Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review
Abstract
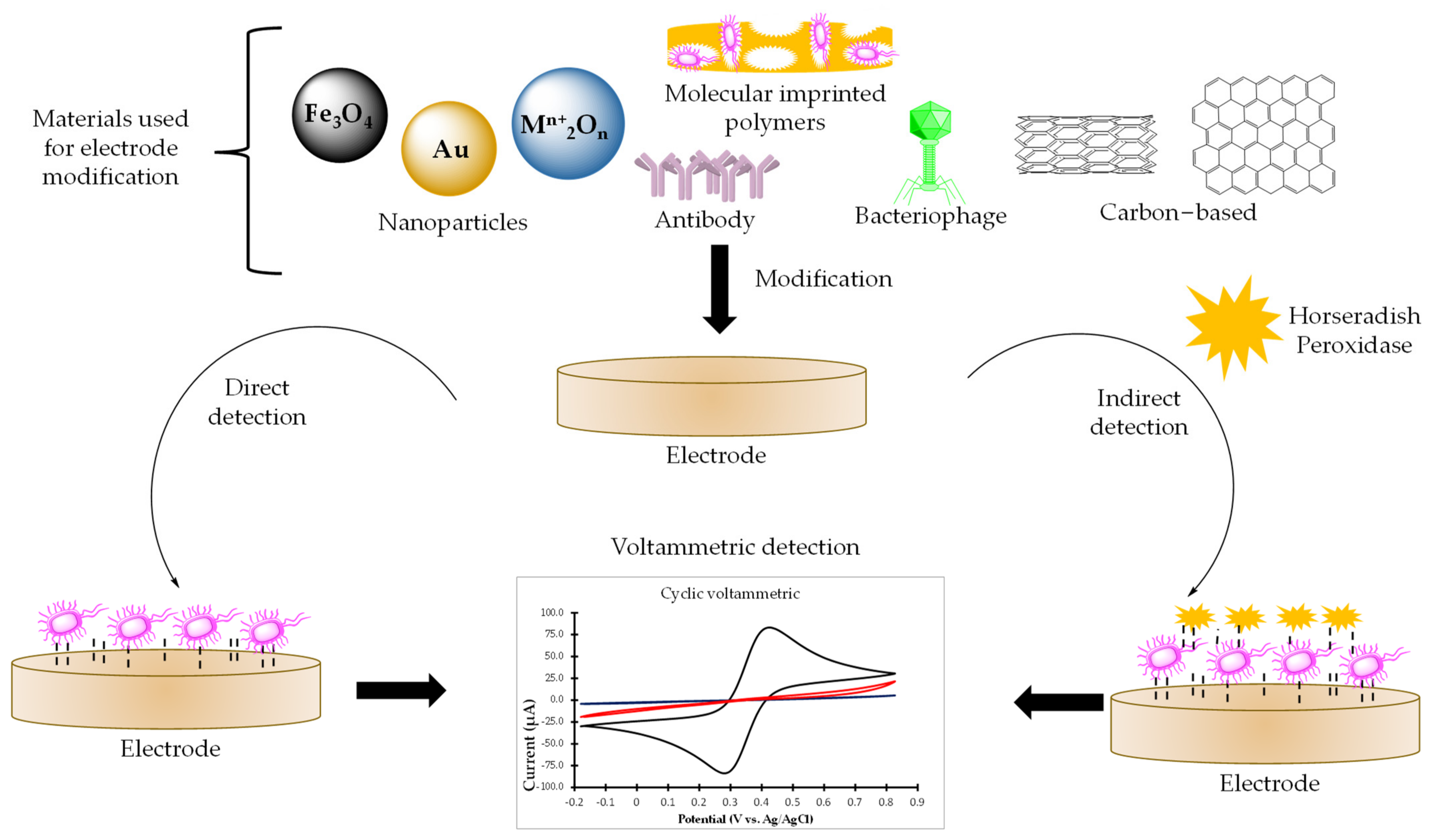
:1. Introduction
2. Voltammetric Microbial Detection
2.1. Sensors for Bacterial Detection
2.2. Biosensor for Direct Bacteria Detection
2.3. Sensors for Indirect Methods
| Sensor | Related Compound | Bacteria | Matrix | Detection | LOD (µM) | REF |
|---|---|---|---|---|---|---|
| LPS-MIP-SPCE | Lipopolysaccharides | P. aeruginosa and E. coli | Water | CV | −−− | [54] |
| Paper-based 3D Nafion-Ppy-GO-SPCE cell | Lipopolysaccharides | S. enterica serotype | Fruit juice | DPV | 1.17 × 10−4 (NO) | [56] |
| Poly-e-lysine-nanocarbon film electrode | Lipopolysaccharides | E. coli | −−− | CV | 2.0 ng mL−1 | [65] |
| SPE | PQS, HHQ, PCH, HQNO, PYO | P. aeruginosa | −−− | CV and SWV | −−− | [58] |
| Paper-based graphene sensor | Pyocianin | P. aeruginosa | Water | SWV | 0.3 | [20] |
| SPE | Pyocianin | P. aeruginosa | Human serum, blood, saliva | SWV | 0.1–25.0 | [66] |
| Boron-doped diamond | Pyocianin | P. aeruginosa | Spiked sputum | DPV | 0.05 | [65] |
| AuSPE | Pyocianin | P. aeruginosa | Human saliva | CV | 2.0 | [59] |
| SPE | Pyocianin | P. aeruginosa | Biological fluids | SWV | 0.1–1.8 | [67] |
| PANI-AuNP-ITO | Pyocianin | P. aeruginosa | Corneal Ulcer | SWV | 0.5 | [68] |
| Screen-printed sensing glove | Pyocianin Pyoverdine | P. aeruginosa | Contaminated surfaces | SWV | 3.3 × 10−3 1.6 | [60] |
| Fe3O4@SiO2-MMIP | AHLs | P. aeruginosa, aeromonas strains 128 and 130 and Aeromonas hydrophila | Bacteria supernatant samples | DPV | 8.0 × 10−4 | [62] |
| Poly-L-lysine-GCE | Mycolic acids | Mycobacterium smegmatis | Water | SWV | 59.0 CFU mL−1 | [63] |
| Two-electrode multiplexer (Au SPE) | Resazurin | E. coli | Lysogeny broth | DPV | −−− | [64] |
2.4. DNA-Based Sensors
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balloux, F.; van Dorp, L. Q&A: What are pathogens, and what have they done to and for us? BMC Biol. 2017, 15, 1–6. [Google Scholar]
- Acke, S.; Couvreur, S.; Bramer, W.M.; Schmickler, M.-N.; de Schryver, A.; Haagsma, J.A. Global infectious disease risks associated with occupational exposure among non-healthcare workers: A systematic review of the literature. Occup. Environ. Med. 2022, 79, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.; Cardoso, J.; Guerra, N.; Ribeiro, E.; Viegas, C.; Verde, S.C.; Sousa-Uva, A. Exposure and health effects of bacteria in healthcare units: An overview. Appl. Sci. 2022, 12, 1958. [Google Scholar] [CrossRef]
- Lu, R.; Frederiksen, M.W.; Uhrbrand, K.; Li, Y.; Østergaard, C.; Madsen, A.M. Wastewater treatment plant workers’ exposure and methods for risk evaluation of their exposure. Ecotoxicol. Environ. Saf. 2020, 205, 111365. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef] [Green Version]
- Lazcka, O.; Del Campo, F.J.; Muñoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef]
- Canciu, A.; Tertis, M.; Hosu, O.; Cernat, A.; Cristea, C.; Graur, F. Modern analytical techniques for detection of bacteria in surface and wastewaters. Sustainability 2021, 13, 7229. [Google Scholar] [CrossRef]
- Shin, D.J.; Andini, N.; Hsieh, K.; Yang, S.; Wang, T.-H. Emerging analytical techniques for rapid pathogen identification and susceptibility testing. Annu. Rev. Anal. Chem. 2019, 12, 41–67. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.S.; Ramasamy, R. Fluorescence in situ hybridization (FISH) tests for identifying protozoan and bacterial pathogens in infectious diseases. Diagnostics 2022, 12, 1286. [Google Scholar] [CrossRef]
- Jagannath, A.; Cong, H.; Hassan, J.; Gonzalez, G.; Gilchrist, M.D.; Zhang, N. Pathogen detection on microfluidic platforms: Recent advances, challenges, and prospects. Biosens. Bioelectron. 2022, 10, 100134. [Google Scholar] [CrossRef]
