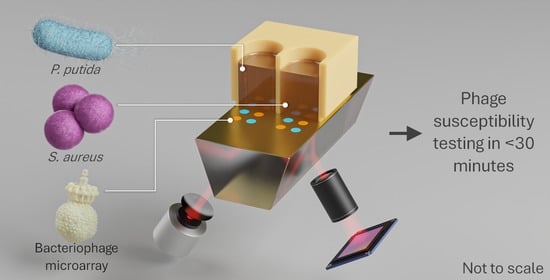
Ultrafast and Multiplexed Bacteriophage Susceptibility Testing by Surface Plasmon Resonance and Phase Imaging of Immobilized Phage Microarrays
Abstract
1. Introduction
1.1. Antimicrobial Resistance
1.2. Phage Susceptibility Testing
1.3. Surface Plasmon Resonance
1.4. Phase Imaging
2. Materials and Methods
2.1. Materials
2.2. Host Bacterium and Bacteriophage Preparation
2.3. Substrate Preparation
2.4. Bacteriophage Immobilization
2.5. Phage–Bacteria Interaction Monitoring by Surface Plasmon Resonance Imaging
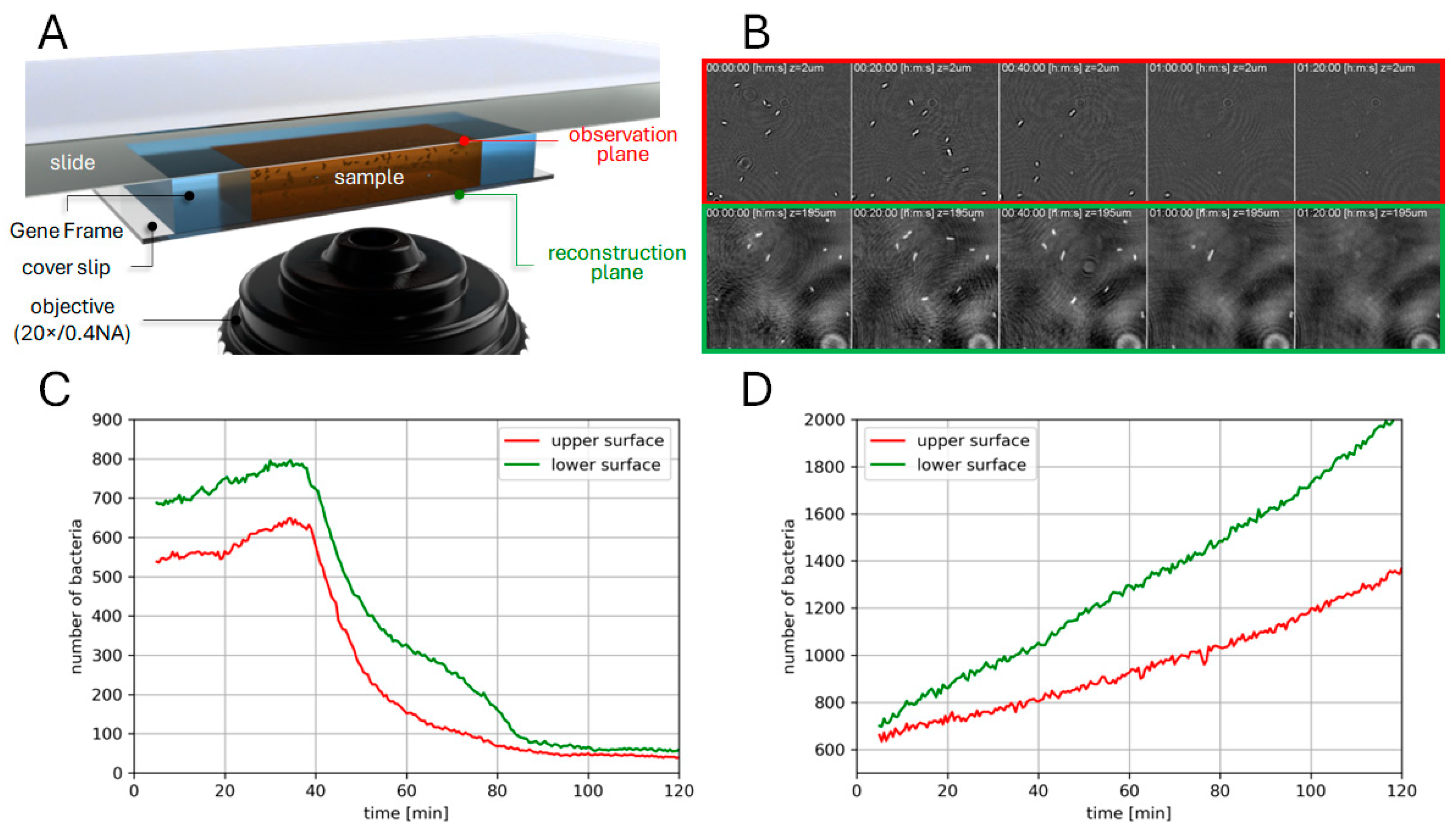
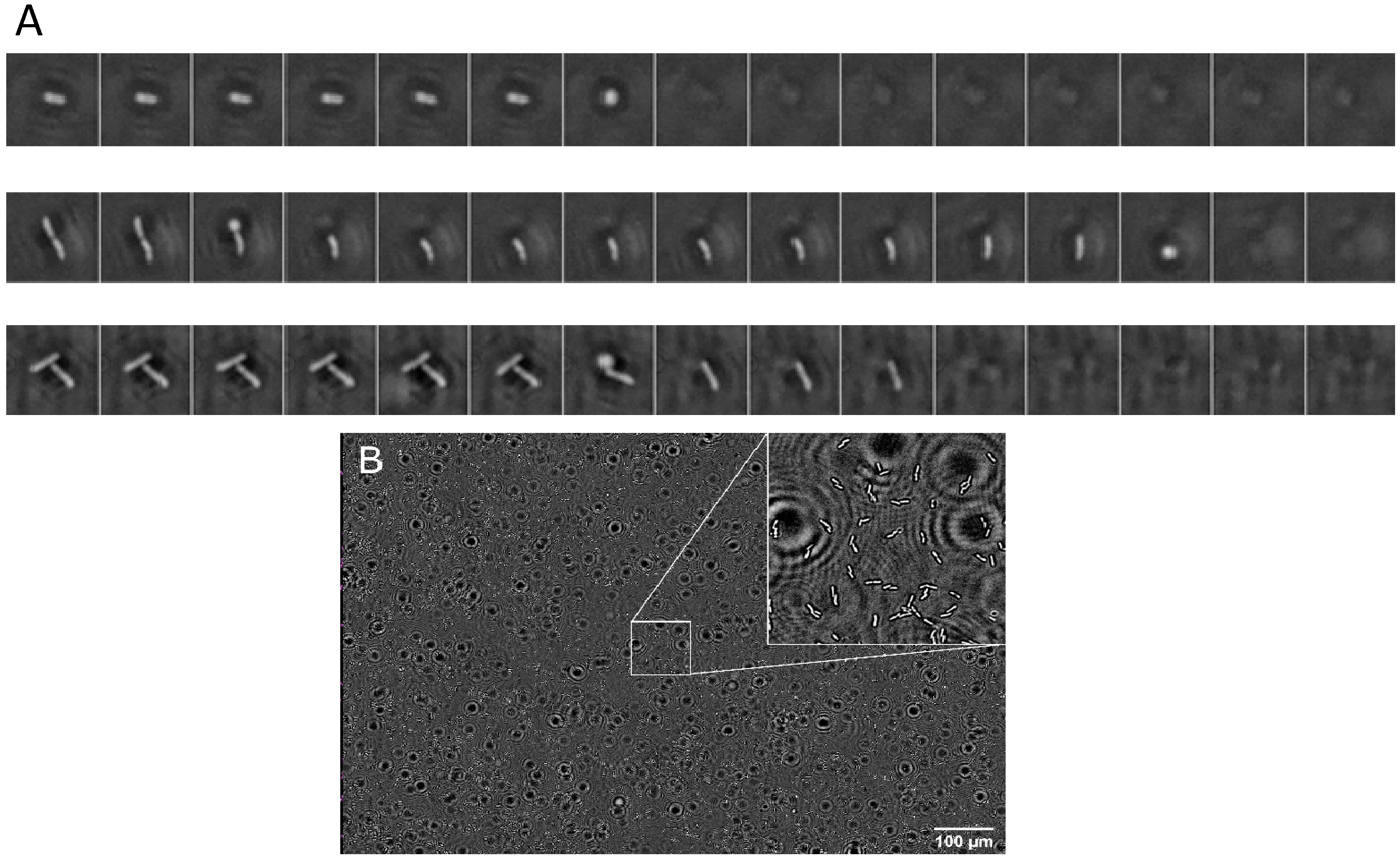
2.6. Phase Imaging
2.7. Data Analysis
3. Results
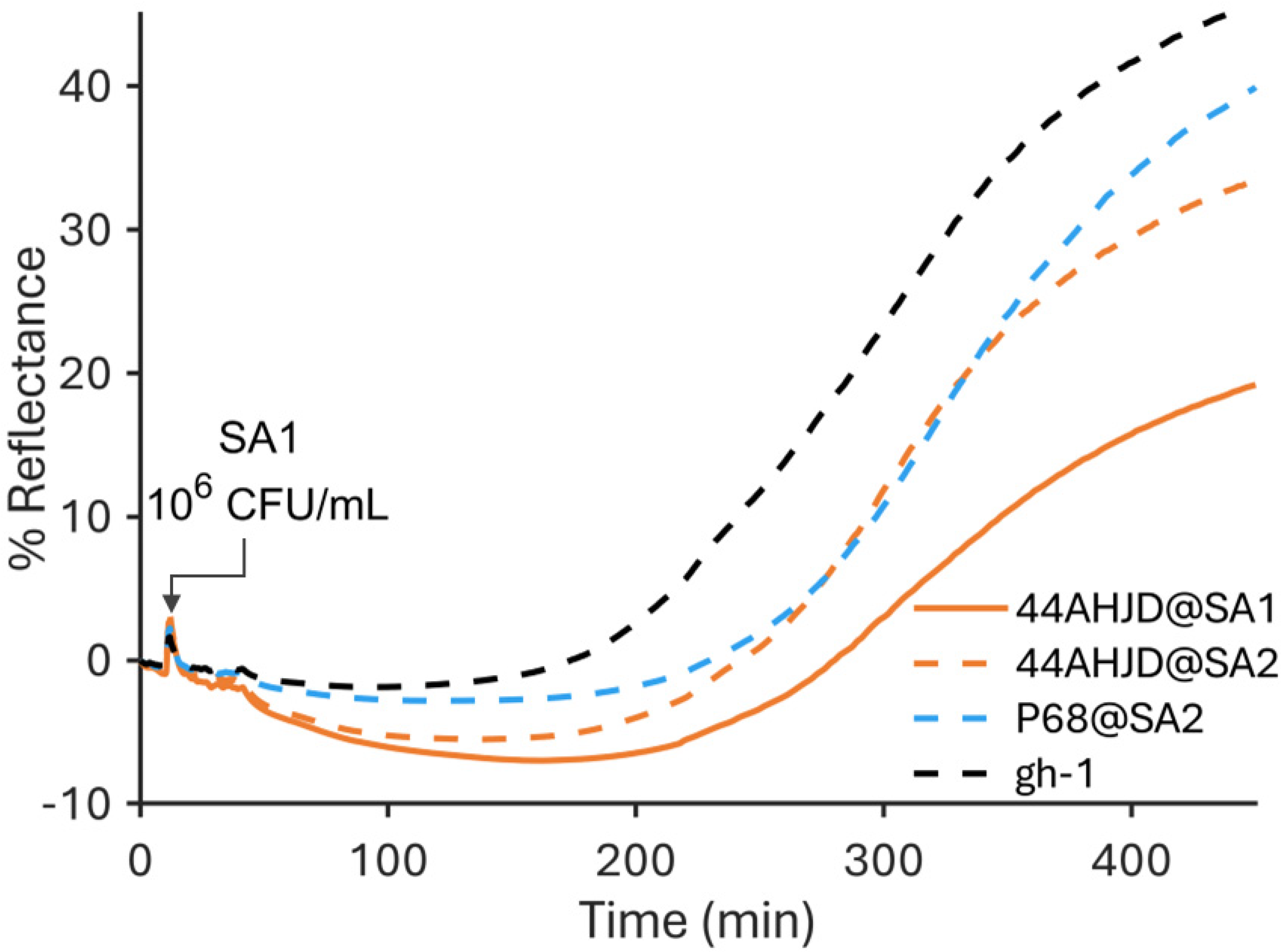
3.1. SPR
3.2. Phase Imaging
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kakasis, A.; Panitsa, G. Bacteriophage Therapy as an Alternative Treatment for Human Infections. A Comprehensive Review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H. Efficacy and Tolerability of a Cocktail of Bacteriophages to Treat Burn Wounds Infected by Pseudomonas Aeruginosa ( PhagoBurn ): A Randomised, Controlled, Double-Blind Phase 1/2 Trial. Lancet Infect. Dis. 2018, 3099, 1–11. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage Host Range and Bacterial Resistance, 1st ed.; Elsevier Inc.: Philadelphia, PA, USA, 2010; Volume 70. [Google Scholar]
- Loc-Carrillo, C.; Abedon, S.T. Pros and Cons of Phage Therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.; Hyman, P. Phage Choice, Isolation, and Preparation for Phage Therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Resch, G. Personalized Bacteriophage Therapy to Treat Pandrug-Resistant Spinal P. Aeruginosa Infection; pp. 1–7. Available online: https://www.researchsquare.com/article/rs-1231457/v1 (accessed on 15 April 2022).
