Aptamer-Based Sensors for Thrombin Detection Application
Abstract
:1. Introduction
2. Thrombin
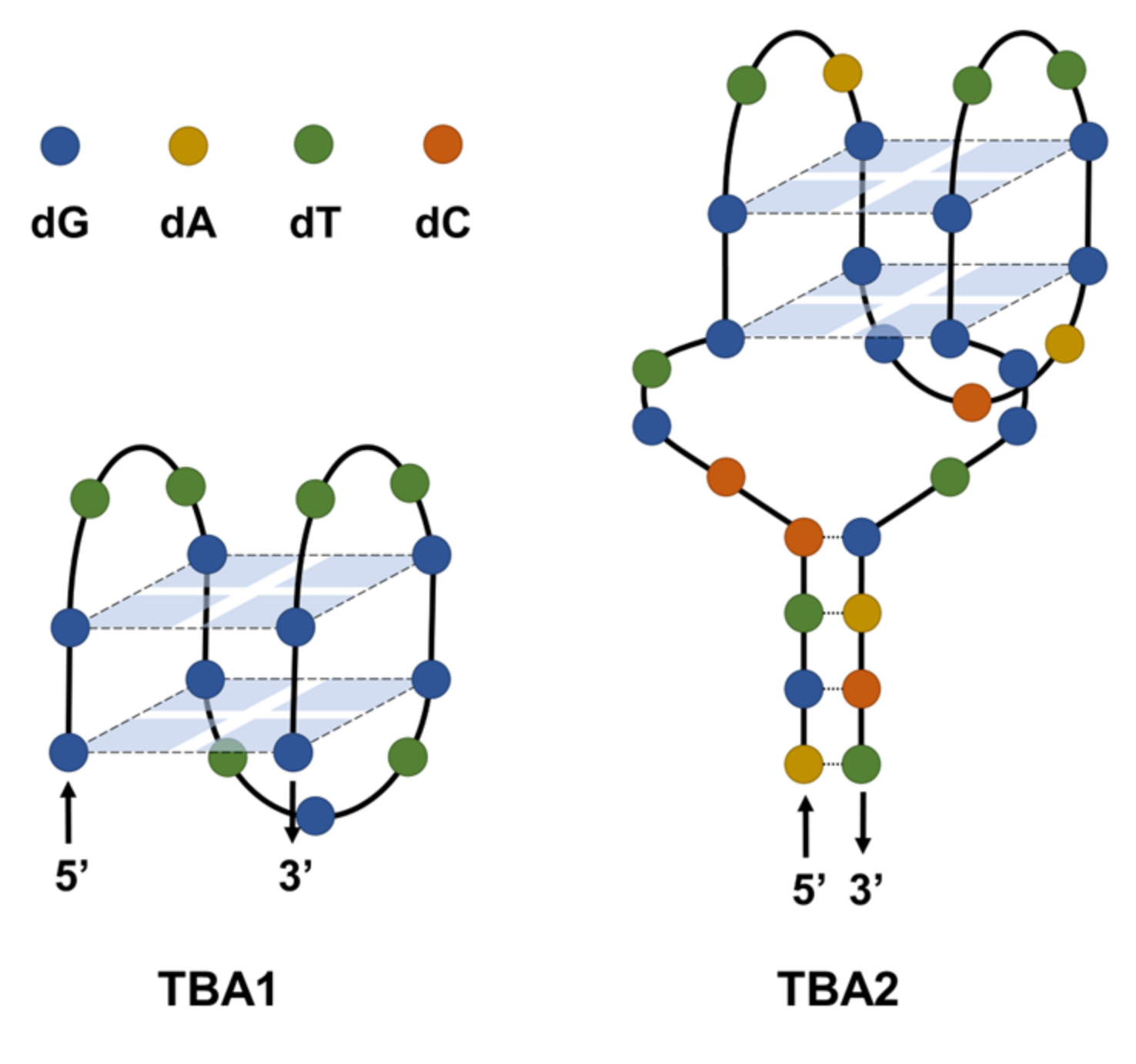
3. Thrombin Binding Aptamer (TBA)
4. Optical Aptamer Sensors for Thrombin Detection
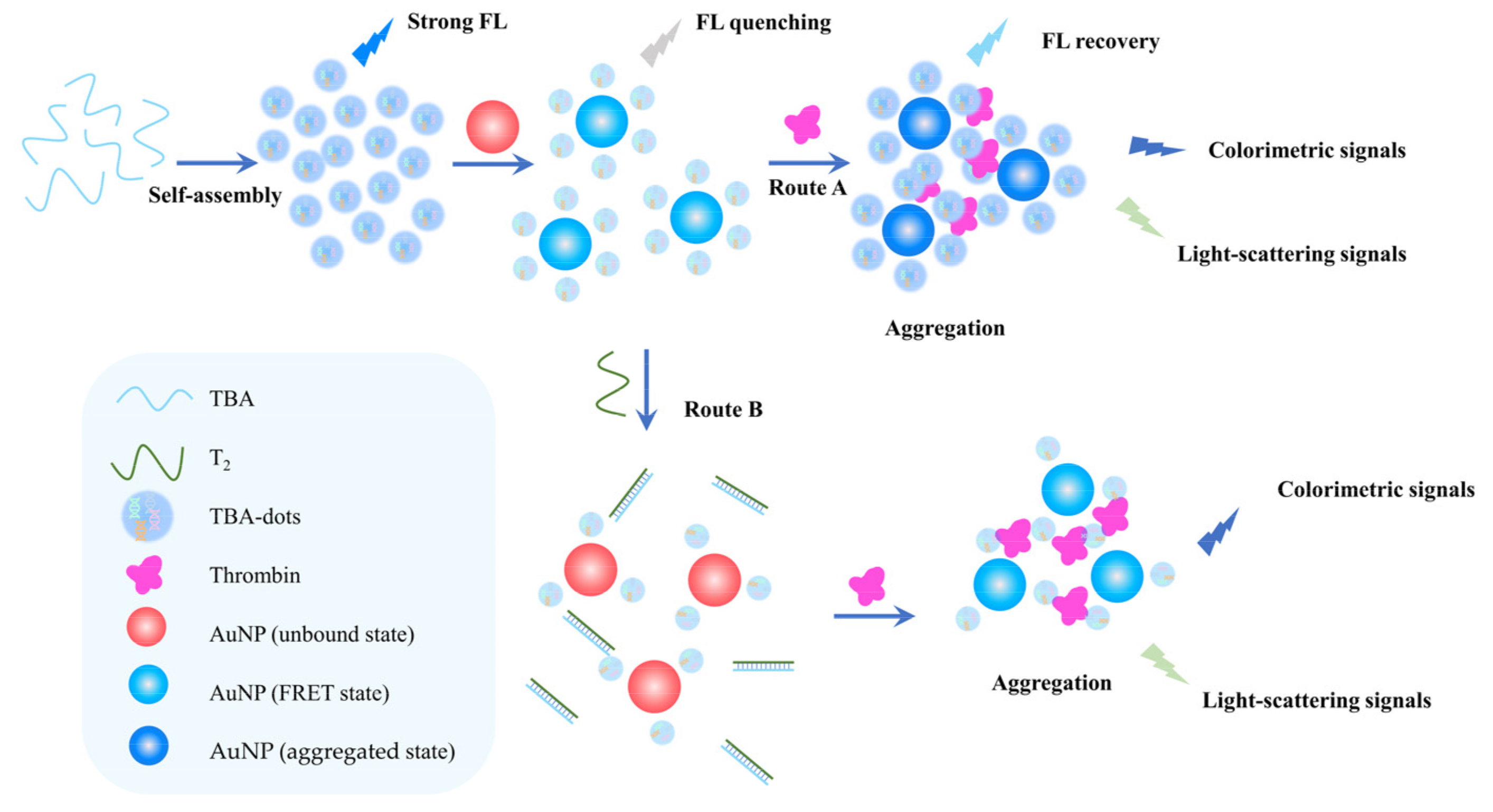
4.1. Fluorescent Sensors
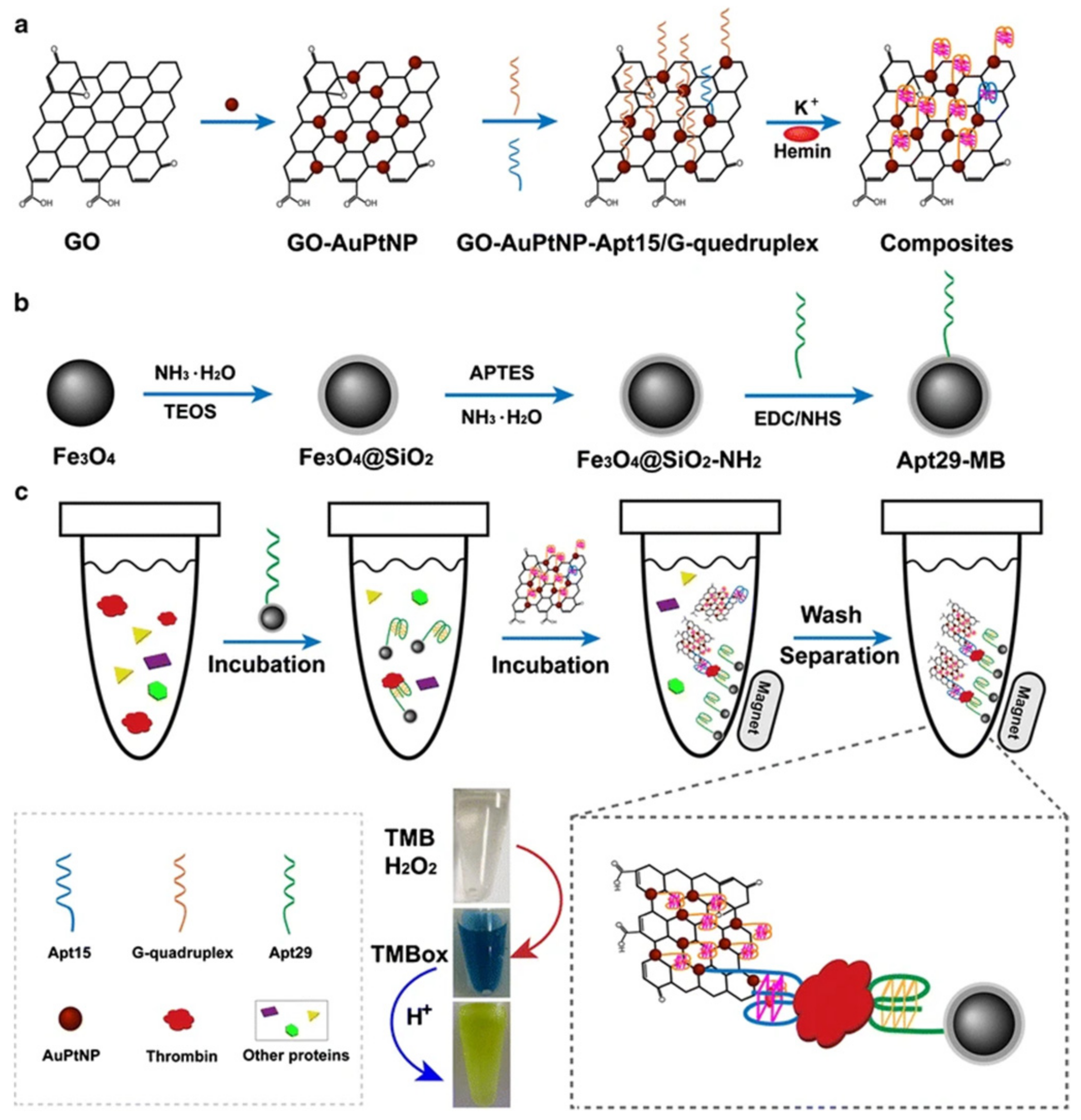
4.2. Colorimetric Sensors
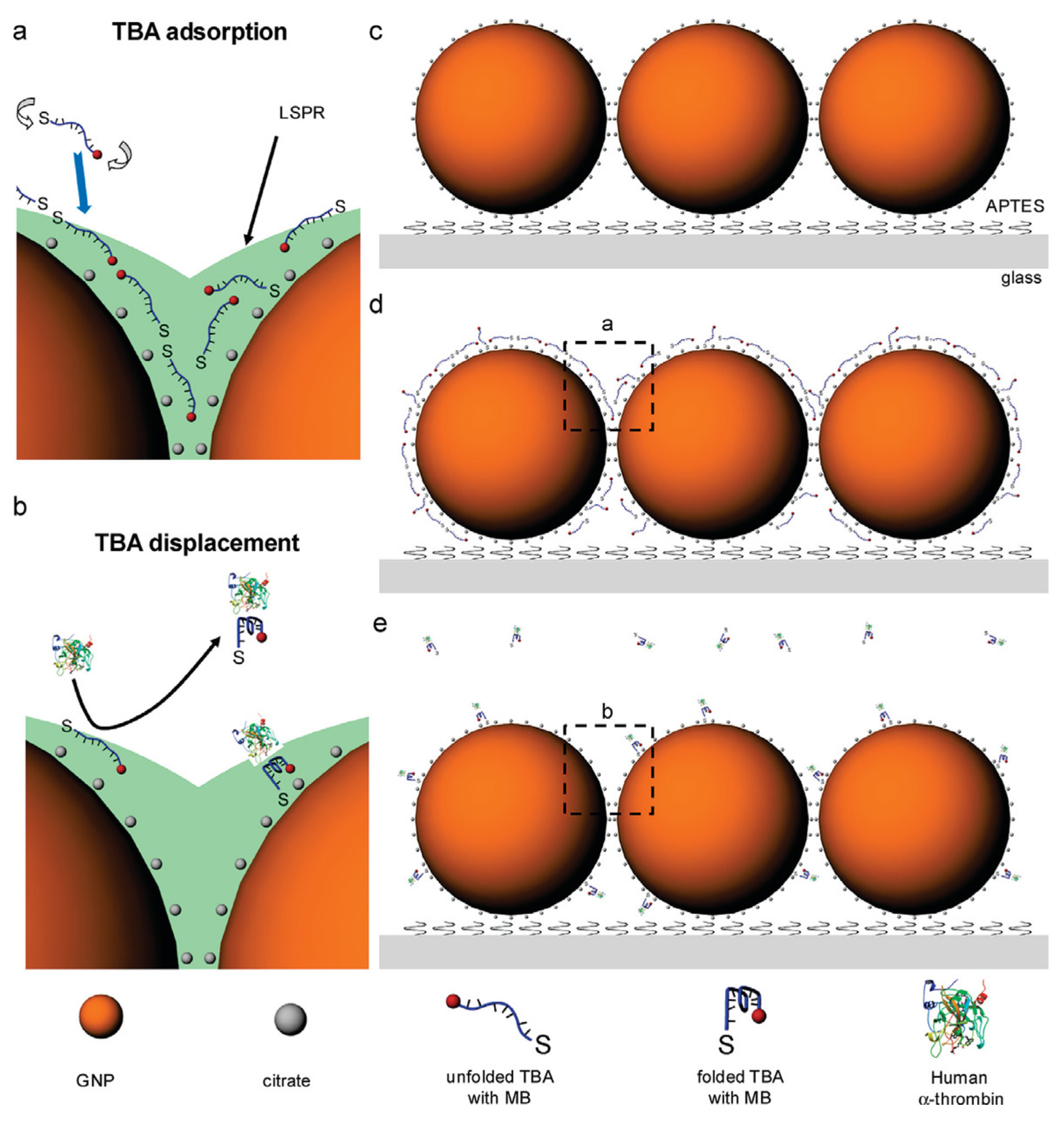
4.3. SERS-Based Sensors
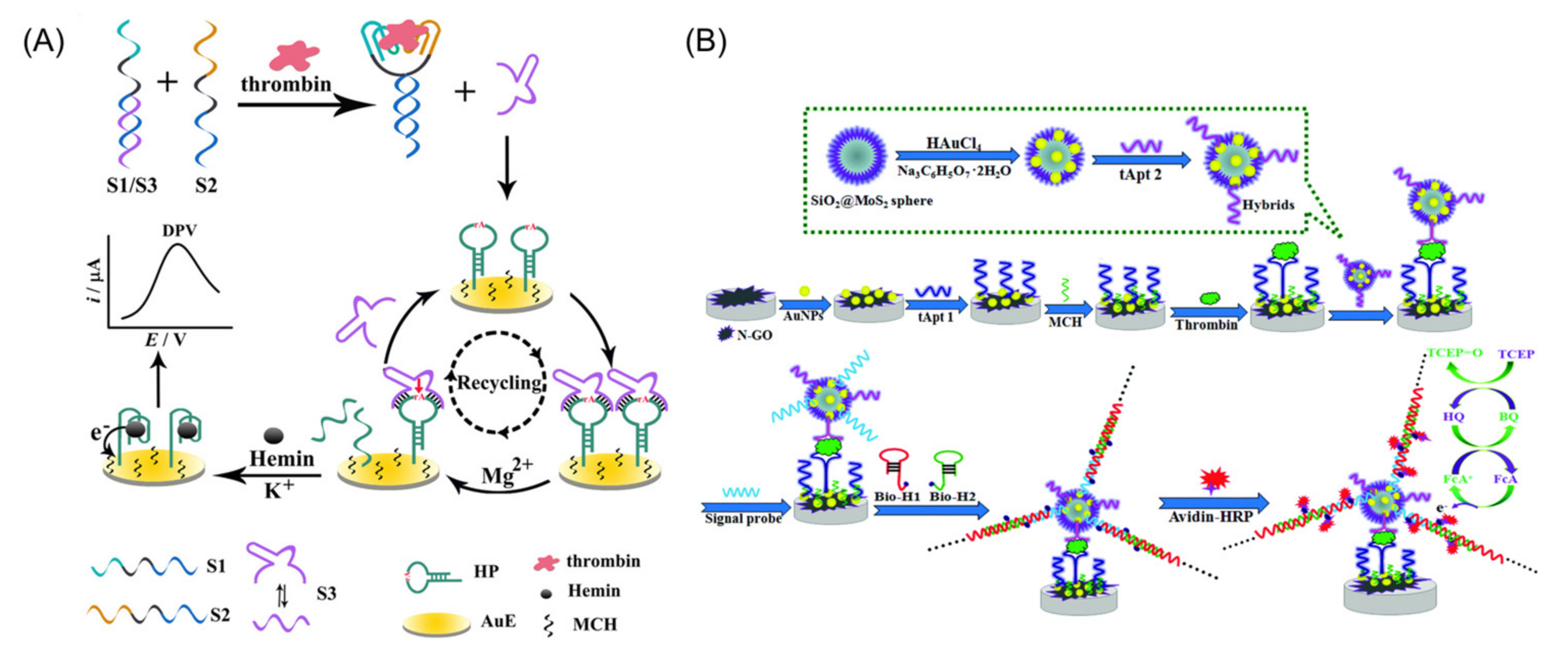
5. Electrochemical Aptamer Sensors for Thrombin Detection
6. Other Aptamer Sensors for Thrombin Detection
7. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Posma, J.J.N.; Posthuma, J.J.; Spronk, H.M.H. Coagulation and non-coagulation effects of thrombin. J. Thromb. Haemost. 2016, 14, 1908–1916. [Google Scholar] [CrossRef] [Green Version]
- Negrier, C.; Shima, M.; Hoffman, M. The central role of thrombin in bleeding disorders. Blood Rev. 2019, 38, 100582. [Google Scholar] [CrossRef] [PubMed]
- Krenzlin, H.; Lorenz, V.; Alessandri, B. The involvement of thrombin in the pathogenesis of glioblastoma. J. Neurosci. Res. 2017, 95, 2080–2085. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wright, J.; Wall, T.; Grammas, P. Brain Endothelial Cells Synthesize Neurotoxic Thrombin in Alzheimer’s Disease. Am. J. Pathol. 2010, 176, 1600–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsopanoglou, N.E.; Maragoudakis, M.E. Role of Thrombin in Angiogenesis and Tumor Progression. Semin. Thromb. Hemost. 2004, 30, 63–69. [Google Scholar]
- Mannironi, C.; Scerch, C.; Fruscoloni, P.; Tocchini-Valentini, G.P. Molecular recognition of amino acids by RNA aptamers: The evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA 2000, 6, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Huizenga, D.E.; Szostak, J.W. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef]
- Xu, W.; Ellington, A.D. Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc. Natl. Acad. Sci. USA 1996, 93, 7475–7480. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Shangguan, D.; Cao, Z.; Fang, X.; Tan, W. Cell-Specific Internalization Study of an Aptamer from Whole Cell Selection. Chem. Eur. J. 2008, 14, 1769–1775. [Google Scholar] [CrossRef]
- Gopinath, S.C.B. Methods developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef]
- Mukherjee, M.; Sistla, S.; Veerabhadraiah, S.R.; Bettadaiah, B.; Thakur, M.; Bhatt, P. DNA aptamer selection and detection of marine biotoxin 20 Methyl Spirolide G. Food Chem. 2021, 363, 130332. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Dong, Y.; Han, X.; Wang, S.; Liang, X. Aptamers and Aptasensors for Highly Specific Recognition and Sensitive Detection of Marine Biotoxins: Recent Advances and Perspectives. Toxins 2018, 10, 427. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Zhang, C.; Ma, T.; Liu, X.; Chen, Z.; Li, S.; Deng, Y. Advances in aptamer screening and aptasensors’ detection of heavy metal ions. J. Nanobiotechnol. 2021, 19, 166. [Google Scholar] [CrossRef]
- Jolly, P.; Formisano, N.; Estrela, P. DNA aptamer-based detection of prostate cancer. Chem. Pap. 2015, 69, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Bakhtiari, H.; Palizban, A.A.; Khanahmad, H.; Mofid, M.R. Aptamer-based approaches for in vitro molecular detection of cancer. Res. Pharm. Sci. 2020, 15, 107–122. [Google Scholar]