- Mocan, T.; Matea, C.T.; Pop, T.; Mosteanu, O.; Buzoianu, A.D.; Puia, C.; Iancu, C.; Mocan, L. Development of nanoparticle-based optical sensors for pathogenic bacterial detection. J. Nanobiotechnol. 2017, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical methodologies for the detection of pathogens. ACS Sens. 2018, 3, 1069–1086. [Google Scholar] [CrossRef]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of pathogenic microorganisms by microfluidics based analytical methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef]
- Simoska, O.; Stevenson, K.J. Electrochemical sensors for rapid diagnosis of pathogens in real time. Analyst 2019, 144, 6461–6478. [Google Scholar] [CrossRef]
- Tertis, M.; Hosu, O.; Feier, B.; Cernat, A.; Florea, A.; Cristea, C. Electrochemical peptide-based sensors for foodborne pathogens detection. Molecules 2021, 26, 3200. [Google Scholar] [CrossRef]
- Fatema, K.N.; Liu, Y.; Cho, K.Y.; Oh, W.C. Comparative study of electrochemical biosensors based on highly efficient mesoporous ZrO2-Ag-G-SiO2 and In2O3-G-SiO2 for rapid recognition of E. coli O157:H7. ACS Omega 2020, 5, 22719–22730. [Google Scholar] [CrossRef]
- Fan, Y.J.; Hsu, Y.C.; Gu, B.C.; Wu, C.C. Voltammetric measurement of Escherichia coli concentration through p-APG hydrolysis by endogenous β-galactosidase. Microchem. J. 2020, 154, 104641. [Google Scholar] [CrossRef]
- Alatraktchi, F.A. Rapid measurement of the waterborne pathogen Pseudomonas aeruginosa in different spiked water sources using electrochemical sensing: Towards on-site applications. Measurement 2022, 195, 111124. [Google Scholar] [CrossRef]
- Riu, J.; Giussani, B. Electrochemical biosensors for the detection of pathogenic bacteria in food. TrAC Trends Anal. Chem. 2020, 126, 115863. [Google Scholar] [CrossRef]
- Castle, L.M.; Schuh, D.A.; Reynolds, E.E.; Furst, A.L. Electrochemical sensors to detect bacterial foodborne pathogens. ACS Sens. 2021, 6, 1717–1730. [Google Scholar] [CrossRef]
- Han, S.; Li, X.; Guo, G.; Sun, Y.; Yuan, Z. Voltammetric measurement of microorganism populations. Anal. Chim. Acta 2000, 405, 115–121. [Google Scholar] [CrossRef]
- Al-Fandi, M.G.; Oweis, R.J.; Hayajneh, R.H.; Alhamdan, I.R.; Alabed, R.A.; Al-Rawi, O.F. Direct electrochemical bacterial sensor using ZnO nanorods disposable electrode. Sens. Rev. 2018, 38, 326–334. [Google Scholar] [CrossRef]
- Panhwar, S.; Hassan, S.S.; Mahar, R.B.; Carlson, K.; Talpur, M.Y. Highly Sensitive and Selective Electrochemical Sensor for Detection of Escherichia coli by Using L-Cysteine Functionalized Iron Nanoparticles. J. Electrochem. Soc. 2019, 166, B227–B235. [Google Scholar] [CrossRef]
- Dar, K.K.; Shao, S.; Tan, T.; Lv, Y. Molecularly imprinted polymers for the selective recognition of microorganisms. Biotechnol. Adv. 2020, 45, 107640. [Google Scholar] [CrossRef]
- Zheng, X.; Khaoulani, S.; Ktari, N.; Lo, M.; Khalil, A.M.; Zerrouki, C.; Chehimi, M.M. Towards clean and safe water: A review on the emerging role of imprinted polymer-based electrochemical sensors. Sensors 2021, 21, 4300. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; Grinsven, B.V.; Cleij, T.J.; Peeters, M. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, Y.; Lu, X. Molecular imprinting technology for sensing foodborne pathogenic bacteria. Anal. Bioanal. Chem. 2021, 413, 4581–4598. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef] [Green Version]
- Khalid, S.A.; Hassan, R.Y.; El Nashar, R.M.; El-Sherbiny, I.M. Voltammetric determination of Salmonella Typhimurium in minced beef meat using a chip-based imprinted sensor. RSC Adv. 2022, 12, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Lakshmi, G.B.V.S.; Kumar, A.; Solanki, P. Polypyrrole based molecularly imprinted polymer platform for Klebsiella pneumonia detection. ECS Sensors Plus 2022, 1, 010603. [Google Scholar] [CrossRef]