- Dougherty, P.F.; Yotter, D.W.; Matthews, T.R. Microdilution Transfer Plate Technique for Determining in Vitro Synergy of Antimicrobial Agents. Antimicrob. Agents Chemother. 1977, 11, 225–228. [Google Scholar] [CrossRef][Green Version]
- Agún, S.; Fernández, L.; González-Menéndez, E.; Martínez, B.; Rodríguez, A.; García, P. Study of the Interactions between Bacteriophage PhiIPLA-RODI and Four Chemical Disinfectants for the Elimination of Staphylococcus Aureus Contamination. Viruses 2018, 10, 103. [Google Scholar] [CrossRef]
- Cooper, C.J.; Denyer, S.P.; Maillard, J.Y. Rapid and Quantitative Automated Measurement of Bacteriophage Activity against Cystic Fibrosis Isolates of Pseudomonas Aeruginosa. J. Appl. Microbiol. 2011, 110, 631–640. [Google Scholar] [CrossRef]
- Henry, M.; Biswas, B.; Vincent, L.; Mokashi, V.; Schuch, R.; Bishop-Lilly, K.A.; Sozhamannan, S. Development of a High Throughput Assay for Indirectly Measuring Phage Growth Using the OmniLog TM System. Bacteriophage 2012, 2, 159–167. [Google Scholar] [CrossRef]
- Estrella, L.A.; Quinones, J.; Henry, M.; Hannah, R.M.; Pope, R.K.; Hamilton, T.; Teneza-mora, N.; Hall, E.; Biswajit, B. Characterization of Novel Staphylococcus Aureus Lytic Phage and Defining Their Combinatorial Virulence Using the OmniLog® System. Bacteriophage 2016, 6, e1219440. [Google Scholar] [CrossRef]
- Rajnovic, D.; Muñoz-Berbel, X.; Mas, J. Fast Phage Detection and Quantification: An Optical Density-Based Approach. PLoS ONE 2019, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Perlemoine, P.; Marcoux, P.R.; Picard, E.; Hadji, E.; Zelsmann, M.; Mugnier, G.; Marchet, A.; Resch, G.; O’Connell, L.; Lacot, E. Phage