Henley, W.H.; Alarie, J.P.; Ramsey, J.M. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva. Lab Chip 2013, 13, 1325–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, F.; Hashsham, S.A. Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: A review. Anal. Chim. Acta 2012, 733, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mauk, M.G.; Chen, D.; Liu, C.; Bau, H.H. A large volume, portable, real-time PCR reactor. Lab Chip 2010, 10, 3170–3177. [Google Scholar] [CrossRef]
- Li, Z.; Ju, R.; Sekine, S.; Zhang, D.; Zhuang, S.; Yamaguchi, Y. All-in-one microfluidic device for on-site diagnosis of pathogens based on an integrated continuous flow PCR and electrophoresis biochip. Lab Chip 2019, 19, 2663–2668. [Google Scholar] [CrossRef]
- Krishnan, M.; Ugaz, V.M.; Burns, M.A. PCR in a Rayleigh-Bénard convection cell. Science 2002, 298, 793. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, V.K.; Bakthavathsalam, P.; Bergquist, P.L.; Sunna, A. A portable nucleic acid detection system using natural convection combined with a smartphone. Biosens. Bioelectron. 2019, 134, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.H.; Park, S.H.; Choi, Y.H. A palmtop PCR system with a disposable polymer chip operated by the thermosiphon effect. Lab Chip 2010, 10, 202–210. [Google Scholar] [CrossRef]
- Priye, A.; Hassan, Y.A.; Ugaz, V.M. Education: DNA replication using microscale natural convection. Lab Chip 2012, 12, 4946–4954. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, Y.; Zhang, D.; Zhuang, S.; Yamaguchi, Y. The development of a portable buoyancy-driven PCR system and its evaluation by capillary electrophoresis. Sens. Actuators B Chem. 2016, 230, 779–784. [Google Scholar] [CrossRef]
- Priye, A.; Wong, S.; Bi, Y.; Carpio, M.; Chang, J.; Coen, M.; Cope, D.; Harris, J.; Johnson, J.; Keller, A. Lab-on-a-drone: Toward pinpoint deployment of smartphone-enabled nucleic acid-based diagnostics for mobile health care. Anal. Chem. 2016, 88, 4651–4660. [Google Scholar] [CrossRef]
- Qiu, X.; Ge, S.; Gao, P.; Li, K.; Yang, S.; Zhang, S.; Ye, X.; Xia, N.; Qian, S. A Low-Cost and Fast Real-Time PCR System Based on Capillary Convection. Slas Technol. Transl. Life Sci. Innov. 2017, 23, 2951–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.; Zhang, S.; Mei, L.; Wu, D.; Guo, Q.; Li, K.; Ge, S.; Ye, X.; Xia, N.; Mauk, M.G. Characterization and analysis of real-time capillary convective PCR toward commercialization. Biomicrofluidics 2017, 11, 024103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.; Zhang, S.; Xiang, F.; Wu, D.; Guo, M.; Ge, S.; Li, K.; Ye, X.; Xia, N.; Qian, S. Instrument-free point-of-care molecular diagnosis of H1N1 based on microfluidic convective PCR. Sens. Actuators B Chem. 2017, 243, 738–744. [Google Scholar] [CrossRef]
- Qiu, X.; Shu, J.I.; Baysal, O.; Wu, J.; Qian, S.; Ge, S.; Li, K.; Ye, X.; Xia, N.; Yu, D. Real-time capillary convective PCR based on horizontal thermal convection. Microfluid. Nanofluidics 2019, 23, 39. [Google Scholar] [CrossRef]
- Shu, B.; Zhang, C.; Xing, D. A sample-to-answer, real-time convective polymerase chain reaction system for point-of-care diagnostics. Biosens. Bioelectron. 2017, 97, 360–368. [Google Scholar] [CrossRef]
- Shu, J.; Baysal, O.; Qian, S.; Qiu, X.; Wang, F. Performance of convective polymerase chain reaction by doubling time. Int. J. Heat Mass Transf. 2019, 133, 1230–1239. [Google Scholar] [CrossRef]
- Liu, C.; Mauk, M.G.; Hart, R.; Qiu, X.; Bau, H.H. A self-heating cartridge for molecular diagnostics. Lab Chip 2011, 11, 2686–2692. [Google Scholar] [CrossRef]
- Gan, W.; Zhuang, B.; Zhang, P.; Han, J.; Li, C.X.; Liu, P. A filter paper-based microdevice for low-cost, rapid, and automated DNA extraction and amplification from diverse sample types. Lab Chip 2014, 14, 3719–3728. [Google Scholar] [CrossRef]
- Qiu, X.; Mauk, M.G. An integrated, cellulose membrane-based PCR chamber. Microsyst. Technol. 2015, 21, 841–850. [Google Scholar] [CrossRef]
- Kim, J.; Jang, S.H.; Jia, G.; Zoval, J.V.; Da Silva, N.A.; Madou, M.J. Cell lysis on a microfluidic CD (compact disc). Lab Chip 2004, 4, 516–522. [Google Scholar] [CrossRef]
- Mitsakakis, K.; Zengerle, R.; Czilwik, G.; Zhao, Y.; Klein, V. C-reactive protein and interleukin 6 microfluidic immunoassays with on-chip pre-stored reagents and centrifugo-pneumatic liquid control. Lab Chip 2017, 17, 1666–1677. [Google Scholar]
- Zhang, S.; Lin, Y.; Wang, J.; Wang, P.; Chen, J.; Xue, M.; He, S.; Zhou, W.; Xu, F.; Liu, P.; et al. A convenient nucleic acid test on the basis of the capillary convective PCR for the on-site detection of Enterovirus 71. J. Mol. Diagn. 2014, 16, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xue, M.; Zhang, J.; Chen, Q.; Chen, J.; Wang, Z.; Zhou, W.; Chen, P.; Xia, N.; Ge, S. A one-step dipstick assay for the on-site detection of nucleic acid. Clin. Biochem. 2013, 46, 1852–1856. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Wan Abas, W.A.B.; Pingguan-Murphy, B.; et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2016, 16, 611–621. [Google Scholar] [CrossRef]
- Du, K.; Cai, H.; Park, M.; Wall, T.A.; Stott, M.A.; Alfson, K.J.; Griffiths, A.; Carrion, R.; Patterson, J.L.; Hawkins, A.R.; et al. Multiplexed efficient on-chip sample preparation and sensitive amplification-free detection of Ebola virus. Biosens. Bioelectron. 2017, 91, 489–496. [Google Scholar] [CrossRef]
- Qiu, X.; Ge, S.; Gao, P.; Li, K.; Yang, S.; Zhang, S.; Ye, X.; Xia, N.; Qian, S. A smartphone-based point-of-care diagnosis of H1N1 with microfluidic convection PCR. Microsyst. Technol. 2017, 23, 2951–2956. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).