- Pintavirooj, C.; Vongmanee, N.; Sukjee, W.; Sangma, C.; Visitsattapongse, S. Biosensors for Klebsiella pneumoniae with molecularly imprinted polymer (MIP) technique. Sensors 2022, 22, 4638. [Google Scholar] [CrossRef]
- Lopez-Tellez, J.; Sanchez-Ortega, I.; Hornung-Leoni, C.T.; Santos, E.M.; Miranda, J.M.; Rodriguez, J.A. Impedimetric biosensor based on a Hechtia argentea lectin for the detection of Salmonella spp. Chemosensors 2020, 8, 115. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, J.; Du, X. Electrochemical biosensors for detection of foodborne pathogens. Micromachines 2019, 10, 222. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.R.; Seenath, R.; Kulak, M.R.; Lipkowski, J. Characterization of a self-assembled monolayer of 1-thio-β-D-glucose with electrochemical surface enhanced raman spectroscopy using a nanoparticle modified gold electrode. Langmuir 2015, 31, 10076–10086. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M. Electrochemical biosensors for rapid pathogen detection. Curr. Opin. Electrochem. 2021, 29, 100750. [Google Scholar] [CrossRef]
- Svalova, T.S.; Medvedeva, M.V.; Kozitsina, A.N. A “Clickable” electrodeposited polymer films based on 3-ethynylthiophene for the covalent immobilization of proteins. application to a label-free electrochemical immunosensor for Escherichia coli and Staphylococcus aureus determination. Electroanalysis 2021, 33, 2469–2475. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Tang, H.; Qian, Y.; Gu, H.; He, H. A sandwich-type electrochemical immunosensor for the sensitive determination of Salmonella Typhimurium in food. Electroanalysis 2022, 34, 911–918. [Google Scholar] [CrossRef]
- Vu, Q.K.; Tran, Q.H.; Vu, N.P.; Anh, T.L.; Le Dang, T.T.; Matteo, T.; Nguyen, T.H.H. A label-free electrochemical biosensor based on screen-printed electrodes modified with gold nanoparticles for quick detection of bacterial pathogens. Mater. Today Commun. 2021, 26, 101726. [Google Scholar] [CrossRef]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luan, T.; Yang, X.; Wang, S.; Zheng, Y.; Huang, T.; Yan, X. Trace detection of specific viable bacteria using tetracysteine-tagged bacteriophages. Anal. Chem. 2014, 86, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, C.; Chau, Y.; Lee, Y.K. The synergy of chemical immobilization and electrical orientation of T4 bacteriophage on a micro electrochemical sensor for low-level viable bacteria detection via differential pulse voltammetry. Biosens. Bioelectron. 2020, 151, 111914. [Google Scholar] [CrossRef]
- Fei, J.; Dou, W.; Zhao, G. A sandwich electrochemical immunosensor for Salmonella pullorum and Salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Microchim Acta 2015, 182, 2267–2275. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Liu, S.; Yu, J.; Wang, H.; Cui, M.; Huang, J. Electrochemical immunosensor assay (EIA) for sensitive detection of E. coli O157:H7 with signal amplification on a SG–PEDOT–AuNPs electrode interface. Analyst 2015, 140, 551–559. [Google Scholar] [CrossRef]
- Setterington, E.B.; Alocilja, E.C. Electrochemical biosensor for rapid and sensitive detection of magnetically extracted bacterial pathogens. Biosensors 2012, 1, 15–31. [Google Scholar] [CrossRef] [Green Version]
- Cho, I.H.; Choi, K.J.; Kim, J.H.; Lee, K.; Ly, S.Y. Analysis of Staphylococcus aureus molecules in non-treated blood using mercury immobilized carbon nanotube sensor. Molecules 2022, 27, 1837. [Google Scholar] [CrossRef]
- Zhao, G.; Zhan, X.; Dou, W. A disposable immunosensor for Shigella flexneri based on multiwalled carbon nanotube/sodium alginate composite electrode. Anal. Biochem. 2011, 408, 53–58. [Google Scholar] [CrossRef]
- Güner, A.; Çevik, E.; Şenel, M.; Alpsoy, L. An electrochemical immunosensor for sensitive detection of Escherichia coli O157: H7 by using chitosan, MWCNT, polypyrrole with gold nanoparticles hybrid sensing platform. Food Chem. 2017, 229, 358–365. [Google Scholar] [CrossRef]