Susceptibility Testing and Infectious Titer Determination through Wide-Field Lensless Monitoring of Phage Plaque Growth. PLoS ONE 2021, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M. Handbook of Surface Plasmon Resonance, 2nd ed.; Schasfoort, R.B.M., Ed.; Royal Society of Chemistry: London, UK, 2017; ISBN 978-1-78262-730-2. [Google Scholar]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- Hurot, C.; Brenet, S.; Buhot, A.; Barou, E.; Belloir, C.; Briand, L.; Hou, Y. Highly Sensitive Olfactory Biosensors for the Detection of Volatile Organic Compounds by Surface Plasmon Resonance Imaging. Biosens. Bioelectron. 2019, 123, 230–236. [Google Scholar] [CrossRef]
- Pardoux, É.; Roux, A.; Mathey, R.; Boturyn, D.; Roupioz, Y. Antimicrobial Peptide Arrays for Wide Spectrum Sensing of Pathogenic Bacteria. Talanta 2019, 203, 322–327. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- O’Connell, L.; Roupioz, Y.; Marcoux, P. Optical Bacteriophage Susceptibility Testing by SPR (Surface Plasmon Resonance). In Plasmonics in Biology and Medicine XVIII; International Society for Optics and Photonics: Bellingham, WA, USA, 2021; Volume 11661, p. 116610O. [Google Scholar] [CrossRef]
- O’Connell, L.; Marcoux, P.R.; Roupioz, Y. Strategies for Surface Immobilization of Whole Bacteriophages: A Review. ACS Biomater. Sci. Eng. 2021, 7, 1987–2014. [Google Scholar] [CrossRef]
- Genuer, V.; Gal, O.; Méteau, J.; Marcoux, P.; Schultz, E.; Lacot, É.; Maurin, M.; Dinten, J.-M. Optical Elastic Scattering for Early Label-Free Identification of Clinical Pathogens. Adv. Biomed. Clin. Diagnostic Surg. Guid. Syst. XIV 2016, 9698, 96980A. [Google Scholar] [CrossRef]