- Zhao, X.; Dai, X.; Zhao, S.; Cui, X.; Gong, T.; Song, Z.; Meng, H.; Zhang, X.; Yu, B. Aptamer-based fluorescent sensors for the detection of cancer biomarkers. Spectrochim. Acta Part A 2021, 247, 119038. [Google Scholar] [CrossRef]
- Ronda, L.; Tonelli, A.; Sogne, E.; Autiero, I.; Spyrakis, F.; Pellegrino, S.; Abbiati, G.; Maffioli, E.; Schulte, C.; Piano, R.; et al. Rational Design of a User-Friendly Aptamer/Peptide-Based Device for the Detection of Staphylococcus Aureus. Sensors 2020, 20, 4977. [Google Scholar] [CrossRef]
- Cunha, I.; Biltes, R.; Sales, M.; Vasconcelos, V. Aptamer-Based Biosensors to Detect Aquatic Phycotoxins and Cyanotoxins. Sensors 2018, 18, 2367. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.Y.; Kim, S.T.; Han, H.-S.; Kim, K.; Han, J.S. Optical Aptamer Probes of Fluorescent Imaging to Rapid Monitoring of Circulating Tumor Cell. Sensors 2016, 16, 1909. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, X.; Feng, P.; Han, Q.; Liu, W.; Lu, Y.; Song, C.; Li, F. Photodriven Regeneration of G-Quadruplex Aptasensor for Sensitively Detecting Thrombin. Anal. Chem. 2020, 92, 7419–7424. [Google Scholar] [CrossRef]
- Malecka, K.; Ferapontova, E.E. Femtomolar Detection of Thrombin in Serum and Cerebrospinal Fluid via Direct Electrocatalysis of Oxygen Reduction by the Covalent G4-Hemin-Aptamer Complex. ACS Appl. Mater. Interfaces 2021, 13, 37979–37988. [Google Scholar] [CrossRef]
- Qiu, F.; Gan, X.; Jiang, B.; Yuan, R.; Xiang, Y. Electrode immobilization-free and sensitive electrochemical sensing of thrombin via magnetic nanoparticle-decorated DNA polymers. Sens. Actuators B Chem. 2021, 331, 129395. [Google Scholar] [CrossRef]
- Motta, J.-P.; Palese, S.; Giorgio, C.; Chapman, K.; Denadai-Souza, A.; Rousset, P.; Sagnat, D.; Guiraud, L.; Edir, A.; Seguy, C.; et al. Increased mucosal thrombin is associated with Crohn’s disease and causes inflammatory damage through protease-activated receptors activation. J. Crohn’s Colitis 2021, 15, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.; Jaberi, N.; Avan, A.; Ryzhikov, M.; Keramati, M.R.; Parizadeh, M.R.; Hassanian, S.M. Role of thrombin in the pathogenesis of central nervous system inflammatory diseases. J. Cell. Physiol. 2017, 232, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Reddel, C.J.; Tan, C.W.; Chen, V.M. Thrombin Generation and Cancer: Contributors and Consequences. Cancers 2019, 11, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. Pathologies at the nexus of blood coagulation and inflammation: Thrombin in hemostasis, cancer, and beyond. J. Mol. Med. 2013, 91, 1257–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, G.; Sørensen, B.; Dargaud, Y.; Negrier, C.; Brummel-Ziedins, K.; Key, N.S. Thrombin generation and whole blood viscoelastic assays in the management of hemophilia: Current state of art and future perspectives. Blood 2013, 121, 1944–1950. [Google Scholar] [CrossRef] [Green Version]
- Monroe, D.M.; Hoffman, M. What does it take to make the perfect clot? Arterioscler. Thromb. Vasc. Biol. 2006, 26, 41–48. [Google Scholar] [CrossRef]