- Xiang, C.; Li, R.; Adhikari, B.; She, Z.; Li, Y.; Kraatz, H.B. Sensitive electrochemical detection of Salmonella with chitosan–gold nanoparticles composite film. Talanta 2015, 140, 122–127. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Devarakonda, S.; Kumar, S.; Jang, J. Development of a paper-based electrochemical immunosensor using an antibody-single walled carbon nanotubes bio-conjugate modified electrode for label-free detection of foodborne pathogens. Sens. Actuators B Chem. 2017, 253, 115–123. [Google Scholar] [CrossRef]
- Bu, S.; Wang, K.; Li, Z.; Wang, C.; Hao, Z.; Liu, W.; Wan, J. An electrochemical biosensor based on methylene blue-loaded nanocomposites as signal-amplifying tags to detect pathogenic bacteria. Analyst 2020, 145, 4328–4334. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Li, T.; Ye, C.; Gao, X.; Yang, T.; Liang, X.; Zhang, J. A label-free electrochemical immunosensor for rapid detection of salmonella in milk by using CoFe-MOFs-graphene modified electrode. Food Control 2021, 130, 108357. [Google Scholar] [CrossRef]
- Stoica, B.E.; Gavrila, A.; Sarbu, A.; Lovu, H.; Brisset, H.; Miron, H.; Iordache, T. Un-covering the behavior of screen-printed carbon electrodes modified with polymers molecularly imprinted with lipopolysaccharide. Electrochem. Commun. 2021, 124, 106965. [Google Scholar] [CrossRef]
- Kato, D.; Oda, A.; Tanaka, M.; Iijima, S.; Kamata, T.; Todokoro, M.; Yoshimi, Y.; Niwa, O. Poly-e-lysine modified nanocarbon film electrodes for LPS detection. Electroanalysis 2014, 26, 618–624. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, J.; Wan, K.; Jiang, D.; Jin, C. Miniaturized paper-supported 3D cell-based electrochemical sensor for bacterial lipopolysaccharide detection. ACS Sens. 2020, 5, 1325–1335. [Google Scholar] [CrossRef]
- Hicks, J.M.; Silman, N.J.; Jackson, S.K.; Aylott, J.W.; Rawson, F.J. Mass transport of lipopolysaccharide induced H2O2 detected by an intracellular carbon nanoelectrode sensor. Bioelectrochemistry 2020, 135, 107547. [Google Scholar] [CrossRef]
- Schneider, S.; Ettenauer, J.; Pap, I.-J.; Aspöck, C.; Walochnik, J.; Brandl, M. Main metabolites of Pseudomonas aeruginosa: A study of electrochemical properties. Sensors 2022, 22, 4694. [Google Scholar] [CrossRef]
- Alatraktchi, F.A.; Andersen, S.B.; Johansen, H.K.; Molin, S.; Svendsen, W.E. Fast selective detection of pyocyanin using cyclic voltammetry. Sensors 2016, 16, 408. [Google Scholar] [CrossRef]
- Ciui, B.; Tertis, M.; Cernat, A.; Săndulescu, R.; Wang, J.; Cristea, C. Finger-based printed sensors integrated on a glove for on-site screening of Pseudomonas aeruginosa virulence factors. Anal. Chem. 2018, 90, 7761–7768. [Google Scholar] [CrossRef]
- Rumbaugh, K.P. Convergence of hormones and autoinducers at the host/pathogen interface. Anal. Bioanal. Chem. 2007, 387, 425–435. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, D.; Shao, J.; Sun, X. Magnetic molecularly imprinted polymer nanoparticles based electrochemical sensor for the measurement of Gram-negative bacterial quorum signaling molecules (N-acyl-homoserine-lactones). Biosens. Bioelectron. 2016, 75, 411–419. [Google Scholar] [CrossRef]
- Brugnera, M.F.; Miyata, M.; Fujimura Leite, C.Q.; Boldrin Zanoni, M.V. Sensitive Voltammetric Sensor for Fast Detection of Mycolic Acids Present in Mycobacteria. Int. J. Electrochem. Sci. 2013, 8, 6591–6602. [Google Scholar]
- Nagar, B.; Silva, W.O.; Girault, H. Voltammetry in two-electrode mode for rapid electrochemical screening using a fully printed and flexible multiplexer sensor. ChemElectroChem 2021, 8, 3700–3706. [Google Scholar] [CrossRef]
- Cernat, A.; Canciu, A.; Tertis, M.; Graur, F.; Cristea, C. Synergic action of thermosensitive hydrogel and Au/Ag nanoalloy for sensitive and selective detection of pyocyanin. Anal. Bioanal. Chem. 2019, 411, 3829–3838. [Google Scholar] [CrossRef]