- Tardif, M.; Picard, E.; Gaude, V.; Jager, J.-B.; Peyrade, D.; Hadji, E.; Marcoux, P.R. On-Chip Optical Nano-Tweezers for Culture-Less Fast Bacterial Viability Assessment. Small 2022, 18, 2103765. [Google Scholar] [CrossRef]
- Le Galudec, J.; Dupoy, M.; Rebuffel, V.; Marcoux, P.R. Mid-Infrared Multispectral Lensless Imaging for Wide-Field and Label-Free Microbial Identification. In Biomedical Spectroscopy, Microscopy, and Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11359. [Google Scholar] [CrossRef]
- Marcoux, P.R.; Dupoy, M.; Cuer, A.; Kodja, J.L.; Lefebvre, A.; Licari, F.; Louvet, R.; Narassiguin, A.; Mallard, F. Optical Forward-Scattering for Identification of Bacteria within Microcolonies. Appl. Microbiol. Biotechnol. 2014, 98, 2243–2254. [Google Scholar] [CrossRef]
- Tardif, M.; Jager, J.B.; Marcoux, P.R.; Uchiyamada, K.; Picard, E.; Hadji, E.; Peyrade, D. Single-Cell Bacterium Identification with a SOI Optical Microcavity. Appl. Phys. Lett. 2016, 109, 1–6. [Google Scholar] [CrossRef]
- Rosenblum, E.D.; Tyrone, S. Serology, Density, and Morphology of Staphylococcal Phages. J. Bacteriol. 1964, 88, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Boezi, J.A. Characterization of Bacteriophage Gh-1 for Pseudomonas Putida. J. Bacteriol. 1966, 92, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Hosseinidoust, Z.; Olsson, A.L.J.; Tufenkji, N. Going Viral: Designing Bioactive Surfaces with Bacteriophage. Colloids Surfaces B Biointerfaces 2014, 124, 2–16. [Google Scholar] [CrossRef]
- Tang, Z.; Huang, X.; Baxi, S.; Chambers, J.R.; Sabour, P.M.; Wang, Q. Whey Protein Improves Survival and Release Characteristics of Bacteriophage Felix O1 Encapsulated in Alginate Microspheres. Food Res. Int. 2013, 52, 460–466. [Google Scholar] [CrossRef]