- Monroe, D.M.; Hoffman, M.; Roberts, H.R. Platelets and Thrombin Generation. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1381–1389. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Garton, H.J.L.; Hua, Y.; Keep, R.F.; Xi, G. The Role of Thrombin in Brain Injury After Hemorrhagic and Ischemic Stroke. Transl. Stroke Res. 2021, 12, 496–511. [Google Scholar] [CrossRef]
- Ito, K.; Hongo, K.; Date, T.; Ikegami, M.; Hano, H.; Owada, M.; Morimoto, S.; Kashiwagi, Y.; Katoh, D.; Yoshino, T.; et al. Tissue thrombin is associated with the pathogenesis of dilated cardiomyopathy. Int. J. Cardiol. 2017, 228, 821–827. [Google Scholar] [CrossRef]
- Hayosh, Z.I.; Abu Bandora, E.; Shelestovich, N.; Nulman, M.; Bakon, M.; Yaniv, G.; Khaitovitch, B.; Balan, S.; Gerasimova, A.; Drori, T.; et al. In-thrombus thrombin secretion: A new diagnostic marker of atrial fibrillation in cryptogenic stroke. J. Neurointerv. Surg. 2021, 13, 799–804. [Google Scholar] [CrossRef]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Feistritzer, C.; Mosheimer, B.A.; Kaneider, N.C.; Riewald, M.; Patsch, J.R.; Wiedermann, C.J. Thrombin Affects Eosinophil Migration via Protease-Activated Receptor-1. Int. Arch. Allergy Immunol. 2004, 135, 12–16. [Google Scholar] [CrossRef]
- Martorell, L.; Martínez-González, J.; Rodriguez, C.; Gentile, M.; Calvayrac, O.; Badimon, L. Thrombin and protease-activated receptors (PARs) in atherothrombosis. Thromb. Haemost. 2008, 99, 305–315. [Google Scholar] [CrossRef]
- Ossovskaya, V.S.; Bunnett, N.W. Protease-Activated Receptors: Contribution to Physiology and Disease. Physiol. Rev. 2004, 84, 579–621. [Google Scholar] [CrossRef] [Green Version]
- Wojtukiewicz, M.Z.; Hempel, D.; Sierko, E.; Tucker, S.C.; Honn, K.V. Thrombin—Unique coagulation system protein with multifaceted impacts on cancer and metastasis. Cancer Metastasis Rev. 2016, 35, 213–233. [Google Scholar] [CrossRef]
- Owen, W.G. PAR-3 is a low-affinity substrate, high affinity effector of thrombin. Biochem. Biophys. Res. Commun. 2003, 305, 166–168. [Google Scholar] [CrossRef]
- Di Cera, E. Thrombin interactions. Chest 2003, 124, 11S–17S. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Kubik, M.F.; Stephens, A.W.; Schneider, D.; Marlar, R.A.; Tasset, D. High-affinity RNA ligands to human α-thrombin. Nucleic Acids Res. 1994, 22, 2619–2626. [Google Scholar] [CrossRef] [Green Version]
- Macaya, R.; Waldron, J.A.; Beutel, B.A.; Gao, H.; Joesten, M.E.; Yang, M.; Patel, R.; Bertelsen, A.H.; Cook, A.F. Structural and Functional Characterization of Potent Antithrombotic Oligonucleotides Possessing Both Quadruplex and Duplex Motifs. Biochemistry 1995, 34, 4478–4492. [Google Scholar] [CrossRef]
- Krauss, I.R.; Spiridonova, V.; Pica, A.; Napolitano, V.; Sica, F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016, 44, 983–991. [Google Scholar]
- Wakui, K.; Yoshitomi, T.; Yamaguchi, A.; Tsuchida, M.; Saito, S.; Shibukawa, M.; Furusho, H.; Yoshimoto, K. Rapidly Neutralizable and Highly Anticoagulant Thrombin-Binding DNA Aptamer Discovered by MACE SELEX. Mol. Ther. Nucleic Acids 2019, 16, 348–359. [Google Scholar] [CrossRef] [Green Version]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Krauss, I.R.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, 46, 12177–12185. [Google Scholar] [CrossRef] [Green Version]