- Buzid, A.; Shang, F.; Reen, F.J.; Muimhneacháin, E.; Clarke, S.L.; Zhou, L.; Luong, J.H.T.; O’Gara, F.; McGlacken, G.P.; Glennon, J.D. Molecular signature of Pseudomonas aeruginosa with simultaneous nanomolar detection of quorum sensing signaling molecules at a boron-doped diamond electrode. Sci. Rep. 2016, 6, 30001. [Google Scholar] [CrossRef] [Green Version]
- Webster, T.A.; Sismaet, H.J.; Conte, J.L.; Chan, I.J.; Goluch, E.D. Electrochemical detection of Pseudomonas aeruginosa in human fluid samples via pyocyanin. Biosens. Bioelectron. 2014, 60, 265–270. [Google Scholar] [CrossRef]
- Khalifa, M.M.; Elkhawaga, A.A.; Hassan, M.A.; Zahran, A.M.; Fathalla, A.M.; El-Said, W.A.; El-Badawy, O. Highly specific Electrochemical sensing of Pseudomonas aeruginosa in patients suffering from corneal ulcers: A comparative study. Sci. Rep. 2019, 9, 18320. [Google Scholar] [CrossRef] [Green Version]
- Razmi, N.; Hasanzadeh, M.; Willander, M.; Nur, O. Electrochemical genosensor based on gold nanostars for the detection of Escherichia coli O157:H7 DNA. Anal. Methods 2022, 14, 1562–1570. [Google Scholar] [CrossRef]
- Wu, T.; Wang, C.; Wu, M.; Wang, P.; Feng, Q. Novel integrating polymethylene blue nanoparticles with dumbbell hybridization chain reaction for electrochemical detection of pathogenic bacteria. Food Chem. 2022, 382, 132501. [Google Scholar] [CrossRef]
- Giallo, M.L.D.; Ariksoysal, D.O.; Marrazza, G.; Mascini, M.; Ozsoz, M. Disposable electrochemical enzyme-amplified genosensor for Salmonella bacteria detection. Anal. Lett. 2005, 38, 2509–2523. [Google Scholar] [CrossRef]
- Farabullini, F.; Lucarelli, F.; Palchetti, I.; Marrazza, G.; Mascini, M. Disposable electrochemical genosensor for the simultaneous analysis of different bacterial food contaminants. Biosens. Bioelectron. 2007, 22, 1544–1549. [Google Scholar] [CrossRef]
- Kara, P.; Meric, B.; Ozsoz, M. Application of impedimetric and voltammetric genosensor for detection of a biological warfare: Anthrax. Electroanalysis 2008, 20, 2629–2634. [Google Scholar] [CrossRef]
- Martínez-Paredes, G.; González-García, M.B.; Costa-García, A. Genosensor for detection of four pneumoniae bacteria using gold nanostructured screen-printed carbon electrodes as transducers. Sens. Actuators B Chem. 2010, 149, 329–335. [Google Scholar] [CrossRef]
- Dash, S.K.; Sharma, M.; Khare, S.; Kumar, A. Carbon-Mercaptooctadecane/Carboxylated Multi-walled Carbon Nanotubes Composite Based Genosensor for Detection of Bacterial Meningitis. Indian J. Microbiol. 2014, 54, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Bizid, S.; Blili, S.; Mlika, R.; Said, A.H.; Korri-Youssoufi, H. Direct Electrochemical DNA Sensor based on a new redox oligomer modified with ferrocene and carboxylic acid: Application to the detection of Mycobacterium tuberculosis mutant strain. Anal. Chim. Acta 2017, 994, 10–18. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Babaie, P.; Mokhtarzadeh, A.; Hajizadeh, N.; Mahboob, S. A novel DNA based bioassay toward ultrasensitive detection of Brucella using gold nanoparticles supported histidine: A new platform for the assay of bacteria in the cultured and human biofluids with and without polymerase chain reactions (PCR). Int. J. Biol. Macromol. 2018, 120, 422–430. [Google Scholar] [CrossRef]
- Flauzino, J.M.; Peres, R.C.; Alves, L.M.; Vieira, J.G.; Dos Santos, J.G.; Brito-Madurro, A.G.; Madurro, J.M. DNA electrochemical biosensor for detection of Alicyclobacillus acidoterrestris utilizing Hoechst 33258 as indicator. Bioelectrochemistry 2021, 140, 107801. [Google Scholar] [CrossRef]
- Hu, J.; Shen, Z.; Tan, L.; Yuan, J.; Gan, N. Electrochemical aptasensor for simultaneous detection of foodborne pathogens based on a double stirring bars-assisted signal amplification strategy. Sens. Actuators B Chem. 2021, 345, 130337. [Google Scholar] [CrossRef]