- Mandula, O.; Kleman, J.-P.; Lacroix, F.; Allier, C.; Fiole, D.; Hervé, L.; Blandin, P.; Kraemer, D.C.; Morales, S. Phase and Fluorescence Imaging with a Surprisingly Simple Microscope Based on Chromatic Aberration. Opt. Express 2020, 28, 2079. [Google Scholar] [CrossRef]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An Open and Extensible Platform for Single-Particle Tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Harwood, C.S.; Fosnaugh, K.; Dispensa, M. Flagellation of Pseudomonas Putida and Analysis of Its Motile Behavior. J. Bacteriol. 1989, 171, 4063–4066. [Google Scholar] [CrossRef]
- Pollitt, E.J.G.; Diggle, S.P. Defining Motility in the Staphylococci. Cell. Mol. Life Sci. 2017, 74, 2943–2958. [Google Scholar] [CrossRef]
- O’Connell, L.; Roupioz, Y.; Marcoux, P.R. Container Material Dictates Stability of Bacteriophage Suspensions: Light Scattering & Infectivity Measurements Reveal Mechanisms of Infectious Titer Decay. J. Appl. Microbiol. 2022, 1–15. [Google Scholar] [CrossRef]
- Richter, Ł.; Księżarczyk, K.; Paszkowska, K.; Janczuk-Richter, M.; Niedziółka-Jönsson, J.; Gapiński, J.; Łoś, M.; Hołyst, R.; Paczesny, J. Adsorption of Bacteriophages on Polypropylene Labware Affects Reproducibility of Phage Research. Sci. Rep. 2021, 11, 7387. [Google Scholar] [CrossRef] [PubMed]
- Kovalyova, I.V.; Kropinski, A.M. The Complete Genomic Sequence of Lytic Bacteriophage Gh-1 Infecting Pseudomonas Putida—Evidence for Close Relationship to the T7 Group. Virology 2003, 311, 305–315. [Google Scholar] [CrossRef]