- Ikebukuro, K.; Okumura, Y.; Sumikura, K.; Karube, I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005, 33, e108. [Google Scholar] [CrossRef] [Green Version]
- Troisi, R.; Balasco, N.; Autiero, I.; Vitagliano, L.; Sica, F. Exosite Binding in Thrombin: A Global Structural/Dynamic Overview of Complexes with Aptamers and Other Ligands. Int. J. Mol. Sci. 2021, 22, 10803. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [Green Version]
- Kelly, J.A.; Feigon, J.; Yeates, T. Reconciliation of the X-ray and NMR Structures of the Thrombin-Binding Aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1996, 256, 417–422. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, X.; Bing, T.; Cao, Z.; Shangguan, D. General Peroxidase Activity of G-Quadruplex−Hemin Complexes and Its Application in Ligand Screening. Biochemistry 2009, 48, 7817–7823. [Google Scholar] [CrossRef]
- Wu, Y.; Zou, L.; Lei, S.; Yu, Q.; Ye, B. Highly sensitive electrochemical thrombin aptasensor based on peptide-enhanced electrocatalysis of hemin/G-quadruplex and nanocomposite as nanocarrier. Biosens. Bioelectron. 2017, 97, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiao, Y.; Liu, Q.; Chen, Z. Colorimetric Detection of Thrombin Based on Intensity of Gold Nanoparticle Oligomers with Dark-Field Microscope. ACS Sustain. Chem. Eng. 2018, 6, 6738–6745. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, Q.; Li, L.; Kong, J.; Zhang, X. Click chemistry-based aptasensor for highly sensitive electrochemical detection of thrombin. Anal. Methods 2017, 9, 3825–3830. [Google Scholar] [CrossRef]
- Wu, S.; Min, H.; Shi, W.; Cheng, P. Multicenter metal–organic framework-based ratiometric fluorescent sensors. Adv. Mater. 2019, 32, 1805871. [Google Scholar] [CrossRef]
- Liu, J.; Li, R.; Yang, B. Carbon Dots: A New Type of Carbon-Based Nanomaterial with Wide Applications. ACS Cent. Sci. 2020, 6, 2179–2195. [Google Scholar] [CrossRef]
- Hajian, R.; Balderston, S.; Tran, T.; DeBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- Ma, L.; Sun, N.; Zhang, J.; Tu, C.; Cao, X.; Duan, D.; Diao, A.; Man, S. Polyethylenimine-coated Fe3O4 nanoparticles effectively quench fluorescent DNA, which can be developed as a novel platform for protein detection. Nanoscale 2017, 9, 17699–17703. [Google Scholar] [CrossRef]
- Du, C.; Hu, Y.; Zhang, Q.; Guo, Z.; Ge, G.; Wang, S.; Zhai, C.; Zhu, M. Competition-derived FRET-switching cationic conjugated polymer-Ir(III) complex probe for thrombin detection. Biosens. Bioelectron. 2018, 100, 132–138. [Google Scholar] [CrossRef]
- Umrao, S.; Jain, V.; Anusha; Chakraborty, B.; Roy, R. Protein-induced fluorescence enhancement as aptamer sensing mechanism for thrombin detection. Sens. Actuators B Chem. 2018, 267, 294–301. [Google Scholar] [CrossRef]
- Zeng, X.; Zhou, Q.; Wang, L.; Zhu, X.; Cui, K.; Peng, X.; Steele, T.W.J.; Chen, H.; Xu, H.; Zhou, Y. A Fluorescence Kinetic-Based Aptasensor Employing Stilbene Isomerization for Detection of Thrombin. Materials 2021, 14, 6927. [Google Scholar] [CrossRef]