- Li, F.; Yu, Z.; Xu, Y.; Ma, H.; Zhang, G.; Song, Y.; Yan, H.; He, X. Using the synergism strategy for highly sensitive and specific electrochemical sensing of Streptococcus pneumoniae Lyt-1 gene sequence. Anal. Chim. Acta 2015, 886, 175–181. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 2016, 34, 941–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vakyly, S.; Sedighian, H.; Jahromi, Z.; Jahangiri, A.; Halabian, R.; Rezaei, A.; Keshmiri, F. A sensitive and selective electrochemical sensor based on gold nanoparticle/multi-walled carbon nanotubes for detection of Staphylococcus aureus Alpha-toxin. Appl. Phys. A 2022, 128, 1–10. [Google Scholar] [CrossRef]
- Al Mamun, M.; Wahab, Y.A.; Hossain, M.M.; Hashem, A.; Johan, M.R. Electrochemical biosensors with aptamer recognition layer for the diagnosis of pathogenic bacteria: Barriers to commercialization and remediation. Trends Anal. Chem. 2021, 145, 116458. [Google Scholar] [CrossRef]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef]
- Chen, J.; Luo, Z.; Sun, C.; Huang, Z.; Zhou, C.; Yin, S.; Duan, Y.; Li, Y. Research progress of DNA walker and its recent applications in biosensor. Trends Anal. Chem. 2019, 120, 115626. [Google Scholar] [CrossRef]
- Wu, T.; Wang, C.; Han, X.; Feng, Q.; Wang, P. Combination of DNA walker and Pb2+-specific DNAzyme-based signal amplification with a signal-off electrochemical DNA sensor for Staphylococcus aureus detection. Anal. Chim. Acta 2022, 1222, 340179. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Huang, H.; Deng, J.; Fang, L.; Luo, J.; Zhang, S.; Huang, J.; Liang, W.; Zheng, J. A sensitive electrochemical strategy via multiple amplification reactions for the detection of E. coli O157:H7. Biosens. Bioelectron. 2020, 147, 111752. [Google Scholar] [CrossRef]
- Zhou, H.; Duan, S.; Huang, J.; He, F. An ultrasensitive electrochemical biosensor for Pseudomonas aeruginosa assay based on a rolling circle amplification-assisted multipedal DNA walker. Chem. Commun. 2020, 56, 6273–6276. [Google Scholar] [CrossRef]
- Wei, W.; Lin, H.; Hao, T.; Wang, S.; Hu, Y.; Guo, Z.; Luo, X. DNA walker-mediated biosensor for target-triggered triple-mode detection of Vibrio parahaemolyticus. Biosens. Bioelectron. 2021, 186, 113305. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, H.; Liu, W.; Wen, H.; He, F. Rapid 16S rDNA electrochemical sensor for detection of bacteria based on the integration of target-triggered hairpin self-assembly and tripedal DNA walker amplification. Anal. Chim. Acta 2022, 1190, 339266. [Google Scholar] [CrossRef]
- Sharifi, S.; Vahed, S.Z.; Ahmadian, E.; Dizaj, S.M.; Eftekhari, A.; Khalilov, R.; Ahmadi, M.; Hamidi-Asl, E.; Labib, M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2020, 150, 111933. [Google Scholar] [CrossRef]
- Ly, S.Y.; Yoo, H.S.; Choa, S.H. Diagnosis of Helicobacter pylori bacterial infections using a voltammetric biosensor. J. Microbiol. Methods 2011, 87, 44–48. [Google Scholar] [CrossRef]
- Abdalhai, M.H.; Fernandes, A.M.; Xia, X.; Musa, A.; Ji, J.; Sun, X. Electrochemical genosensor to detect pathogenic bacteria (Escherichia coli O157:H7) as applied in real food samples (fresh beef) to improve food safety and quality control. J. Agric. Food Chem. 2015, 63, 5017–5025. [Google Scholar] [CrossRef]
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Muniandy, S.; Dinshaw, I.J.; Teh, S.J.; Lai, C.W.; Ibrahim, F.; Thong, K.L.; Leo, B.F. Graphene-based label-free electrochemical aptasensor for rapid and sensitive detection of foodborne pathogen. Anal. Bioanal. Chem. 2017, 409, 6893–6905. [Google Scholar] [CrossRef]
- Ye, Y.; Yan, W.; Liu, Y.; He, S.; Cao, X.; Xu, X.; Zheng, H.; Gunasekaran, S. Electrochemical detection of Salmonella using an invA genosensor on polypyrrole-reduced graphene oxide modified glassy carbon electrode and AuNPs-horseradish peroxidase-streptavidin as nanotag. Anal. Chim. Acta 2019, 1074, 80–88. [Google Scholar] [CrossRef]