- Li, X.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kuhner, P.; Peschel, A.; Xia, G.; Winstel, V. An Accessory Wall Teichoic Acid Glycosyltransferase Protects Staphylococcus Aureus from the Lytic Activity of Podoviridae. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rajaure, M.; Berry, J.; Kongari, R.; Cahill, J.; Young, R. Membrane Fusion during Phage Lysis. Proc. Natl. Acad. Sci. USA 2015, 112, 5497–5502. [Google Scholar] [CrossRef]
- Bradley, D.E. Spheroplast Formation in Cells of Escherichia Coli Infected with a Phi-X 174 Type Bacteriophage. J. Gen. Virol. 1968, 3, 141–142. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O'Connell, L.; Mandula, O.; Leroy, L.; Aubert, A.; Marcoux, P.R.; Roupioz, Y. Ultrafast and Multiplexed Bacteriophage Susceptibility Testing by Surface Plasmon Resonance and Phase Imaging of Immobilized Phage Microarrays. Chemosensors 2022, 10, 192. https://doi.org/10.3390/chemosensors10050192
O'Connell L, Mandula O, Leroy L, Aubert A, Marcoux PR, Roupioz Y. Ultrafast and Multiplexed Bacteriophage Susceptibility Testing by Surface Plasmon Resonance and Phase Imaging of Immobilized Phage Microarrays. Chemosensors. 2022; 10(5):192. https://doi.org/10.3390/chemosensors10050192
Chicago/Turabian StyleO'Connell, Larry, Ondrej Mandula, Loïc Leroy, Axelle Aubert, Pierre R. Marcoux, and Yoann Roupioz. 2022. "Ultrafast and Multiplexed Bacteriophage Susceptibility Testing by Surface Plasmon Resonance and Phase Imaging of Immobilized Phage Microarrays" Chemosensors 10, no. 5: 192. https://doi.org/10.3390/chemosensors10050192
APA StyleO'Connell, L., Mandula, O., Leroy, L., Aubert, A., Marcoux, P. R., & Roupioz, Y. (2022). Ultrafast and Multiplexed Bacteriophage Susceptibility Testing by Surface Plasmon Resonance and Phase Imaging of Immobilized Phage Microarrays. Chemosensors, 10(5), 192. https://doi.org/10.3390/chemosensors10050192