- Li, J.; Zhou, W.; Yuan, R.; Xiang, Y. Aptamer proximity recognition-dependent strand translocation for enzyme-free and amplified fluorescent detection of thrombin via catalytic hairpin assembly. Anal. Chim. Acta 2018, 1038, 126–131. [Google Scholar] [CrossRef]
- Chen, X.; Li, T.; Tu, X.; Luo, L. Label-free fluorescent aptasensor for thrombin detection based on exonuclease I assisted target recycling and SYBR Green I aided signal amplification. Sens. Actuators B Chem. 2018, 265, 98–103. [Google Scholar] [CrossRef]
- Gao, L.; Li, Q.; Deng, Z.; Brady, B.; Xia, N.; Zhou, Y.; Shi, H. Highly sensitive protein detection via covalently linked aptamer to MoS2 and exonuclease-assisted amplification strategy. Int. J. Nanomed. 2017, 12, 7847–7853. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Sun, M.; Luo, F.; Xu, K.; Lin, Z.; Zhang, L. Stimulus-response click chemistry based aptamer-functionalized mesoporous silica nanoparticles for fluorescence detection of thrombin. Talanta 2018, 178, 563–568. [Google Scholar] [CrossRef]
- Li, J.; Hu, X.; Shi, S.; Zhang, Y.; Yao, T. Three label-free thrombin aptasensors based on aptamers and [Ru(bpy)2(o-mopip)]2+. J. Mater. Chem. B 2016, 4, 1361–1367. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Xu, Y.; Cheng, G.; He, P.; Fang, Y. Simultaneously fluorescence detecting thrombin and lysozyme based on magnetic nanoparticle condensation. Talanta 2009, 79, 557–561. [Google Scholar] [CrossRef]
- Qi, Y.; Chen, Y.; Xiu, F.-R.; Hou, J. An aptamer-based colorimetric sensing of acetamiprid in environmental samples: Convenience, sensitivity and practicability. Sens. Actuators B Chem. 2020, 304, 127359. [Google Scholar] [CrossRef]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317, 126459. [Google Scholar] [CrossRef]
- Cheng, R.; Liu, S.; Shi, H.; Zhao, G. A highly sensitive and selective aptamer-based colorimetric sensor for the rapid detection of PCB 77. J. Hazard. Mater. 2018, 341, 373–380. [Google Scholar] [CrossRef]
- Zhu, X.; Tang, L.; Wang, J.; Peng, B.; Ouyang, X.; Tan, J.; Yu, J.; Feng, H.; Tang, J. Enhanced peroxidase-like activity of boron nitride quantum dots anchored porous CeO2 nanorods by aptamer for highly sensitive colorimetric detection of kanamycin. Sens. Actuators B Chem. 2021, 330, 129318. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. G-quadruplex-based DNAzyme for facile colorimetric detection of thrombin. Chem. Commun. 2008, 31, 3654–3656. [Google Scholar] [CrossRef]
- Wang, L.; Yang, W.; Li, T.; Li, D.; Cui, Z.; Wang, Y.; Ji, S.; Song, Q.; Shu, C.; Ding, L. Colorimetric determination of thrombin by exploiting a triple enzyme-mimetic activity and dual-aptamer strategy. Mikrochim. Acta 2017, 184, 3145–3151. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, Y.; Binyam, A.; Liu, M.; Wu, Y.; Li, F. Discovering the enzyme mimetic activity of metal-organic framework (MOF) for label-free and colorimetric sensing of biomolecules. Biosens. Bioelectron. 2016, 86, 432–438. [Google Scholar] [CrossRef]
- Liu, M.; Li, J.; Li, B. A colorimetric aptamer biosensor based on cationic polythiophene derivative as peroxidase mimetics for the ultrasensitive detection of thrombin. Talanta 2017, 175, 224–228. [Google Scholar] [CrossRef]