- Pourmadadi, M.; Shayeh, J.S.; Omidi, M.; Yazdian, F.; Alebouyeh, M.; Tayebi, L. A glassy carbon electrode modified with reduced graphene oxide and gold nanoparticles for electrochemical aptasensing of lipopolysaccharides from Escherichia coli bacteria. Microchim. Acta 2019, 186, 1–8. [Google Scholar] [CrossRef]
- Song, X.; Lv, M.M.; Lv, Q.Y.; Cui, H.F.; Fu, J.; Huo, Y.Y. A novel assay strategy based on isothermal amplification and cascade signal amplified electrochemical DNA sensor for sensitive detection of Helicobacter pylori. Microchem. J. 2021, 166, 106243. [Google Scholar] [CrossRef]


| Bacteria | Electrode | Detection | Lineal Interval (CFU mL−1) | Sample | LOD (CFU mL−1) | REF |
|---|---|---|---|---|---|---|
| E. coli, | Glassy carbon/tetracycline | CV | 2.0 × 104–5.0 × 107 | Food | 2.0 × 104 | [23] |
| B. subtilis, | 2.0 × 104–5.0 × 107 | 2.0 × 104 | ||||
| S. aureus, | 3.0 × 104–9.0 × 107 | 3.0 × 1047 | ||||
| Salmonella and | 3.0 × 104–1.0 × 108 | 3.0 × 104 | ||||
| L. lactis | 5.0 × 104–1.6 × 108 | 5.0 × 104 | ||||
| E. coli O157:H7 | ZnO nanorods | CV | −−− | −−− | 1.0 × 103 | [24] |
| E. coli | L-cysteine | CV | 1.0 × 101–1.0 x 1010 | Tap water | 10.0 | [25] |
| E. coli O157:H7 | ZrO2 and In2O3/graphene | CV | 1.0 × 101–1.0 × 105 | −−− | 10.0 | [18] |
| S. aureus | Mercury/carbon nanotubes | CV | −−− | Human blood and serum | −−− | [47] |
| Salmonella | Poly(dopamine) | CV | 1.0 × 102–1.0 × 109 | Meat | 47.0 | [31] |
| K. pneumoniae | Pyrrol | DPV | 1.0 × 100–1.0 × 105 | Urine | ≈2.0 | [22] |
| K. pneumoniae | Methacrylamide/acrylamide/N-vinylpyrrolidone/graphene | CV | 1.0 × 101–1.0 × 105 | −−− | ≈2.0 | [33] |
| E. coli O157 | Screen-printed carbon/gold nanoparticles/anti-E. coli O157 | CV | 1.0 × 101–1.0 × 106 | −−− | 15.0 | [40] |
| S. aureus and | 3-ethynylthiophene/bovine serum albumin/anti-E. coli or anti-S. aureus | CV | 1.0 × 102–1.0 × 106 | −−− | 15.9 | [38] |
| E. coli | 7.2 | |||||
| B. cereus and | Screen-printed carbon/magnetic nanoparticles/polyaniline anti-B. cereus or anti-E. coli | CV | 4.0 × 100–3.9 × 102 6.0 × 100–5.9 × 104 | −−− | 40.0 | [46] |
| E. coli O157:H7 | 6.0 | |||||
| E. coli B | T4 bacteriophage | DPV | 1.9 × 102–1.9 × 109 | −−− | 14.0 | [43] |
| Shigella flexneri | Multiwalled carbon nanotubes/sodium alginate/anti-S. flexneri HRP | CV | 1.0 × 104–1.0 × 1011 | −−− | 3.1 × 103 | [48] |
| E. coli O157:H7 | Sulfonated graphene/poly-(3,4-ethylenedioxythiophene)/gold nanoparticles/anti-E. coli HRP | DPV | 7.8 × 101–7.8 × 106 | Water and milk | 34.0 | [45] |
| S. gallinarum | Anti-S. gallinarum gold nanoparticles/anti-S. gallinarum HRP | CV | 1.0 × 104–1.0 × 109 | −−− | 3.0 × 103 | [44] |
| E. coli O157:H7 | Pencil graphite/chitosan/multi-walled carbon nanotubes/gold nanoparticles/polypyrrole/anti-E. coli | CV | 3.0 × 101–3.0 × 107 | −−− | 30.0 | [49] |
| S. Typhimurium | Glassy carbon chitosan/S. Typhimurium/anti-S. Typhimurium HRP | CV | 5.6 × 101–5.6 × 108 | Milk and eggs | 35.0 | [39] |
| S. Typhimurium | Chitosan/gold nanoparticles/S. Typhimurium/anti-S. Typhimurium HRP | CV | 1.0 × 101–1.0 × 105 | −−− | 5.0 | [50] |
| S. aureus | Single-walled carbon nanotubes/anti-S. aureus | DPV | 1.0 × 101–1.0 × 107 | −−− | 13.0 | [51] |
| E. coli O157:H7 | Methylene blue nanocomposites/magainin I | DPV | 1.0 × 102–1.0 × 107 | −−− | 32.0 | [52] |
| Salmonella | Glassy carbon/grapheme MOFs/CoFe/gold nanoparticles/anti-Salmonella | CV | 2.4 × 102–2.4 × 108 | Milk | 120.0 | [53] |
| Analyte | Sample | Method | Working Electrode | LOD | Linear Range | REF |
|---|---|---|---|---|---|---|
| Salmonella | Salmonella amplicons | DPV | SPCE | 3.0 × 10−10 M of Salmonella DNA target sequence | 1.0 × 10−8–1.5 × 10−7 M of Salmonella DNA target sequence | [71] |