- Lee, W.; Shaban, S.M.; Pyun, D.G.; Kim, D.-H. Solid-phase colorimetric apta-biosensor for thrombin detection. Thin Solid Films 2019, 686, 137428. [Google Scholar] [CrossRef]
- Kuang, L.; Cao, S.-P.; Zhang, L.; Li, Q.-H.; Liu, Z.-C.; Liang, R.-P.; Qiu, J.-D. A novel nanosensor composed of aptamer bio-dots and gold nanoparticles for determination of thrombin with multiple signals. Biosens. Bioelectron. 2016, 85, 798–806. [Google Scholar] [CrossRef]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Moller, R.; Hoagland, D.; Oishi, K.; Horiuchi, S.; et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat. Biomed. Eng. 2021, 5, 815–829. [Google Scholar] [CrossRef]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.; Li, J.; Qiang, W.; Sun, L.; Li, H.; Xu, D. Microfluidic chip-based silver nanoparticles aptasensor for colorimetric detection of thrombin. Talanta 2016, 150, 81–87. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Cho, H.; Baker, B.; Wachsmann-Hogiu, S.; Pagba, C.V.; Laurence, T.A.; Lane, S.M.; Lee, L.P.; Tok, J.B.-H. Aptamer-Based SERRS Sensor for Thrombin Detection. Nano Lett. 2008, 8, 4386–4390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, T.; Wu, X.; Xu, L.; Liu, L.; Ma, W.; Kuang, H.; Xu, C. Ultrasensitive Detection of Prostate-Specific Antigen and Thrombin Based on Gold-Upconversion Nanoparticle Assembled Pyramids. Small 2017, 13, 1603944. [Google Scholar] [CrossRef] [PubMed]
- del Campo, F.J. Miniaturization of electrochemical flow devices. Electrochem. Commun. 2014, 45, 91–94. [Google Scholar] [CrossRef]
- Shinwari, M.W.; Zhitomirsky, D.; Deen, I.A.; Selvaganapathy, P.R.; Deen, M.J.; Landheer, D. Microfabricated reference electrodes and their biosensing applications. Sensors 2010, 10, 1679–1715. [Google Scholar] [CrossRef]
- Yang, J.; Dou, B.; Yuan, R.; Xiang, Y. Aptamer/protein proximity binding-triggered molecular machine for amplified electrochemical sensing of thrombin. Anal. Chem. 2017, 89, 5138–5143. [Google Scholar] [CrossRef]
- Chung, S.; Moon, J.-M.; Choi, J.; Hwang, H.; Shim, Y.-B. Magnetic force assisted electrochemical sensor for the detection of thrombin with aptamer-antibody sandwich formation. Biosens. Bioelectron. 2018, 117, 480–486. [Google Scholar] [CrossRef]
- Fan, T.; Du, Y.; Yao, Y.; Wu, J.; Meng, S.; Luo, J.; Zhang, X.; Yang, D.; Wang, C.; Qian, Y.; et al. Rolling circle amplification triggered poly adenine-gold nanoparticles production for label-free electrochemical detection of thrombin. Sens. Actuators B Chem. 2018, 266, 9–18. [Google Scholar] [CrossRef]
- Gao, X.; Cai, Q.; Li, H.; Jie, G. Supersandwich Nanowire/Quantum Dots Sensitization Structure-Based Photoelectrochemical “Signal-On” Platform for Ultrasensitive Detection of Thrombin. Anal. Chem. 2020, 92, 6734–6740. [Google Scholar] [CrossRef]
- Guo, W.-J.; Yang, X.-Y.; Wu, Z.; Zhang, Z.-L. A colorimetric and electrochemical dual-mode biosensor for thrombin using a magnetic separation technique. J. Mater. Chem. B 2020, 8, 3574–3581. [Google Scholar] [CrossRef]
- Liang, X.; Zhao, J.; Ma, Z. Improved binding induced self-assembled DNA to achieve ultrasensitive electrochemical proximity assay. Sens. Actuators B Chem. 2020, 304, 127278. [Google Scholar] [CrossRef]
- Yang, J.; Dou, B.; Yuan, R.; Xiang, Y. Proximity Binding and Metal Ion-Dependent DNAzyme Cyclic Amplification-Integrated Aptasensor for Label-Free and Sensitive Electrochemical Detection of Thrombin. Anal. Chem. 2016, 88, 8218–8223. [Google Scholar] [CrossRef]
- Shuai, H.-L.; Wu, X.; Huang, K.-J. Molybdenum disulfide sphere-based electrochemical aptasensors for protein detection. J. Mater. Chem. B 2017, 5, 5362–5372. [Google Scholar] [CrossRef]
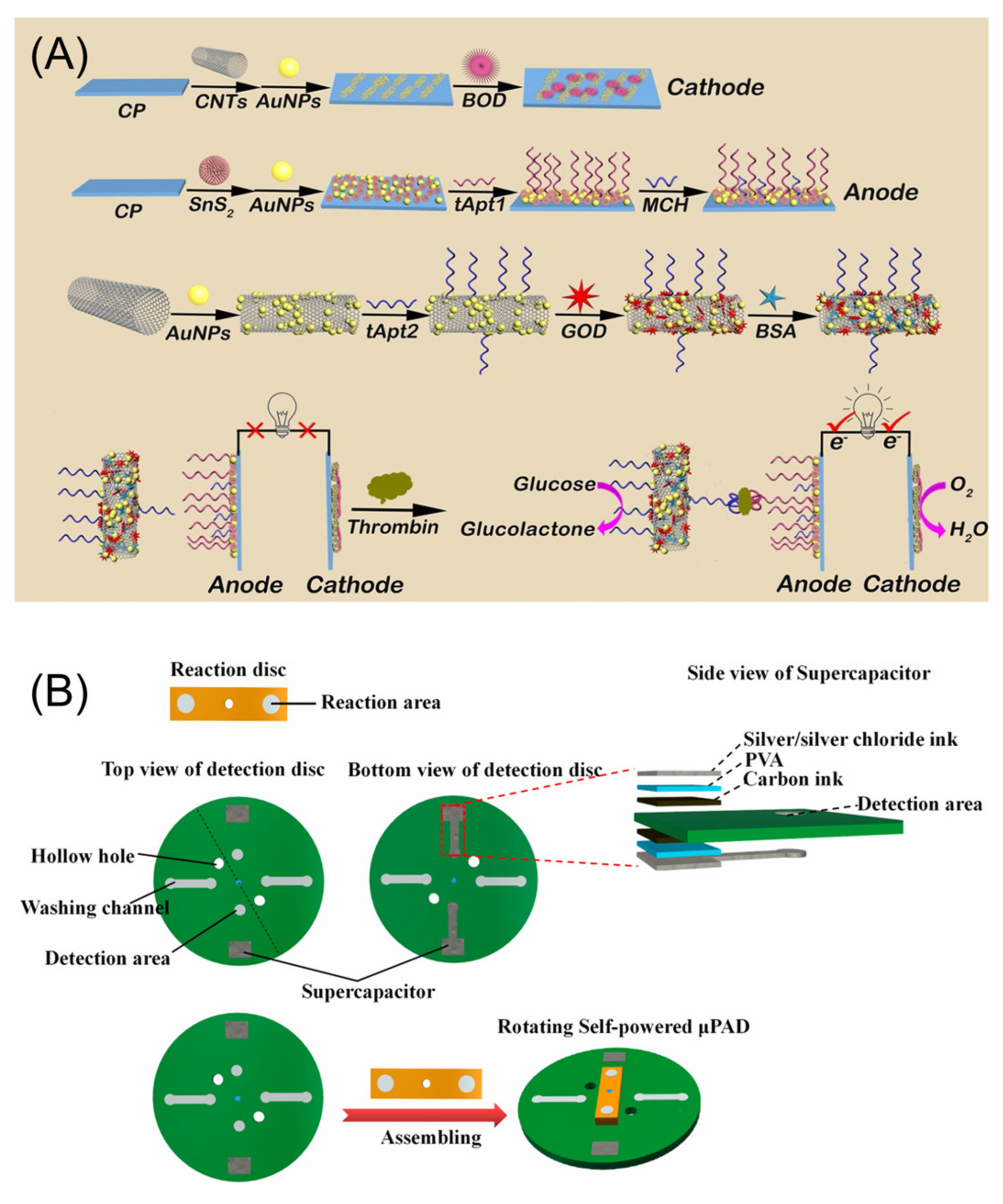
- Wang, Y.; Wang, F.; Han, Z.; Huang, K.; Wang, X.; Liu, Z.; Wang, S.; Lu, Y. Construction of sandwiched self-powered biosensor based on smart nanostructure and capacitor: Toward multiple signal amplification for thrombin detection. Sens. Actuators B Chem. 2020, 304, 127418. [Google Scholar] [CrossRef]
- Li, Q.; Xu, Y.; Qi, J.; Zheng, X.; Liu, S.; Lin, D.; Zhang, L.; Liu, P.; Li, B.; Chen, L. A self-powered rotating paper-based analytical device for sensing of thrombin. Sens. Actuators B Chem. 2022, 351, 130917. [Google Scholar] [CrossRef]
- Zafar, M.S.; Dastgeer, G.; Kalam, A.; Al-Sehemi, A.G.; Imran, M.; Kim, Y.H.; Chae, H. Precise and Prompt Analyte Detection via Ordered Orientation of Receptor in WSe2-Based Field Effect Transistor. Nanomaterials 2022, 12, 1305. [Google Scholar] [CrossRef]
- Martínez-Pérez, P.; Gómez-Gómez, M.; Angelova, T.; Griol, A.; Hurtado, J.; Bellieres, L.; Garcia-Ruperez, J. Continuous detection of increasing concentrations of thrombin employing a label-free photonic crystal aptasensor. Micromachines 2020, 11, 464. [Google Scholar] [CrossRef]
- Huang, Y.; Cheng, Z.; Han, R.; Gao, X.; Qian, L.; Wen, Y.; Zhang, X.; Liu, G. Target-induced molecular-switch on triple-helix DNA-functionalized carbon nanotubes for simultaneous visual detection of nucleic acids and proteins. Chem. Commun. 2020, 56, 13657–13660. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Niu, J.; Jiang, D.; Xiao, D.; Zhou, C. One-step sensitive thrombin detection based on a nanofibrous sensing platform. J. Mater. Chem. B 2019, 7, 5161–5169. [Google Scholar] [CrossRef]
- Ma, H.; Lu, S.; Xie, Q.; Wang, T.; Lu, H.; Yu, L. A stable liquid crystals sensing platform decorated with cationic surfactant for detecting thrombin. Microchem. J. 2021, 170, 106698. [Google Scholar] [CrossRef]
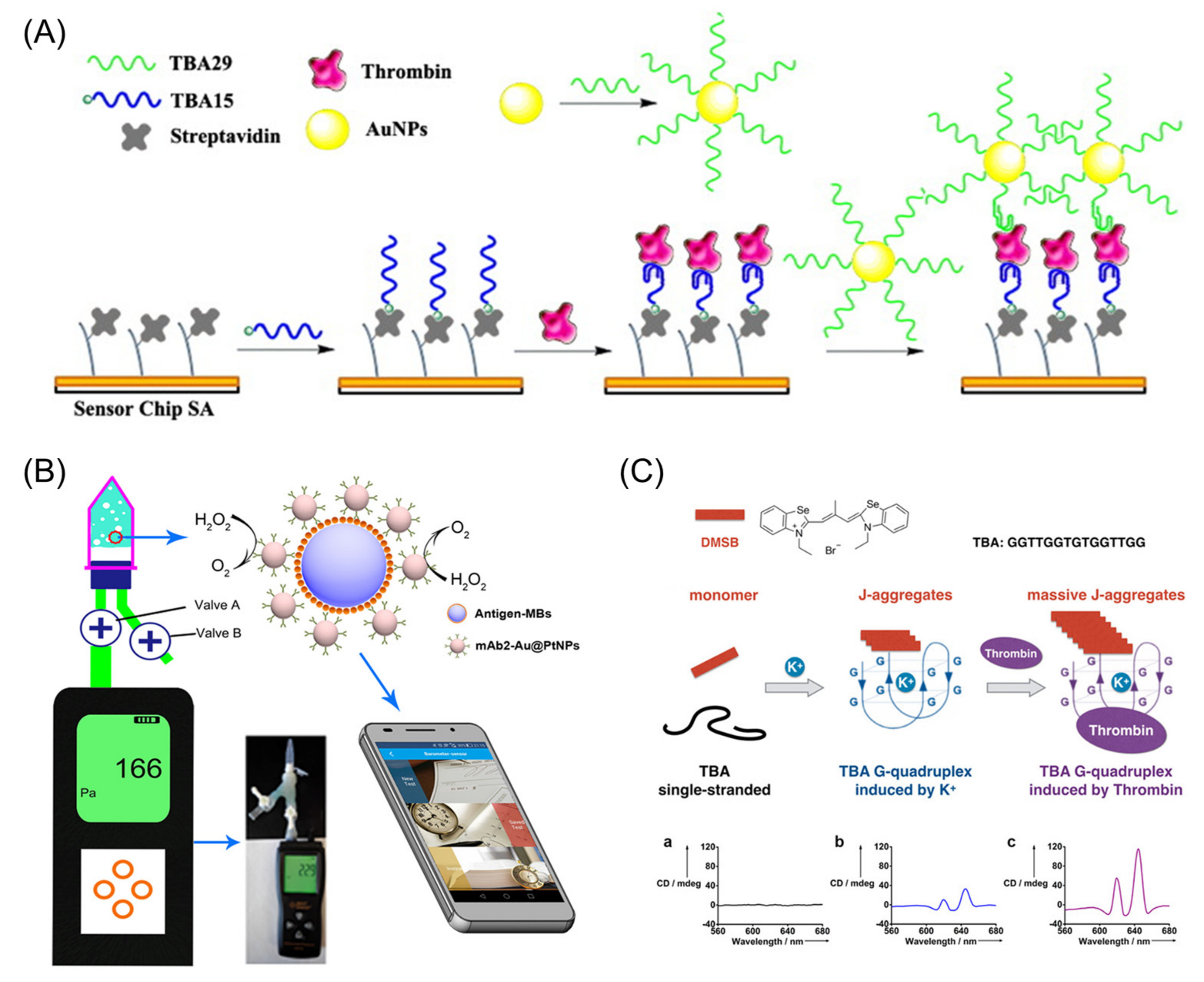
- Bai, Y.; Feng, F.; Zhao, L.; Wang, C.; Wang, H.; Tian, M.; Qin, J.; Duan, Y.; He, X. Aptamer/thrombin/aptamer-AuNPs sandwich enhanced surface plasmon resonance sensor for the detection of subnanomolar thrombin. Biosens. Bioelectron. 2013, 47, 265–270. [Google Scholar] [CrossRef]
- Fu, Q.; Wu, Z.; Du, D.; Zhu, C.; Lin, Y.; Tang, Y. Versatile Barometer Biosensor Based on Au@Pt Core/Shell Nanoparticle Probe. ACS Sens. 2017, 2, 789–795. [Google Scholar] [CrossRef]
- Shen, G.; Zhang, H.; Yang, C.; Yang, Q.; Tang, Y. Thrombin Ultrasensitive Detection Based on Chiral Supramolecular Assembly Signal-Amplified Strategy Induced by Thrombin-Binding Aptamer. Anal. Chem. 2017, 89, 548–551. [Google Scholar] [CrossRef] [Green Version]
- Sypabekova, M.; Aitkulov, A.; Blanc, W.; Tosi, D. Reflector-less nanoparticles doped optical fiber biosensor for the detection of proteins: Case thrombin. Biosens. Bioelectron. 2020, 165, 112365. [Google Scholar] [CrossRef]