| B. anthracis | −−− | DPV | Gold and graphite electrode | 2.0 × 10−11 M of B. anthracis target sequence | 6.6 × 10−6–3.3 × 10−4 M of B. anthracis target sequence | [73] |
| Helicobacter pylori | Human gastric tissues | CV SWV | BCNE | 6.0 × 10−8 g mL−1 of H. pylori DNA | 7.0 × 10−7–7.9 × 10−6 g mL−1 of H. pylori DNA | [92] |
| N. meningitidis | Cerebrospinal fluid | CV DPV | COO−MWCNT-SPCE | 2.0 × 10−6 g of N. meningitidis ssG-DNA | 2.5 × 10−6–1.0 × 10−5 g 6 μL−1 of N. meningitidis ssG-DNA | [75] |
| E. coli | Beef meat | CV DPV | MWCNT-Chi-GCE-Bi | 2.0 × 10−14 M of E. coli tDNA | 2.0×10−14–1.9×10−13 M of E. coli tDNA | [93] |
| Streptococcus pneumoniae Lyt-1 gene sequence | Clinical sample | CV ACV | DNA probe-modified gold disk electrode | ~5.0 × 10−16 M of S. pneumoniae Lyt-1 gene sequence | 1.0 × 10−14–1.0 × 10−10 M of S. pneumoniae Lyt-1 gene sequence | [80] |
| M. tuberculosis | Clinical sample | CV | Fc-acid-OMPA deposited on gold electrode | 2.0 × 10−16 M of M. tuberculosis DNA | 1.0 × 10−15–1.0 × 10−10 M of M. tuberculosis DNA | [76] |
| S. typhimurium | Raw chicken meat | CV DPV | Glassy carbon electrode | 1.0 × 101 CFU mL−1 | 1.0 × 101–1 × 106 CFU mL−1 | [94] |
| S. typhimurium | Chicken meat | DPV | ssDNA-rGO-AP-GCE | 1.0 × 101 CFU mL−1 | 1.0 × 101–1 × 108 CFU mL−1 | [95] |
| Brucella | Cultured and human samples | CV SWV | Gold electrode | −−− | 1.0 × 10−16–1.0 ×10−7 M | [77] |
| Salmonella | −−− | DPV | PPy-rGO-GCE-AuNP-HRP-SA | 4.7×10−17 M/ 8.1 CFU mL−1 | 1.0 × 10−16–1.0 × 10−10 M/ 9.6–9.6×104 CFU mL−1 | [96] |
| E. coli 055:B5 | Serum | CV SWV | RGO-AuNP-GCE | 3.0 × 10−14 g mL−1 of lipopolysaccharides from E. coli | 1.0 × 10−13–9.0 × 10−13 g mL−1 of lipopolysaccharides from E. coli | [97] |
| A. acidoterrestris | Orange juice | DPV | GE-ERGO-poly(3-HBA) | 1.2 × 10−8 g mL−1 of A. acidoterrestris | 1.2 × 10−8–1.2 × 10−4 g mL−1 of A. acidoterrestris | [78] |
| V. parahaemolyticus S. typhimurium | Shrimp and fish | SWV | SPCE | 4.0 CFU mL−1 7.0 CFU mL−1 | 1.0 × 101–1.0 × 108 CFU mL−1 | [79] |
| Helicobacter pylori | −−− | DPV | RNA cleaving DNAzyme-G-quadruplex DNAzyme gold electrode | 3.4 × 10−17 M of H. pylori tDNA/1.3 × 10−12 g of H. pylori genomic DNA | 1.0 × 10−16–1.0 ×10−11 M of H. pylori tDNA/2.1 × 10−12–6.7 × 10−11 g of H. pylori genomic DNA | [98] |
| E. coli O157:H7 | Water | CV SWV | Au-GNS | 1.0 × 10−23 M of E. coli target DNA | 1.0 × 10−17–7.3 × 10−17 M of E. coli target DNA | [69] |
| S. aureus alpha-toxin | Human serum | SWV | Au-MWCNT-BMIM-PF6-CPE | 1.0 × 10−9 M of alpha-toxin | 3.0 × 10−9–2.5 × 10−7 M of alpha-toxin | [82] |
| S. aureus | Human serum, milk and pear juice | DPV | DNA-Au NC-CS-GCE | 1.0 CFU mL−1 | 1.0 × 101–1.0 × 108 CFU mL−1 | [70] |
| S. aureus | Raw milk, beer and apple juice | DPV | T-H-MB-MWCNT-CS-GCE | 1.0 CFU mL−1 | 1.0 × 101–1.0 × 107 CFU mL−1 | [86] |
| S. aureus | Sputum | DPV | Ferrocene-planar Au electrode | 20.0 CFU mL−1 | 1.0 × 102–1.0 × 108 CFU mL−1 | [90] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Tellez, J.; Ramirez-Montes, S.; Ferreira, T.A.; Santos, E.M.; Rodriguez, J.A. Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review. Chemosensors 2022, 10, 424. https://doi.org/10.3390/chemosensors10100424
Lopez-Tellez J, Ramirez-Montes S, Ferreira TA, Santos EM, Rodriguez JA. Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review. Chemosensors. 2022; 10(10):424. https://doi.org/10.3390/chemosensors10100424
Chicago/Turabian StyleLopez-Tellez, Jorge, Sandra Ramirez-Montes, T. Alexandra Ferreira, Eva M. Santos, and Jose A. Rodriguez. 2022. "Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review" Chemosensors 10, no. 10: 424. https://doi.org/10.3390/chemosensors10100424
APA StyleLopez-Tellez, J., Ramirez-Montes, S., Ferreira, T. A., Santos, E. M., & Rodriguez, J. A. (2022). Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review. Chemosensors, 10(10), 424. https://doi.org/10.3390/chemosensors10100424