| Analytical Method | Aptamer | Linear Range | LOD | Serum Sample | Ref. |
|---|---|---|---|---|---|
| Fluorescence | TBA2 | 1–300 nM | 200 pM | No | [57] |
| TBA2 | 0.05–200 pM | 0.05 pM | Yes | [58] | |
| TBA1 | 0.25 pM–25 nM | 8.9 pM | Yes | [59] | |
| TBA1, TBA2 and HD22 | 0.3–7.5 μM | 0.56 μM | No | [60] | |
| TBA1 and TBA2 | 20 pM–1 nM | 8.3 pM | Yes | [61] | |
| TBA1 and TBA2 | 0.28–86 nM | 30 pM | Yes | [62] | |
| TBA2 | 0.04–140 pM | 6 fM | Yes | [63] | |
| TBA2 | 14–285 nM | 8.11 nM | No | [64] | |
| TBA1 and TBA2 | 3.7–612.7 nM | 0.76 nM | Yes | [65] | |
| TBA1 and TBA2 | 0.13–4 nM | 0.06 nM | Yes | [66] | |
| TBA2 | 8–160 nM | 6.6 nM | Yes | [76] | |
| TBA1 and TBA2 | 50 pM–5 nM | 1.0 pM | Yes | [99] | |
| Colorimetry | TBA1 and TBA2 | 0.02–0.2 μM | 20 nM | No | [71] |
| TBA1 and TBA2 | 0.3–100 nM | 0.15 nM | Yes | [72] | |
| TBA2 | 10–80 nM | 0.8 nM | Yes | [73] | |
| TBA1 | 0.01–0.10 nM | 4 pM | No | [74] | |
| TBA1 and TBA2 | 0.267–2.67 pM | 0.356 pM | Yes | [75] | |
| TBA2 | 0–35 nM | 0.59 nM | Yes | [76] | |
| TBA1 and TBA2 | 20–5000 pM | 20 pM | No | [79] | |
| Surface-enhanced Raman Scattering | TBA1 | 100 pM–1 μM | 100 pM | Yes | [82] |
| TBA1 | 0.1–10 fM | 0.057 fM | No | [83] | |
| Electrochemistry | TBA1 | 2.48–20.26 nM | 3 pM | No | [20] |
| TBA1 and TBA2 | 5 pM–1 nM | 1.7 pM | Yes | [86] | |
| TBA1 | 1.0–500 nM | 0.49 nM | Yes | [87] | |
| TBA1 and TBA2 | 0.1 pM–10 nM | 35 fM | Yes | [88] | |
| TBA1 | 10 fM–1 μM | 1.41 fM | Yes | [89] | |
| TBA1 | 1 nM–10 mM | 0.35 nM | Yes | [90] | |
| TBA1 and TBA2 | 1 fM–100 nM | 53.70 aM | Yes | [91] | |
| TBA1 and TBA2 | 10 pM–50 nM | 5.6 pM | Yes | [92] | |
| TBA1 | 0.1 fM–0.1 nM | 27 aM | Yes | [93] | |
| TBA1 and TBA2 | 0.6 pM–0.1 nM | 0.22 pM | Yes | [94] | |
| TBA2 | 3–1350 nM | 0.9 nM | Yes | [95] | |
| Resonance Shift | TBA1 | 270 pM–27 nM | 33.5 pM | No | [97] |
| Lateral Flow Strip | TBA1 | 0.25–5 nM | 0.216 nM | No | [98] |
| Polarized Light Microscope | TBA1 | / | 136 nM | Yes | [100] |
| Surface Plasmon Resonance | TBA1 and TBA2 | 0.1–75 nM, | 0.1 nM | Yes | [101] |
| Barometer | TBA1 and TBA2 | 4–128 U/L | 2.4 U/L | Yes | [102] |
| Circular Dichroism | TBA1 | / | 2 pM | Yes | [103] |
| Scattering Spectra | TBA1 | 0.167–5.35 pM | 0.167 pM | No | [104] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Wang, N.; Zhang, L.; Meng, H.; Li, Z. Aptamer-Based Sensors for Thrombin Detection Application. Chemosensors 2022, 10, 255. https://doi.org/10.3390/chemosensors10070255
Sun H, Wang N, Zhang L, Meng H, Li Z. Aptamer-Based Sensors for Thrombin Detection Application. Chemosensors. 2022; 10(7):255. https://doi.org/10.3390/chemosensors10070255
Chicago/Turabian StyleSun, Hongzhi, Nannan Wang, Lin Zhang, Hongmin Meng, and Zhaohui Li. 2022. "Aptamer-Based Sensors for Thrombin Detection Application" Chemosensors 10, no. 7: 255. https://doi.org/10.3390/chemosensors10070255
APA StyleSun, H., Wang, N., Zhang, L., Meng, H., & Li, Z. (2022). Aptamer-Based Sensors for Thrombin Detection Application. Chemosensors, 10(7), 255. https://doi.org/10.3390/chemosensors10070255







